Qualitative and Quantitative Analytical Techniques of Nucleic Acid Modification Based on Mass Spectrometry for Biomarker Discovery
Abstract
:1. Introduction
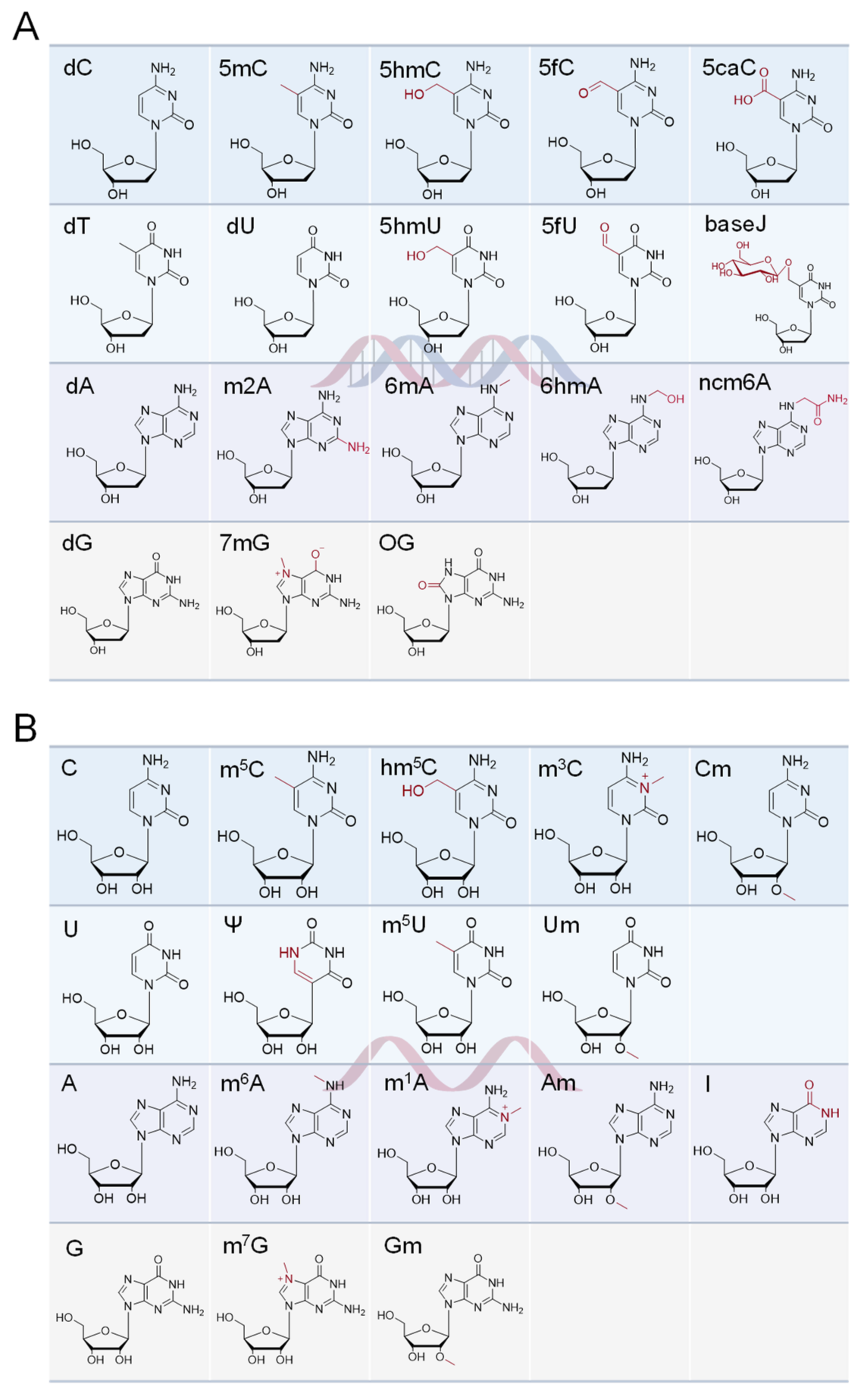
2. Nucleic Acid Modifications
2.1. Modifications of DNA
2.2. Modifications of RNA
3. Qualitative and Quantitative Analysis
3.1. Preparation of Biological Samples
3.1.1. Hydrolysis
3.1.2. Nucleoside Extraction
3.1.3. Chemical Labeling
| Labeling Reagent | Structure | Target Nucleoside | Reaction Condition | LOD | Sensitivity Increase Fold | Sample Consumption | Ref. |
|---|---|---|---|---|---|---|---|
| MSTFA |  | all | 10 μL of methoxyamine hydrochloride as 20 mg/mL solution in pyridine, 90 μL of MSTFA | ND. | ND. | metabolites in 25 μL blood plasma, 5 × 106 cells, or 5 mg tissues | [30] |
| acetone | 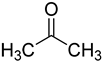 | ribonucleosides | 400 μL of acetone with p-toluene sulfonic acid (1 mg/mL), 50 °C, 2 h | 0.6–6.5 fmol | 7–30 folds | metabolites in 100 μL of urine | [71] |
| BSTFA | 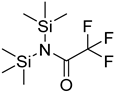 | all | 120 μL of BSTFA, 70 °C, 2 h | ND. | ND. | 10 mg of freeze-dried leaves | [79] |
| acetone | 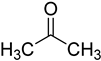 | ribonucleosides | 600 μL of acetone, 6 μL of HClO4, vortex for 30 s, −20 °C for 30 min | 0.026–0.16 ng/mL, 10 μL | ND. | metabolites in 100 μL urine | [80] |
| iodomethane-d3 | 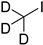 | all | iodomethane-d3, on beads, room temperature, 10 min | 10 fmol/μL, 2 μL | ND. | 1 µg of purified DNA or RNA | [81] |
| 8-DMQ | 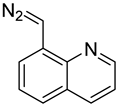 | nucleoside triphosphates | 50/1 molar ratio of 8-DMQ/analyte in 50 mM borate buffer (pH 6.9, 160 μL) with DMSO (40 μL), 25 °C,10 min | 0.4–1.3 fmol | 56–137 folds | metabolites in 1.0 × 107 cells | [82] |
| DMPA |  | nucleotide | molar ratios of DMPA and EDC over nucleotides were set as 40,000 and 5000, with 100 μL of imidazole solution (1 mM, pH 6.0), 50 °C, 1.5 h | 0.12–0.47 fmol | 88–372 folds | metabolites in urine, tissue and cell line samples | [83] |
| 2-DMBA, d5-2DMBA |  | nucleotides, nucleoside diphosphates, nucleoside triphosphates | 200 μL of 250 mg/L 2-DMBA in pH 7.0 borate buffer, 30 °C, 30 min | 0.07–0.39 fmol | 17–174 folds | metabolites in 20 mg of tissue and cell line samples | [84] |
| BDAPE | 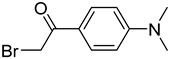 | 5mC, 5hmC, 5fC, 5caC | 4 mM of BDAPE in 200 μL of ACN using 4 mM Et3N as the catalyst, 60 °C, 6 h | 0.06–0.23 fmol | 35–123 folds | 10 μg of genomic DNA | [85] |
| BDMOPE, BMOPE, BDEPE |  | m5Cm, hm5Cm, f5Cm, ca5Cm | 6 mM BDMOPE and 6 mM triethylamine, 60 °C, 6 h | 0.06–0.22 fmol by BDMOPE labeling | 46–462 folds | 10 μg of total RNA and small RNA | [86] |
| BrDPE | 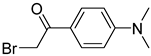 | C, dC, A, dA, G, dG, T, dT, U | BrDPE/analyte ratio 200/1 and 4 mM triethylamine in 125 μL solvent, 40 °C, 3 h | 0.3–12.5 fmol | 31–107 folds | metabolites in 0.2 g of dry sample | [87] |
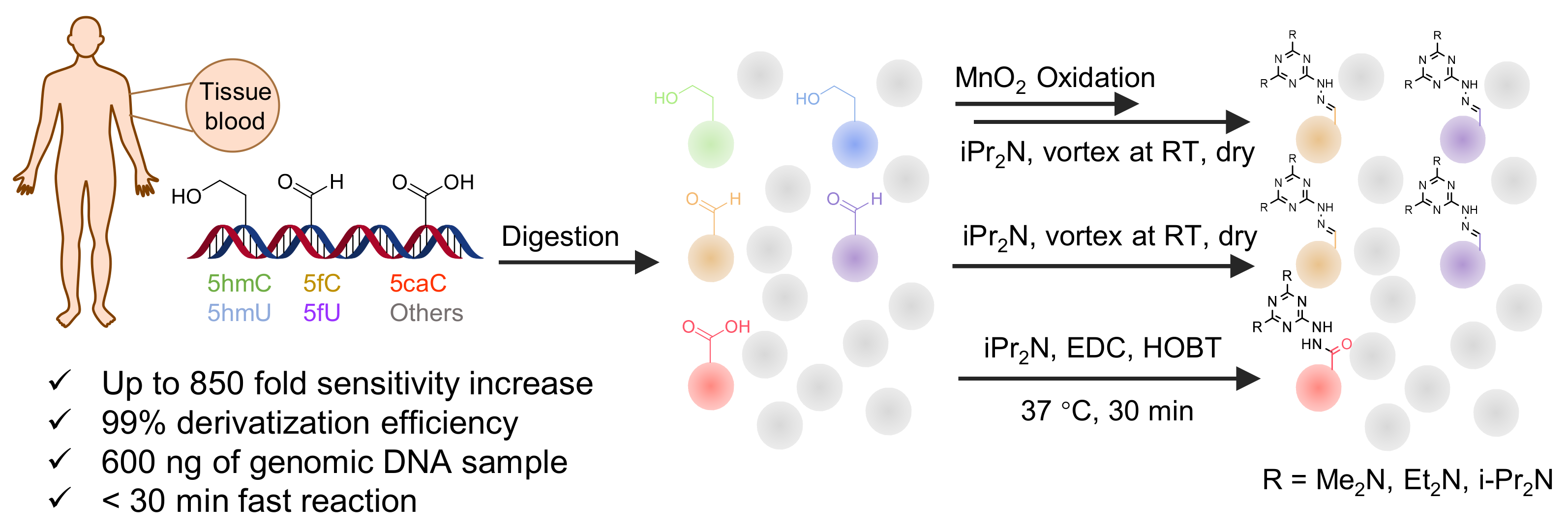
| Me2N, Et2N, and i-Pr2N | 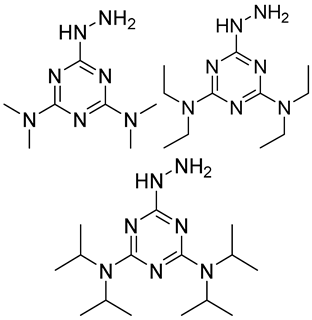 | 5fC, 5caC | 5 mM labeling reagents with 1% HAc in 20% MeOH votex for 10 s for 5fC; labeling reagents in 20 μL 50% ACN, 10 μL 4 mg/mL HOBT and 10 μL 50 mg/mL EDC, 37 °C, 30 min for 5caC | 10–25 amol | 100–125 folds | 600 ng of genomic DNA | [90] |
| i-Pr2N |  | 5hmC | 5 mg of MnO2 in 20 μL reaction volume, 50 °C, 1 h; 5 μL of oxidation product, 1 μL of HAc, 14 μL of 50 mM i-Pr2N solution, vortex and dry | 14 amol | 178 folds | 0.6–2.4 ng of cell-free DNA | [91] |
| i-Pr2N | 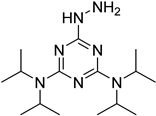 | 5fU, 5hmU, 5fC, 5hmC | 5 mg of MnO2, 2 μL FA, 50 °C, 1 h; 1 mg/mL i-Pr2N and 2 µL HAc, vortex | 26.0–44.4 amol | 275–850 folds | 2 μg of genomic DNA | [92] |
| GirP, GirT and 4-APC |  | 5fdC, 5frC, 5fdU, 5frU, 5frCm, 5frUm | GirP/analyte ratio 50/1, 30 °C, 5 min | 0.03–0.05 fmol | 115–880 folds | mixture of 10 μg genomic DNA and 10 μg total RNA | [93] |
| GirP, GirT and GirD | 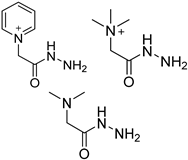 | 5fC, 5caC | GirD/analyte ratio 50/1–150/1, 40 °C, 5–40 min | 0.03–0.42 fmol | 52–260 folds | 20 μg of genomic DNA | [94] |
| rhodamine B hydrazine | 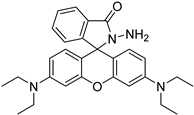 | 5fC | 10 μL of 5 mM labeling reagent in MeOH, 0.2 μL HAc, vortex and dry | 3 amol | 300 folds | total RNA in cell line sample | [95] |
| CAX-B | 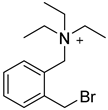 | bases that have active hydrogen | CAX-B in 50% ACN (20 mg/mL), with Et3N(20 μL/mL), was mixed 1:1 with the sample solution, 45 °C, 2 h | 160 amol thymidine | ND. | ND. | [96] |
| Dns-Cl, Dens-Cl | 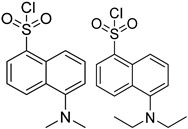 | C, dC, 5mdC, m5C, A, m1A, m6A | 100 μL of reaction buffer (pH 11) and 100 μL of Dns-Cl, 30 °C, 1 h | 0.001–0.01 μg/mL, 5 μL | 1.6–400 folds | metabolites in 106 cells | [97] |
| hydroxyl amine |  | AP sites, βE sites | 1.5 mM derivatization reagents in HEPES (20 mM, pH = 7.5) and Na2EDTA (0.1 mM), 37 °C, 40 min | 0.11 fmol | ND. | 5–20 μg of genomic DNA | [98] |
| CMCT | 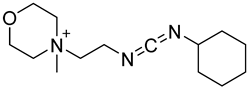 | Ψ, U, m5U, m6U, mcm5U, hm5U, m1Ψ, mo5U | 50 mM CMCT in borate buffer (50 mM, pH 8.5), 40 °C, 14 h | 0.29–2.20 fmol | 6–1408 folds | 500 ng of mRNA | [99] |
3.2. Chromatography-Coupled Mass Spectrometry Technique
3.2.1. LC–MS
3.2.2. CE–MS
3.2.3. Other Mass Spectrometry-Based Techniques
4. Data Analysis
5. Disease Diagnoses Based on Nucleic Acid Modifications
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, H.; Zhang, Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell 2014, 156, 45–68. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Raiber, E.A.; Hardisty, R.; van Delft, P.; Balasubramanian, S. Mapping and elucidating the function of modified bases in DNA. Nat. Rev. Chem. 2017, 1, 0069. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.D. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948, 175, 315–332. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Riggs, A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 1975, 14, 9–25. [Google Scholar] [CrossRef]
- Sood, A.J.; Viner, C.; Hoffman, M.M. DNAmod: The DNA modification database. J. Cheminformatics 2019, 11, 30. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.H.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Vilkaitis, G.; Merkiene, E.; Serva, S.; Weinhold, E.; Klimasauskas, S. The mechanism of DNA cytosine-5 methylation: Kinetic and mutational dissection of HhaI methyltransferase. J. Biol. Chem. 2001, 276, 20924–20934. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhao, B.; He, C.A. Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol. 2016, 23, 74–85. [Google Scholar] [CrossRef] [PubMed]
- He, Y.F.; Li, B.Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.Y.; Ding, J.P.; Jia, Y.Y.; Chen, Z.C.; Li, L.; et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Kriaucionis, S.; Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.F. Assessment of DNA epigenetic modifications. Chem. Res. Toxicol. 2020, 33, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Globisch, D.; Münzel, M.; Müller, M.; Michalakis, S.; Wagner, M.; Koch, S.; Brückl, T.; Biel, M.; Carell, T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 2010, 5, 15367. [Google Scholar] [CrossRef]
- Pfaffeneder, T.; Spada, F.; Wagner, M.; Brandmayr, C.; Laube, S.K.; Eisen, D.; Truss, M.; Steinbacher, J.; Hackner, B.; Kotljarova, O.; et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014, 10, 574–581. [Google Scholar] [CrossRef]
- Gedik, C.M.; Collins, A.; Dubois, J.; Duez, P.; Kouegnigan, L.; Rees, J.F.; Loft, S.; Moller, P.; Jensen, A.; Poulsen, H.; et al. Escodd, Establishing the background level of base oxidation in human lymphocyte DNA: Results of an interlaboratory validation study. FASEB J. 2005, 19, 82–84. [Google Scholar]
- Mangal, D.; Vudathala, D.; Park, J.H.; Lee, S.H.; Penning, T.M.; Blair, I.A. Analysis of 7,8-dihydro-8-oxo-2′-deoxyguanosine in cellular DNA during oxidative stress. Chem. Res. Toxicol. 2009, 22, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Puig, R.; Bueno-Costa, A.; Esteller, M. Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. 2020, 474, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Zhu, P.; Ma, S.Q.; Song, J.H.; Bai, J.Y.; Sun, F.F.; Yi, C.Q. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015, 11, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.X.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Milanowska, K.; Oglou, O.O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways-2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef] [PubMed]
- Basanta-Sanchez, M.; Temple, S.; Ansari, S.A.; D’Amico, A.; Agris, P.F. Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic Acids Res. 2016, 44, e26. [Google Scholar] [CrossRef] [PubMed]
- Ranogajec, A.; Beluhan, S.; Smit, Z. Analysis of nucleosides and monophosphate nucleotides from mushrooms with reversed-phase HPLC. J. Sep. Sci. 2010, 33, 1024–1033. [Google Scholar] [CrossRef]
- Sotgia, S.; Zinellu, A.; Pisanu, E.; Murgia, L.; Pinna, G.A.; Gaspa, L.; Deiana, L.; Carru, C. A hydrophilic interaction ultraperformance liquid chromatography (HILIC-UPLC) method for genomic DNA methylation assessment by UV detection. Anal. Bioanal. Chem. 2010, 396, 2937–2941. [Google Scholar] [CrossRef]
- Lai, Z.J.; Kind, T.; Fiehn, O. Using accurate mass gas chromatography-mass spectrometry with the MINE database for epimetabolite annotation. Anal. Chem. 2017, 89, 10171–10180. [Google Scholar] [CrossRef]
- Morla-Folch, J.; Xie, H.N.; Gisbert-Quilis, P.; Gómez-de Pedro, S.; Pazos-Perez, N.; Alvarez-Puebla, R.A.; Guerrini, L. Ultrasensitive direct quantification of nucleobase modifications in DNA by surface-enhanced Raman scattering: The case of cytosine. Angew. Chem.-Int. Ed. 2015, 54, 13650–13654. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.Y.; Zhou, Q.Y.; Dong, J.H.; Yu, Y.; Zhou, Y.L.; Zhang, X.X. Detection of average methylation level of specific genes by binary-probe hybridization. Talanta 2021, 234, 122630. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zou, G.R.; Tang, J.; Guo, J.Y.; Wang, F.; Chen, Z.L. Probe-labeled electrochemical approach for highly selective detection of 5-carboxycytosine in DNA. Anal. Chim. Acta 2023, 1273, 341521. [Google Scholar] [CrossRef]
- Jiang, S.; Cai, Y.B.; Zhang, Q.Y.; Liu, Q.; Wang, Z.Y.; Zhang, C.Y. Bioorthogonal reaction-mediated enzymatic elongation-driven dendritic nanoassembly for genome-wide analysis of 5-hydroxymethyluracil in breast tissues. Nano Lett. 2023, 23, 10625–10632. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wu, J.; Wu, W.X.; Luo, N.N.; Huang, H.; Chen, Y.H.; Sun, J.; Yu, Q.; Ao, H.; Xu, Q.Q.; et al. AuNPs@MoSe2 heterostructure as a highly efficient coreaction accelerator of electrocheluminescence for amplified immunosensing of DNA methylation. Biosens. Bioelectron. 2023, 222, 114976. [Google Scholar] [CrossRef]
- Guo, J.Y.; Zhao, M.; Chen, C.; Wang, F.; Chen, Z.L. A laser-induced graphene-based electrochemical immunosensor for nucleic acid methylation detection. Analyst 2023, 149, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Kojima, N.; Suda, T.; Fujii, S.; Hirano, K.; Namihira, M.; Kurita, R. Quantitative analysis of global 5-methyl- and 5-hydroxymethylcytosine in TET1 expressed HEK293T cells. Biosens. Bioelectron. 2020, 167, 112472. [Google Scholar] [CrossRef]
- Chowdhury, B.; Cho, I.H.; Hahn, N.; Irudayaraj, J. Quantification of 5-methylcytosine, 5-hydroxymethylcytosine and 5-carboxylcytosine from the blood of cancer patients by an enzyme-based immunoassay. Anal. Chim. Acta 2014, 852, 212–217. [Google Scholar] [CrossRef]
- Crain, P.F. Preparation and enzymatic-hydrolysis of DNA and RNA for mass-spectrometry. Methods Enzymol. 1990, 193, 782–790. [Google Scholar]
- Dong, M.; Wang, C.; Deen, W.M.; Dedon, P.C. Absence of 2′-deoxyoxanosine and presence of abasic sites in DNA exposed to nitric oxide at controlled physiological concentrations. Chem. Res. Toxicol. 2003, 16, 1044–1055. [Google Scholar] [CrossRef]
- Liu, Z.F.; Liu, S.J.; Xie, Z.L.; Blum, W.; Perrotti, D.; Paschka, P.; Kilsovic, R.; Byrd, J.; Chan, K.K.; Marcucci, G. Characterization of in vitro and in vivo hypomethylating effects of decitabine in acute myeloid leukemia by a rapid, specific and sensitive LC-MS/MS method. Nucleic Acids Res. 2007, 35, e31. [Google Scholar] [CrossRef]
- Breyer, V.; Frischmann, M.; Bidmon, C.; Schemm, A.; Schiebel, K.; Pischetsrieder, M. Analysis and biological relevance of advanced glycation end-products of DNA in eukaryotic cells. FEBS J. 2008, 275, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Quinlivan, E.P.; Gregory, J.F. DNA digestion to deoxyribonucleo side: A simplified one-step procedure. Anal. Biochem. 2008, 373, 383–385. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Chan, C.T.Y.; Gu, C.; Lim, K.S.; Chionh, Y.H.; McBee, M.E.; Russell, B.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014, 9, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.T.; Zhang, L.J.; Zhou, K.Y.; Ye, X.X.; Zhang, J.J.; Xie, A.M.; Chen, L.Y.; Kang, J.X.; Cai, C. Simultaneous determination of global DNA methylation and hydroxymethylation levels by hydrophilic interaction liquid chromatography-tandem mass spectrometry. J. Biomol. Screen. 2012, 17, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Rossella, F.; Polledri, E.; Bollati, V.; Baccarelli, A.; Fustinoni, S. Development and validation of a gas chromatography/mass spectrometry method for the assessment of genomic DNA methylation. Rapid Commun. Mass Spectrom. 2009, 23, 2637–2646. [Google Scholar] [CrossRef] [PubMed]
- Rastegar, L.; Mighani, H.; Ghassempour, A. A comparison and column selection of hydrophilic interaction liquid chromatography and reversed-phase high-performance liquid chromatography for detection of DNA methylation. Anal. Biochem. 2018, 557, 123–130. [Google Scholar] [CrossRef]
- Weinfeld, M.; Soderlind, K.J.M.; Buchko, G.W. Influence of nucleic-acid base aromaticity on substrate reactivity with enzymes acting on single-stranded-DNA. Nucleic Acids Res. 1993, 21, 621–626. [Google Scholar] [CrossRef]
- Yuan, F.; Bi, Y.; Zhang, J.Y.; Zhou, Y.L.; Zhang, X.X.; Song, C.X. 5-Carboxylcytosine is resistant towards phosphodiesterase I digestion: Implications for epigenetic modification quantification by mass spectrometry. RSC Adv. 2019, 9, 29010–29014. [Google Scholar] [CrossRef]
- Chu, J.M.; Ye, T.T.; Ma, C.J.; Lan, M.D.; Liu, T.; Yuan, B.F.; Feng, Y.Q. Existence of Internal N7-methylguanosine modification in mRNA determined by differential enzyme treatment coupled with mass spectrometry analysis. ACS Chem. Biol. 2018, 13, 3243–3250. [Google Scholar] [CrossRef]
- Kaiser, S.; Byrne, S.R.; Ammann, G.; Atoi, P.A.; Borland, K.; Brecheisen, R.; DeMott, M.S.; Gehrke, T.; Hagelskamp, F.; Heiss, M.; et al. Strategies to avoid artifacts in mass spectrometry-based epitranscriptome analyses. Angew. Chem.-Int. Ed. 2021, 60, 23885–23893. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chew, B.L.A.; Lai, Y.; Dong, H.P.; Xu, L.; Liu, Y.; Fu, X.Y.; Lin, Z.G.; Shi, P.Y.; Lu, T.K.; et al. A systems-level mass spectrometry-based technique for accurate and sensitive quantification of the RNA cap epitranscriptome. Nat. Protoc. 2023, 18, 2671–2698. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Hurley, L.H. The importance of negative superhelicity in inducing the formation of g-quadruplex and i-motif structures in the c-Myc promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 2009, 52, 2863–2874. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.X.; Zhu, J.B.; Zhang, L.B.; Du, Y.; Dong, S.J.; Wang, E.K. G-quadruplex-based fluorescent assay of S1 nuclease activity and K+. Anal. Chem. 2013, 85, 2431–2435. [Google Scholar] [CrossRef] [PubMed]
- Matuszewski, M.; Wojciechowski, J.; Miyauchi, K.; Gdaniec, Z.; Wolf, W.M.; Suzuki, T.; Sochacka, E. A hydantoin isoform of cyclic N6-threonylcarbamoyladenosine (ct6A) is present in tRNAs. Nucleic Acids Res. 2017, 45, 2137–2149. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh, K.; McFaline, J.L.; Pang, B.; Sullivan, M.; Dong, M.; Plummer, E.; Dedon, P.C. Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nat. Protoc. 2008, 3, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Jora, M.; Borland, K.; Abernathy, S.; Zhao, R.X.; Kelley, M.; Kellner, S.; Addepalli, B.; Limbach, P.A. Chemical amination/imination of carbonothiolated nucleosides during RNA hydrolysis. Angew. Chem.-Int. Ed. 2021, 60, 3961–3966. [Google Scholar] [CrossRef]
- Macon, J.B.; Wolfenden, R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry 1968, 7, 3453–3458. [Google Scholar] [CrossRef]
- Lai, W.Y.; Lyu, C.; Wang, H.L. Vertical ultrafiltration-facilitated DNA digestion for rapid and sensitive UHPLC-MS/MS detection of DNA modifications. Anal. Chem. 2018, 90, 6859–6866. [Google Scholar] [CrossRef]
- Song, X.R.; Song, X.Y.; Lai, W.Y.; Wang, H.L. Hyperactive DNA cutting for unbiased UHPLC-MS/MS quantification of epigenetic DNA marks by engineering DNase I mutants. Anal. Chem. 2022, 94, 17670–17676. [Google Scholar] [CrossRef]
- Delfino, D.; Mori, G.; Rivetti, C.; Grigoletto, A.; Bizzotto, G.; Cavozzi, C.; Malatesta, M.; Cavazzini, D.; Pasut, G.; Percudani, R. Actin-resistant DNase1L2 as a potential therapeutics for CF lung disease. Biomolecules 2021, 11, 410. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.Q.; Lazarus, R.A. Engineering hyperactive variants of human deoxyribonuclease I by altering its functional mechanism. Biochemistry 1997, 36, 6624–6632. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.F.; Chen, S.K.; Zhang, N.; Wang, H.L. Multienzyme cascade bioreactor for a 10 min digestion of genomic DNA into single nucleosides and quantitative detection of structural DNA modifications in cellular genomic DNA. ACS Appl. Mater. Interfaces 2018, 10, 21883–21890. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.C.; Mo, J.Z.; Lu, M.L.; Wang, H.L. Detection of human urinary 5-hydroxymethylcytosine by stable isotope dilution HPLC-MS/MS analysis. Anal. Chem. 2015, 87, 1846–1852. [Google Scholar] [CrossRef] [PubMed]
- He, R.J.; Qiao, J.; Wang, X.X.; Chen, W.L.; Yin, T. A new quantitative method for pseudouridine and uridine in human serum and its clinical application in acute myeloid leukemia. J. Pharm. Biomed. Anal. 2022, 219, 114934. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.Y.; Lin, W.D.; Tsai, Y.H.; Lin, C.T.; Wang, H.C.; Jeng, L.B.; Lee, C.C.; Lin, Y.C.; Lai, C.C.; Tsai, F.J. Analysis of urinary nucleosides as potential tumor markers in human breast cancer by high performance liquid chromatography/electrospray ionization tandem mass spectrometry. Clin. Chim. Acta 2011, 412, 1861–1866. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Hu, Y.Q.; Cao, X.J.; Wang, Y.S. HILIC-MS/MS for the determination of methylated adenine nucleosides in human urine. Anal. Chem. 2021, 93, 17060–17068. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Iida, K.; Oda, Y.; Umemura, T.; Nakajima, H.; Esaka, Y.; Inoue, Y.; Teshima, N. Hydrophilic interaction chromatography-type sorbent prepared by the modification of methacrylate-base resin with polyethyleneimine for solid-phase extraction of polar compounds. Anal. Sci. 2023, 39, 375–381. [Google Scholar] [CrossRef]
- Jiang, H.P.; Chu, J.M.; Lan, M.D.; Liu, P.; Yang, N.; Zheng, F.; Yuan, B.F.; Feng, Y.Q. Comprehensive profiling of ribonucleosides modification by affinity zirconium oxide-silica composite monolithic column online solid-phase microextraction—Mass spectrometry analysis. J. Chromatogr. A 2016, 1462, 90–99. [Google Scholar] [CrossRef]
- Wang, S.T.; Huang, W.; Lu, W.; Yuan, B.F.; Feng, Y.Q. TiO2-based solid phase extraction strategy for highly effective elimination of normal ribonucleosides before detection of 2′-deoxynucleosides/low-abundance 2′-O-modified ribonucleosides. Anal. Chem. 2013, 85, 10512–10518. [Google Scholar] [CrossRef]
- Chu, J.M.; Qi, C.B.; Huang, Y.Q.; Jiang, H.P.; Hao, Y.H.; Yuan, B.F.; Feng, Y.Q. Metal oxide-based selective enrichment combined with stable isotope labeling-mass spectrometry analysis for profiling of ribose conjugates. Anal. Chem. 2015, 87, 7364–7372. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shan, Y.H.; Qiao, L.Z.; Dou, A.; Shi, X.Z.; Xu, G.W. Facile synthesis of boronate-decorated polyethyleneimine-grafted hybrid magnetic nanoparticles for the highly selective enrichment of modified nucleosides and ribosylated metabolites. Anal. Chem. 2013, 85, 11585–11592. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Du, J.; Wang, C.Z.; Wei, Y.M. Fabrication of a dendrimer-modified boronate affinity material for online selective enrichment of cis-diol-containing compounds and its application in determination of nucleosides in urine. RSC Adv. 2015, 5, 106161–106170. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Zou, T.; Feng, S.T.; Wu, F.S.; Zhang, J. Boronic acid-functionalized magnetic porphyrin-based covalent organic framework for selective enrichment of cis-diol-containing nucleosides. Anal. Chim. Acta 2023, 1278, 341691. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; He, Z.D.; Li, W.Z.; Zhao, J.Y.; Chen, T.; Shao, S.M.; Chen, H.M. Reshaping of pipette tip: A facile and practical strategy for sorbent packing-free solid phase extraction. Anal. Chim. Acta 2020, 1100, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Wang, C.Z.; Wei, Y.M. Enhanced binding capacity of boronate affinity fibrous material for effective enrichment of nucleosides in urine samples. RSC Adv. 2016, 6, 28470–28476. [Google Scholar] [CrossRef]
- Ma, Y.F.; Yuan, F.; Yu, Y.; Zhou, Y.L.; Zhang, X.X. Synthesis of a pH-responsive functional covalent organic framework via facile and rapid one-step postsynthetic modification and its application in highly efficient N1-methyladenosine extraction. Anal. Chem. 2020, 92, 1424–1430. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.W.; Bian, H.; Zhu, L.J.; Xia, D.H.; Wang, H.Z. Cyclodextrin porous liquid materials for efficient chiral recognition and separation of nucleosides. ACS Appl. Mater. Interfaces 2020, 12, 45916–45928. [Google Scholar] [CrossRef]
- Becker, M.; Zweckmair, T.; Forneck, A.; Rosenau, T.; Potthast, A.; Liebner, F. Evaluation of different derivatisation approaches for gas chromatographic-mass spectrometric analysis of carbohydrates in complex matrices of biological and synthetic origin. J. Chromatogr. A 2013, 1281, 115–126. [Google Scholar] [CrossRef]
- Li, S.F.; Jin, Y.B.; Tang, Z.; Lin, S.H.; Liu, H.X.; Jiang, Y.Y.; Cai, Z.W. A novel method of liquid chromatography-tandem mass spectrometry combined with chemical derivatization for the determination of ribonucleosides in urine. Anal. Chim. Acta 2015, 864, 30–38. [Google Scholar] [CrossRef]
- Xie, Y.X.; Vitorino, F.N.D.; Chen, Y.; Lempiäinen, J.K.; Zhao, C.F.; Steinbock, R.T.; Lin, Z.T.; Liu, X.Y.; Zahn, E.; Garcia, A.L.; et al. SWAMNA: A comprehensive platform for analysis of nucleic acid modifications. Chem. Commun. 2023, 59, 12499–12502. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.P.; Xiong, J.; Liu, F.L.; Ma, C.J.; Tang, X.L.; Yuan, B.F.; Feng, Y.Q. Modified nucleoside triphosphates exist in mammals. Chem. Sci. 2018, 9, 4160–4167. [Google Scholar] [CrossRef]
- Zeng, H.; Qi, C.B.; Liu, T.; Xiao, H.M.; Cheng, Q.Y.; Jiang, H.P.; Yuan, B.F.; Feng, Y.Q. Formation and determination of endogenous methylated nucleotides in mammals by chemical labeling coupled with mass spectrometry analysis. Anal. Chem. 2017, 89, 4153–4160. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.L.; Qi, C.B.; Cheng, Q.Y.; Ding, J.H.; Yuan, B.F.; Feng, Y.Q. Diazo reagent labeling with mass spectrometry analysis for sensitive determination of ribonucleotides in living organisms. Anal. Chem. 2020, 92, 2301–2309. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zheng, S.J.; Qi, C.B.; Feng, Y.Q.; Yuan, B.F. Sensitive and simultaneous determination of 5-methylcytosine and its oxidation products in genomic DNA by chemical derivatization coupled with liquid chromatography-tandem mass spectrometry analysis. Anal. Chem. 2015, 87, 3445–3452. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ma, C.J.; Ding, J.H.; Qi, C.B.; Xu, X.J.; Yuan, B.F.; Feng, Y.Q. Chemical labeling—Assisted mass spectrometry analysis for sensitive detection of cytidine dual modifications in RNA of mammals. Anal. Chim. Acta 2020, 1098, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.R.; Zhang, L.; Wei, L.J.; Wang, M.L.; Li, B.W.; Guo, B.; Ma, M. One-pot derivatization for wide-scope detection of nucleobases and deoxyribosides in natural medicinal foods with liquid chromatography-tandem mass spectrometry. J. Agric. Food Chem. 2020, 68, 10200–10212. [Google Scholar] [CrossRef]
- Tie, C.; Zhang, X.X. A new labelling reagent for glycans analysis by capillary electrophoresis-mass spectrometry. Anal. Methods 2012, 4, 357–359. [Google Scholar] [CrossRef]
- Zhao, M.Z.; Zhang, Y.W.; Yuan, F.; Deng, Y.; Liu, J.X.; Zhou, Y.L.; Zhang, X.X. Hydrazino-s-triazine based labelling reagents for highly sensitive glycan analysis via liquid chromatography-electrospray mass spectrometry. Talanta 2015, 144, 992–997. [Google Scholar] [CrossRef]
- Yu, Y.; Yuan, F.; Zhang, X.H.; Zhao, M.Z.; Zhou, Y.L.; Zhang, X.X. Ultrasensitive determination of rare modified cytosines based on novel hydrazine labeling reagents. Anal. Chem. 2019, 91, 13047–13053. [Google Scholar] [CrossRef]
- Yuan, F.; Yu, Y.; Zhou, Y.L.; Zhang, X.X. 5hmC-MIQuant: Ultrasensitive quantitative detection of 5-hydroxymethylcytosine in low-input cell-free DNA samples. Anal. Chem. 2020, 92, 1605–1610. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Pan, H.Y.; Zheng, X.; Yuan, F.; Zhou, Y.L.; Zhang, X.X. Ultrasensitive simultaneous detection of multiple rare modified nucleosides as promising biomarkers in low-put breast cancer DNA samples for clinical multi-dimensional diagnosis. Molecules 2022, 27, 7041. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.P.; Liu, T.; Guo, N.; Yu, L.; Yuan, B.F.; Feng, Y.Q. Determination of formylated DNA and RNA by chemical labeling combined with mass spectrometry analysis. Anal. Chim. Acta 2017, 981, 1–10. [Google Scholar] [CrossRef]
- Tang, Y.; Xiong, J.; Jiang, H.P.; Zheng, S.J.; Feng, Y.Q.; Yuan, B.F. Determination of oxidation products of 5-methylcytosine in plants by chemical derivatization coupled with liquid chromatography/tandem mass spectrometry analysis. Anal. Chem. 2014, 86, 7764–7772. [Google Scholar] [CrossRef]
- Chen, Y.N.; Shen, X.Y.; Yu, Y.; Xue, C.Y.; Zhou, Y.L.; Zhang, X.X. In-source fragmentation of nucleosides in electrospray ionization towards more sensitive and accurate nucleoside analysis. Analyst 2023, 148, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.G.; Zhang, Q.; Yao, Y.Y.; Giese, R.W. Cationic xylene tag for increasing sensitivity in mass spectrometry. J. Am. Soc. Mass Spectrom. 2015, 26, 1713–1721. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, P.; Hou, X.Y.; Tang, T.; Li, S.Q.; Sun, R.Q.; Zhang, Z.J.; Xu, F.G. Twins labeling derivatization-based LC-MS/MS strategy for absolute quantification of paired prototypes and modified metabolites. Anal. Chim. Acta 2022, 1193, 339399. [Google Scholar] [CrossRef]
- Rahimoff, R.; Kosmatchev, O.; Kirchner, A.; Pfaffeneder, T.; Spada, F.; Brantl, V.; Müller, M.; Carell, T. 5-Formyl- and 5-carboxydeoxycytidines do not cause accumulation of harmful repair intermediates in stem cells. J. Am. Chem. Soc. 2017, 139, 10359–10364. [Google Scholar] [CrossRef]
- Cheng, Q.Y.; Xiong, J.; Ma, C.J.; Dai, Y.; Ding, J.H.; Liu, F.L.; Yuan, B.F.; Feng, Y.Q. Chemical tagging for sensitive determination of uridine modifications in RNA. Chem. Sci. 2020, 11, 1878–1891. [Google Scholar] [CrossRef]
- Tang, Y.; Chu, J.M.; Huang, W.; Xiong, J.; Xing, X.W.; Zhou, X.; Feng, Y.Q.; Yuan, B.F. Hydrophilic material for the selective enrichment of 5-hydroxymethylcytosine and its liquid chromatography-tandem mass spectrometry detection. Anal. Chem. 2013, 85, 6129–6135. [Google Scholar] [CrossRef]
- Hu, H.; Flynn, N.; Zhang, H.L.; You, C.J.; Hang, R.L.; Wang, X.F.; Zhong, H.; Chan, Z.L.; Xia, Y.J.; Chen, X.M. SPAAC-NAD-seq, a sensitive and accurate method to profile NAD+-capped transcripts. Proc. Natl. Acad. Sci. USA 2021, 118, e2025595118. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Jia, G.F.; Pang, X.Q.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A.; et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef]
- Fu, L.J.; Amato, N.J.; Wang, P.C.; McGowan, S.J.; Niedernhofer, L.J.; Wang, Y.S. Simultaneous quantification of methylated cytidine and adenosine in cellular and tissue RNA by nano-flow liquid chromatography-tandem mass spectrometry coupled with the stable isotope-dilution method. Anal. Chem. 2015, 87, 7653–7659. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, G.; Cui, Y.X.; Wang, P.C.; Wang, Y.S. Normalized retention time for scheduled liquid chromatography-multistage mass spectrometry analysis of epitranscriptomic modifications. J. Chromatogr. A 2020, 1623, 461181. [Google Scholar] [CrossRef]
- Song, L.G.; James, S.R.; Kazim, L.; Karpf, A.R. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal. Chem. 2005, 77, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Wilm, M.; Mann, M. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 1996, 68, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Karas, M.; Dülcks, T. Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: When does ESI turn into nano-ESI? J. Am. Soc. Mass Spectrom. 2003, 14, 492–500. [Google Scholar] [CrossRef]
- Sesták, J.; Moravcová, D.; Kahle, V. Instrument platforms for nano liquid chromatography. J. Chromatogr. A 2015, 1421, 2–17. [Google Scholar] [CrossRef]
- Sun, Y.; Stransky, S.; Aguilan, J.; Brenowitz, M.; Sidoli, S. DNA methylation and hydroxymethylation analysis using a high throughput and low bias direct injection mass spectrometry platform. Methodsx 2021, 8, 338880. [Google Scholar] [CrossRef]
- Embrechts, J.; Lemière, F.; Van Dongen, W.; Esmans, E.L.; Buytaert, P.; Van Marck, E.; Kockx, M.; Makar, A. Detection of estrogen DNA-adducts in human breast tumor tissue and healthy tissue by combined nano LC-nano ES tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2003, 14, 482–491. [Google Scholar] [CrossRef]
- Sarin, L.P.; Kienast, S.D.; Leufken, J.; Ross, R.L.; Dziergowska, A.; Debiec, K.; Sochacka, E.; Limbach, P.A.; Fufezan, C.; Drexler, H.C.A.; et al. Nano LC-MS using capillary columns enables accurate quantification of modified ribonucleosides at low femtomol levels. RNA 2018, 24, 1403–1417. [Google Scholar] [CrossRef] [PubMed]
- Mateos-Vivas, M.; Fanali, S.; Rodríguez-Gonzalo, E.; Carabias-Martínez, R.; Aturki, Z. Rapid determination of nucleotides in infant formula by means of nano-liquid chromatography. Electrophoresis 2016, 37, 1873–1880. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.Y.; Liu, G.Z.; Li, F.; Kang, J.W. Determination of 5-methyldeoxycytosine and oxidized derivatives by nano-liquid chromatography with zwitterionic monolithic capillary column. J. Chromatogr. A 2023, 1693, 463895. [Google Scholar] [CrossRef]
- Lin, X.Y.; Zhang, Q.H.; Qin, Y.C.; Zhong, Q.S.; Lv, D.Z.; Wu, X.P.; Fu, P.C.; Lin, H. Potential misidentification of natural isomers and mass-analogs of modified nucleosides by liquid chromatography-triple quadrupole mass spectrometry. Genes 2022, 13, 878. [Google Scholar] [CrossRef] [PubMed]
- Rackowska, E.; Bobrowska-Korczak, B.; Giebultowicz, J. Development and validation of a rapid LC-MS/MS method for determination of methylated nucleosides and nucleobases in urine. J. Chromatogr. B—Anal. Technol. Biomed. Life Sci. 2019, 1128, 121775. [Google Scholar] [CrossRef] [PubMed]
- Kok, R.M.; Smith, D.E.C.; Barto, R.; Spijkerman, A.M.W.; Teerlink, T.; Gellekink, H.J.; Jakobs, C.; Smulders, Y.M. Global DNA methylation measured by liquid chromatography-tandem mass spectrometry: Analytical technique, reference values and determinants in healthy subjects. Clin. Chem. Lab. Med. 2007, 45, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Xie, C.; Chen, Q.; Cao, X.J.; Guo, M.Z.; Zheng, S.; Wang, Y.S. A novel malic acid-enhanced method for the analysis of 5-methyl-2′-deoxycytidine, 5-hydroxymethyl-2′-deoxycytidine, 5-methylcytidine and 5-hydroxymethylcytidine in human urine using hydrophilic interaction liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 2018, 1034, 110–118. [Google Scholar] [CrossRef]
- Fraga, M.F.; Rodríguez, R.; Cañal, M.J. Rapid quantification of DNA methylation by high performance capillary electrophoresis. Electrophoresis 2000, 21, 2990–2994. [Google Scholar] [CrossRef]
- Sotgia, S.; Carru, C.; Franconi, F.; Fiori, P.B.; Manca, S.; Pettinato, S.; Magliona, S.; Ginanneschi, R.; Deiana, L.; Zinellu, A. Rapid quantification of total genomic DNA methylation degree by short-end injection capillary zone electrophoresis. J. Chromatogr. A 2008, 1185, 145–150. [Google Scholar] [CrossRef]
- Stephen, T.K.L.; Guillemette, K.L.; Green, T.K. Analysis of trinitrophenylated adenosine and inosine by capillary electrophoresis and γ-cyclodextrin-enhanced fluorescence detection. Anal. Chem. 2016, 88, 7777–7785. [Google Scholar] [CrossRef]
- Moini, M. Simplifying CE-MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip. Anal. Chem. 2007, 79, 4241–4246. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, E.J.; Chen, D.D.Y. Twenty years of interface development for capillary electrophoresis-electrospray ionization-mass spectrometry. Anal. Chim. Acta 2008, 627, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, R.; Dada, O.O.; Sadilek, M.; Dovichi, N.J. Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis. Rapid Commun. Mass Spectrom. 2010, 24, 2554–2560. [Google Scholar] [CrossRef] [PubMed]
- Busnel, J.M.; Schoenmaker, B.; Ramautar, R.; Carrasco-Pancorbo, A.; Ratnayake, C.; Feitelson, J.S.; Chapman, J.D.; Deelder, A.M.; Mayboroda, O.A. High capacity capillary electrophoresis-electrospray ionization mass spectrometry: Coupling a porous sheathless interface with transient-isotachophoresis. Anal. Chem. 2010, 82, 9476–9483. [Google Scholar] [CrossRef] [PubMed]
- Faserl, K.; Sarg, B.; Kremser, L.; Lindner, H. Optimization and evaluation of a sheathless capillary electrophoresis-electrospray ionization mass spectrometry platform for peptide analysis: Comparison to liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 2011, 83, 7297–7305. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.L.; Zhu, G.J.; Zhang, Z.B.; Mou, S.; Dovichi, N.J. Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests. J. Proteome Res. 2015, 14, 2312–2321. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Aerts, J.T.; Rubakhin, S.S.; Zhang, X.X.; Sweedler, J.V. Analysis of endogenous nucleotides by single cell capillary electrophoresis-mass spectrometry. Analyst 2014, 139, 5835–5842. [Google Scholar] [CrossRef]
- Liu, C.C.; Huang, J.S.; Tyrrell, D.L.J.; Dovichi, N.J. Capillary electrophoresis-electrospray-mass spectrometry of nucleosides and nucleotides: Application to phosphorylation studies of anti-human immunodeficiency virus nucleosides in a human hepatoma cell line. Electrophoresis 2005, 26, 1424–1431. [Google Scholar] [CrossRef]
- Yuan, F.; Zhang, X.H.; Nie, J.; Chen, H.X.; Zhou, Y.L.; Zhang, X.X. Ultrasensitive determination of 5-methylcytosine and 5-hydroxymethylcytosine in genomic DNA by sheathless interfaced capillary electrophoresis-mass spectrometry. Chem. Commun. 2016, 52, 2698–2700. [Google Scholar] [CrossRef]
- Yu, Y.; Zhu, S.H.; Yuan, F.; Zhang, X.H.; Lu, Y.Y.; Zhou, Y.L.; Zhang, X.X. Ultrasensitive and simultaneous determination of RNA modified nucleotides by sheathless interfaced capillary electrophoresis-tandem mass spectrometry. Chem. Commun. 2019, 55, 7595–7598. [Google Scholar] [CrossRef]
- Lechner, A.; Wolff, P.; Leize-Wagner, E.; François, Y.N. Characterization of post-transcriptional RNA modifications by sheathless capillary electrophoresis-high resolution mass spectrometry. Anal. Chem. 2020, 92, 7363–7370. [Google Scholar] [CrossRef] [PubMed]
- Rouzer, C.A.; Chaudhary, A.K.; Nokubo, M.; Ferguson, D.M.; Reddy, G.R.; Blair, I.A.; Marnett, L.J. Analysis of the malondialdehyde-2′-deoxyguanosine adduct pyrimidopurinone in human leukocyte DNA by gas chromatography electron capture negative chemical ionization mass spectrometry. Chem. Res. Toxicol. 1997, 10, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Jaruga, P.; Rodriguez, H. Measurement of 8-hydroxy-2′-deoxyguanosine in DNA by high-performance liquid chromatography-mass spectrometry: Comparison with measurement by gas chromatography-mass spectrometry. Nucleic Acids Res. 2001, 29, e12. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.L.; Ivanisevic, J.; Benton, H.P.; Johnson, C.H.; Patti, G.J.; Hoang, L.T.; Uritboonthai, W.; Kurczy, M.E.; Siuzdak, G. Thermal degradation of small molecules: A global metabolomic investigation. Anal. Chem. 2015, 87, 10935–10941. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.; Minier, M.A.; Chitranshi, P.; Sparkman, O.D.; Jones, P.R.; Xue, L.A. Direct analysis in real time (DART) mass spectrometry of nucleotides and nucleosides: Elucidation of a novel fragment C5H5O+ and its in-source adducts. J. Am. Soc. Mass Spectrom. 2010, 21, 1371–1381. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.M.; Wan, D.B.; Song, F.R.; Liu, Z.Q.; Liu, S.Y. Argon direct analysis in real time mass spectrometry in conjunction with makeup solvents: A method for analysis of labile compounds. Anal. Chem. 2013, 85, 1305–1309. [Google Scholar] [CrossRef]
- Kanu, A.B.; Hampikian, G.; Brandt, S.D.; Hill, H.H. Ribonucleotide and ribonucleoside determination by ambient pressure ion mobility spectrometry. Anal. Chim. Acta 2010, 658, 91–97. [Google Scholar] [CrossRef]
- Lagies, S.; Schlimpert, M.; Braun, L.M.; Kather, M.; Plagge, J.; Erbes, T.; Wittel, U.A.; Kammerer, B. Unraveling altered RNA metabolism in pancreatic cancer cells by liquid-chromatography coupling to ion mobility mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 6319–6328. [Google Scholar] [CrossRef]
- Quinn, R.; Basanta-Sanchez, M.; Rose, R.E.; Fabris, D. Direct infusion analysis of nucleotide mixtures of very similar or identical elemental composition. J. Mass Spectrom. 2013, 48, 703–712. [Google Scholar] [CrossRef]
- Kenderdine, T.; Nemati, R.; Baker, A.; Palmer, M.; Ujma, J.; FitzGibbon, M.; Deng, L.; Royzen, M.; Langridge, J.; Fabris, D. High-resolution ion mobility spectrometry-mass spectrometry of isomeric/isobaric ribonucleotide variants. J. Mass Spectrom. 2020, 55, e4465. [Google Scholar] [CrossRef]
- Kellner, S.; Neumann, J.; Rosenkranz, D.; Lebedeva, S.; Ketting, R.F.; Zischler, H.; Schneider, D.; Helm, M. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem. Commun. 2014, 50, 3516–3518. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.L.; Yu, N.X.; Zhao, R.X.; Wood, A.; Limbach, P.A. Automated identification of modified nucleosides during hraM-LC-MS/MS using a metabolomics ID workflow with neutral loss detection. J. Am. Soc. Mass Spectrom. 2023, 34, 2785–2792. [Google Scholar] [CrossRef] [PubMed]
- Gosset-Erard, C.; Didierjean, M.; Pansanel, J.; Lechner, A.; Wolff, P.; Kuhn, L.; Aubriet, F.; Leize-Wagner, E.; Chaimbault, P.; François, Y.N. Nucleos’ID: A new search engine enabling the untargeted identification of RNA post-transcriptional modifications from tandem mass of nucleosides. Anal. Chem. 2023, 95, 1608–1617. [Google Scholar] [CrossRef] [PubMed]
- Jora, M.; Corcoran, D.; Parungao, G.G.; Lobue, P.A.; Oliveira, L.F.L.; Stan, G.; Addepalli, B.; Limbach, P.A. Higher-energy collisional dissociation mass spectral networks for the rapid, semi-automated characterization of known and unknown ribonucleoside modifications. Anal. Chem. 2022, 94, 13958–13967. [Google Scholar] [CrossRef] [PubMed]
- Lobue, P.A.; Yu, N.X.; Jora, M.; Abernathy, S.; Limbach, P.A. Improved application of RNAModMapper—An RNA modification mapping software tool—For analysis of liquid chromatography tandem mass spectrometry (LC-MS/MS) data. Methods 2019, 156, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Akiyama, M.; Taoka, M.; Yamauchi, Y.; Nobe, Y.; Ishikawa, H.; Takahashi, N.; Isobe, T. Ariadne: A database search engine for identification and chemical analysis of RNA using tandem mass spectrometry data. Nucleic Acids Res. 2009, 37, e47. [Google Scholar] [CrossRef] [PubMed]
- Caudai, C.; Galizia, A.; Geraci, F.; Le Pera, L.; Morea, V.; Salerno, E.; Via, A.; Colombo, T. AI applications in functional genomics. Comput. Struct. Biotechnol. J. 2021, 19, 5762–5790. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef]
- Sanchez-Mut, J.V.; Heyn, H.; Vidal, E.; Moran, S.; Sayols, S.; Delgado-Morales, R.; Schultz, M.D.; Ansoleaga, B.; Garcia-Esparcia, P.; Pons-Espinal, M.; et al. Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. Transl. Psychiatry 2016, 6, e718. [Google Scholar] [CrossRef]
- Farh, K.K.H.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.H.; Shishkin, A.A.; et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef]
- Ling, C.; Rönn, T. Epigenetics in human obesity and type 2 diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Srivastava, S. Epigenetics in cancer: Implications for early detection and prevention. Lancet Oncol. 2002, 3, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Sendinc, E.; Shi, Y. RNA m6A methylation across the transcriptome. Mol. Cell 2023, 83, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Chu, Q.F.; Zheng, Q.X.; Jiang, S.M.; Bao, Z.Y.; Su, Y.S.; Lu, J.; Li, L.J. Role of main RNA modifications in cancer: N6-methyladenosine, 5-methylcytosine, and pseudouridine. Signal Transduct. Target. Ther. 2022, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- You, X.J.; Zhang, S.; Chen, J.J.; Tang, F.; He, J.G.; Wang, J.; Qi, C.B.; Feng, Y.Q.; Yuan, B.F. Formation and removal of 1,N6-dimethyladenosine in mammalian transfer RNA. Nucleic Acids Res. 2022, 50, 9858–9872. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xiao, H.J.; Yang, Y.H.; Zhang, P.P.; Yuan, J.H.; Zhang, W.; Chen, L.J.; Fan, Y.B.; Zhang, J.Z.; Cheng, H.; et al. Crosstalk between 5-methylcytosine and N6-methyladenosine machinery defines disease progression, therapeutic response and pharmacogenomic landscape in hepatocellular carcinoma. Mol. Cancer 2023, 22, 5. [Google Scholar] [CrossRef]
- Huang, W.; Qi, C.B.; Lv, S.W.; Xie, M.; Feng, Y.Q.; Huang, W.H.; Yuan, B.F. Determination of DNA and RNA methylation in circulating tumor cells by mass spectrometry. Anal. Chem. 2016, 88, 1378–1384. [Google Scholar] [CrossRef]
- Pan, H.Y.; Yu, Y.; Cao, T.; Liu, Y.; Zhou, Y.L.; Zhang, X.X. Systematic profiling of exosomal small RNA epigenetic modifications by high-performance liquid chromatography-mass spectrometry. Anal. Chem. 2021, 93, 14907–14911. [Google Scholar] [CrossRef]
- Clark, K.D.; Rubakhin, S.S.; Sweedler, J.V. Single-neuron RNA modification analysis by mass spectrometry: Characterizing RNA modification patterns and dynamics with single-cell resolution. Anal. Chem. 2021, 93, 14537–14544. [Google Scholar] [CrossRef]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.H.; Song, X.Z.; De Hoff, P.; Morey, R.; Liu, J.S.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef]
- Li, W.Z.; Wang, H.Y.; Zhao, Z.J.; Gao, H.Q.; Liu, C.L.; Zhu, L.; Wang, C.; Yang, Y.L. Emerging nanotechnologies for liquid biopsy: The detection of circulating tumor cells and extracellular vesicles. Adv. Mater. 2019, 31, 1805344. [Google Scholar] [CrossRef]
- Yu, D.; Li, Y.X.; Wang, M.Y.; Gu, J.M.; Xu, W.R.; Cai, H.; Fang, X.J.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.F.; Zhou, X. Labeling and sequencing nucleic acid modifications using bio-orthogonal tools. RSC Chem. Biol. 2022, 3, 994–1007. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Dong, J.-H.; Shen, X.-Y.; Gu, Y.-X.; Zhang, R.-H.; Cui, R.-Y.; Liu, Y.-H.; Zhou, J.; Zhou, Y.-L.; Zhang, X.-X. Qualitative and Quantitative Analytical Techniques of Nucleic Acid Modification Based on Mass Spectrometry for Biomarker Discovery. Int. J. Mol. Sci. 2024, 25, 3383. https://doi.org/10.3390/ijms25063383
Liu Y, Dong J-H, Shen X-Y, Gu Y-X, Zhang R-H, Cui R-Y, Liu Y-H, Zhou J, Zhou Y-L, Zhang X-X. Qualitative and Quantitative Analytical Techniques of Nucleic Acid Modification Based on Mass Spectrometry for Biomarker Discovery. International Journal of Molecular Sciences. 2024; 25(6):3383. https://doi.org/10.3390/ijms25063383
Chicago/Turabian StyleLiu, Ying, Jia-Hui Dong, Xu-Yang Shen, Yi-Xuan Gu, Run-Hong Zhang, Ruo-Yao Cui, Ya-Hong Liu, Jiang Zhou, Ying-Lin Zhou, and Xin-Xiang Zhang. 2024. "Qualitative and Quantitative Analytical Techniques of Nucleic Acid Modification Based on Mass Spectrometry for Biomarker Discovery" International Journal of Molecular Sciences 25, no. 6: 3383. https://doi.org/10.3390/ijms25063383
APA StyleLiu, Y., Dong, J.-H., Shen, X.-Y., Gu, Y.-X., Zhang, R.-H., Cui, R.-Y., Liu, Y.-H., Zhou, J., Zhou, Y.-L., & Zhang, X.-X. (2024). Qualitative and Quantitative Analytical Techniques of Nucleic Acid Modification Based on Mass Spectrometry for Biomarker Discovery. International Journal of Molecular Sciences, 25(6), 3383. https://doi.org/10.3390/ijms25063383








