Systematic Identification of Long Noncoding RNAs during Three Key Organogenesis Stages in Zebrafish
Abstract
:1. Introduction
2. Results
2.1. Quality Assement of High-Throughput Sequencing Data
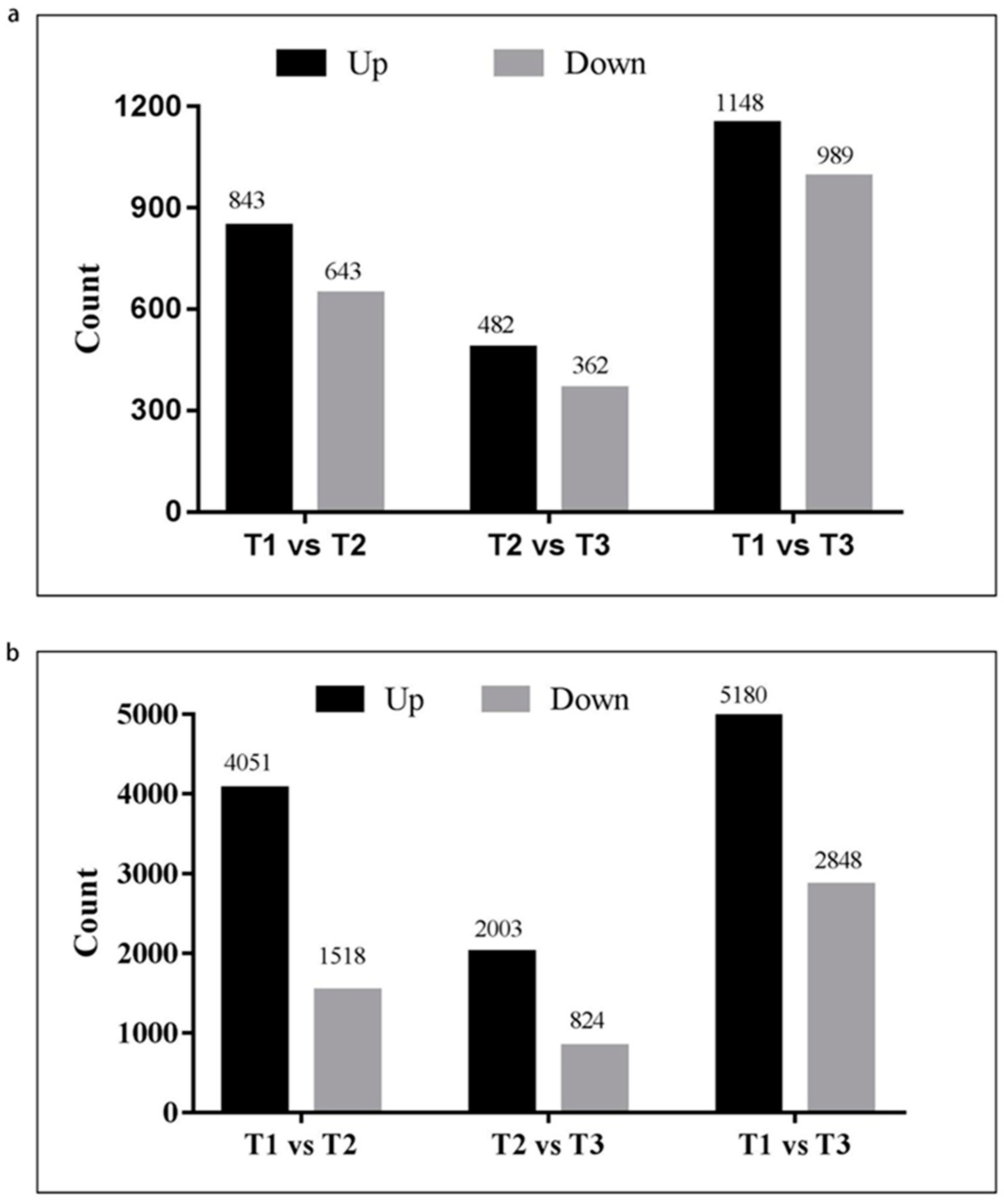
2.2. Identification of Differentially Expressed Genes and lncRNAs at Three Organogenesis Stages
2.3. GO Term and KEGG Pathway Enrichment Analysis of DEGs
2.4. qPCR Validation
2.5. The Spatiotemporal Expression Pattern of lncRNA gas5 at Different Developmental Stages of Zebrafish Embryos
2.6. Expression of lncRNA gas5 in Different Tissues of Adult Zebrafish
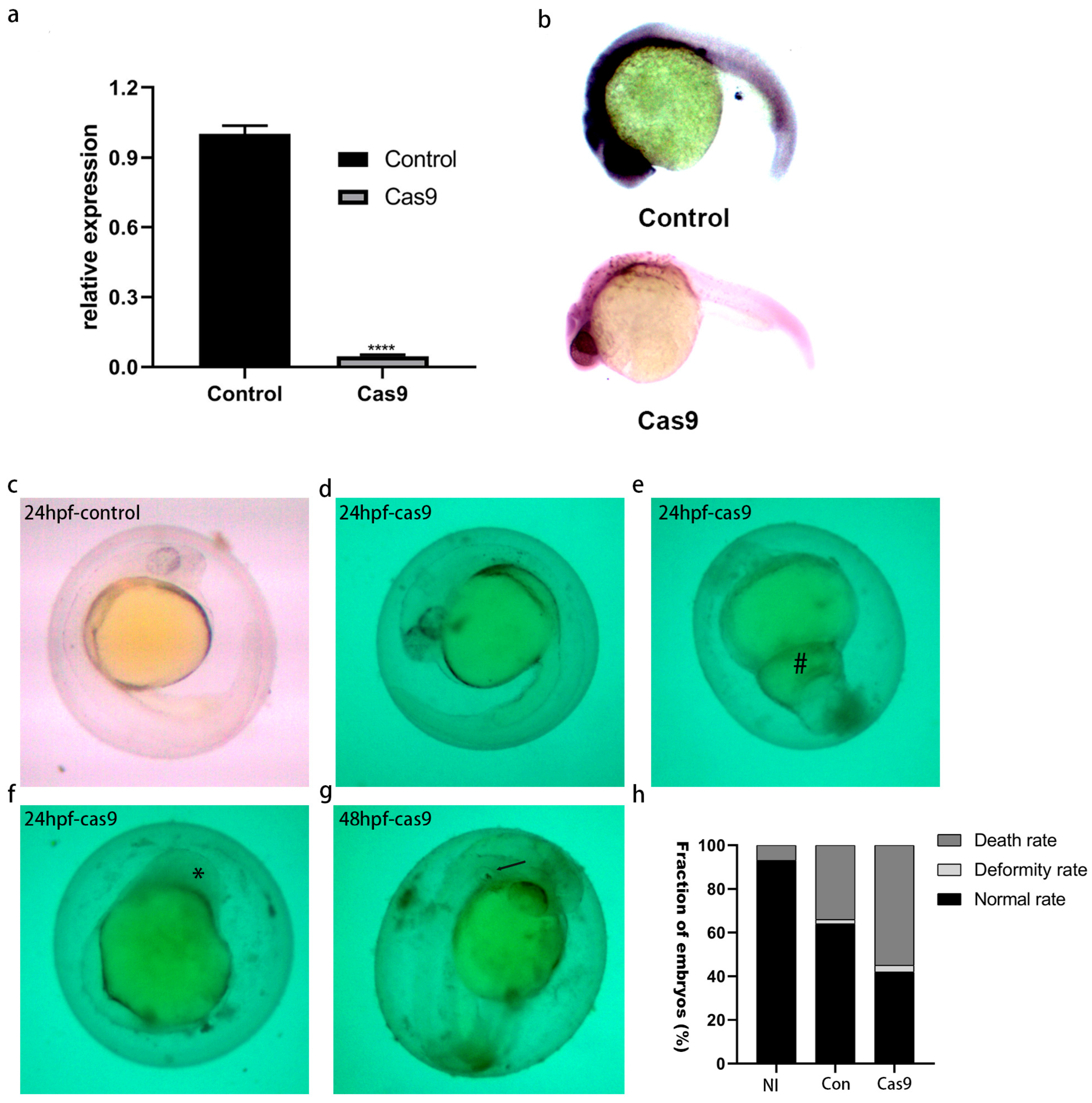
2.7. The Validation of lncRNA gas5 Knockout
2.8. Effect of lncRNA gas5 Knockout on Embryonic Development of Zebrafish
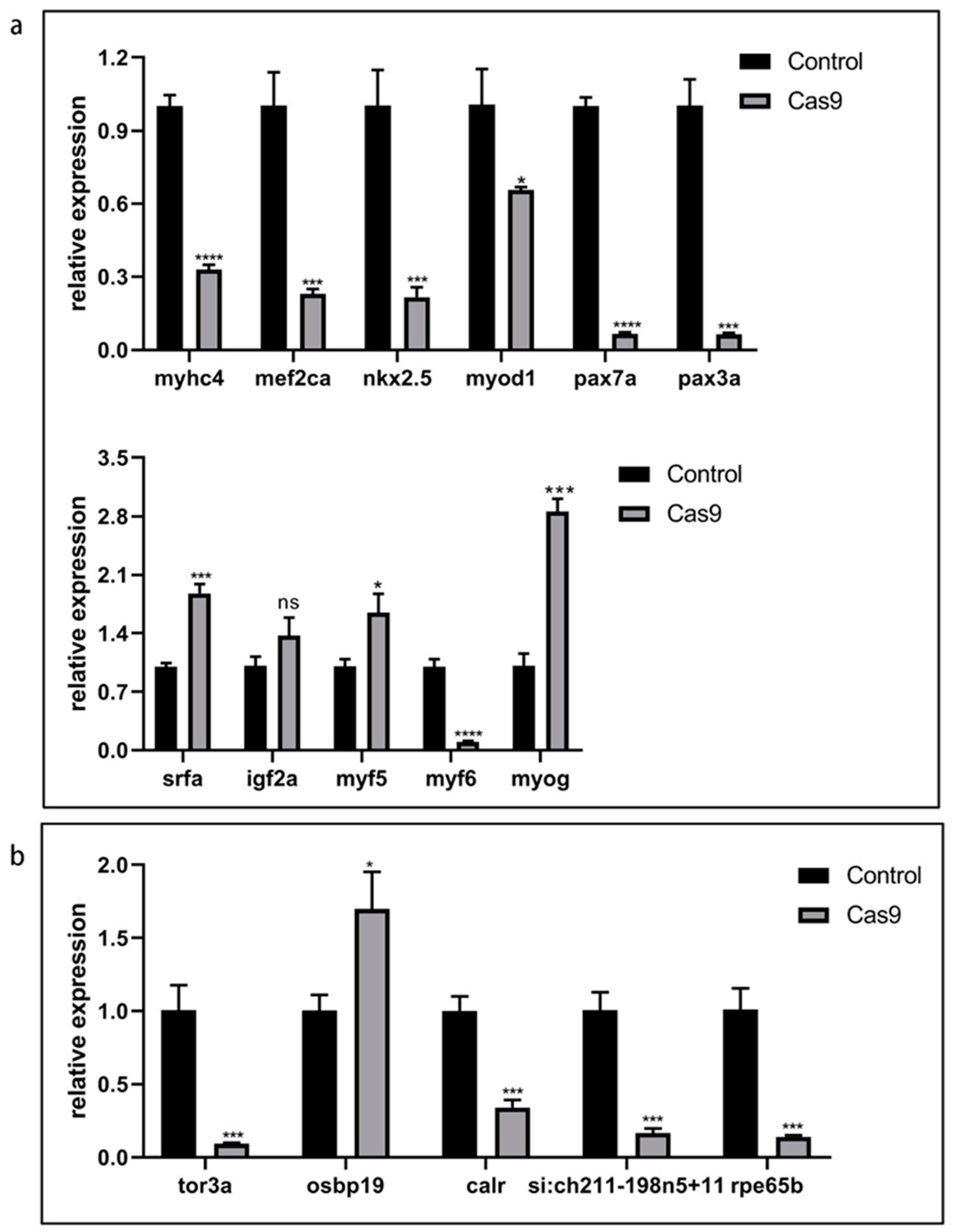
2.9. Effect of lncRNA gas5 Knockout on the Expression of Genes Related to Embryonic Muscle Development in Zebrafish
2.10. Effect of lncRNA gas5 Knockout on the Expression of Its Candidate Target Genes
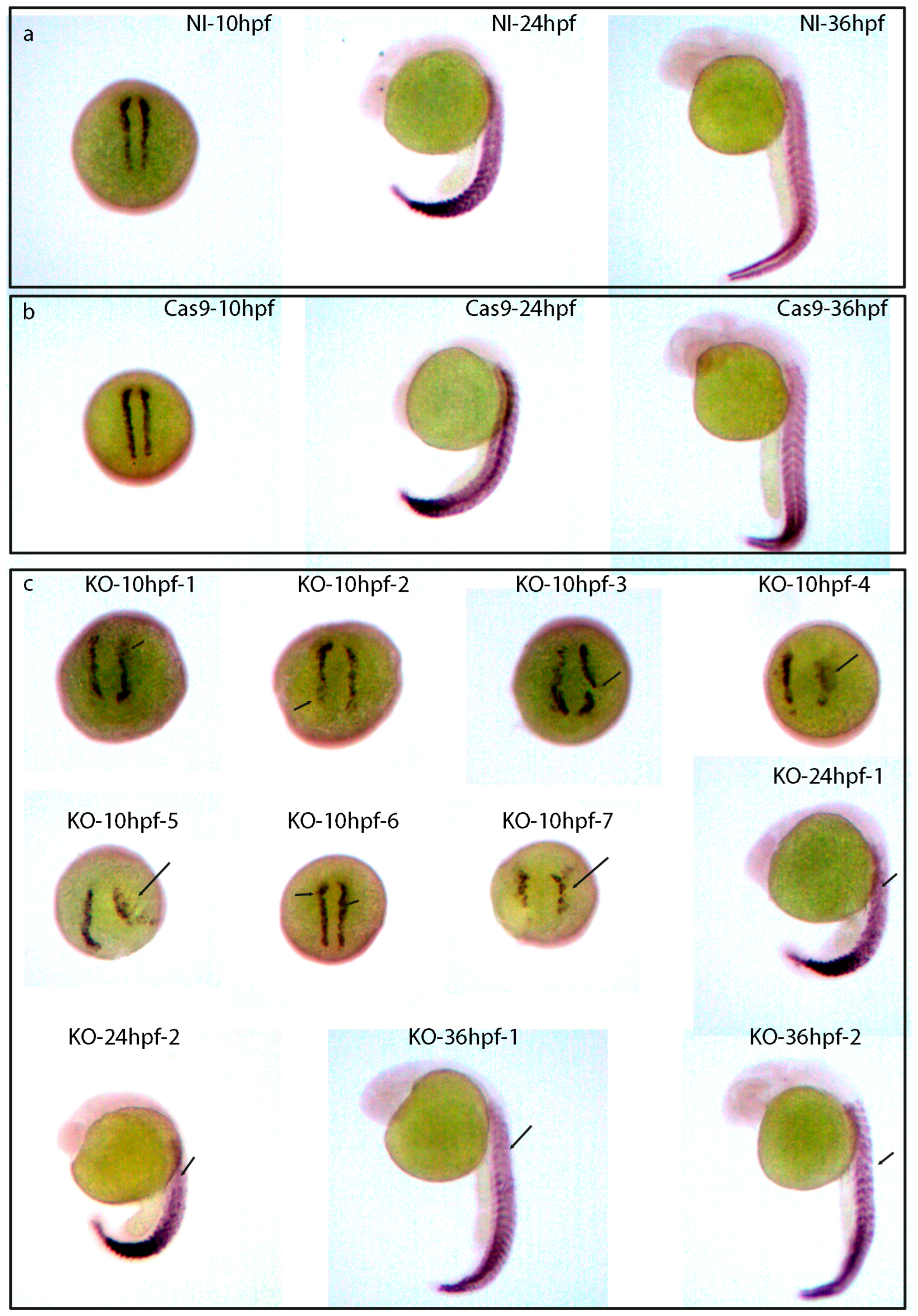
2.11. The Skeletal Muscle Development of Zebrafish Was Affected after lncRNA gas5 Was Knocked Out
3. Discussion
4. Materials and Methods
4.1. Zebrafish Husbandry and Embryo Collection
4.2. cDNA Library Contruction, Transcriptome Sequencing, and Bioinformatic Analysis
4.3. qPCR Detection
4.4. Zebrafish LncRNA gas5
4.5. The Temporal and Spatial Expression Pattern of lncRNA gas5
4.6. LncRNA gas5 Knockout by CRISPR/Cas9
4.7. Prediction of lncRNA gas5 Target Genes of cis Action
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dasgupta, S.; LaDu, J.K.; Garcia, G.R.; Li, S.; Tomono-Duval, K.; Rericha, Y.; Huang, L.; Tanguay, R.L. A CRISPR-Cas9 mutation in sox9b long intergenic noncoding RNA (slincR) affects zebrafish development, behavior, and regeneration. Toxicol. Sci. 2023, 94, 153–166. [Google Scholar] [CrossRef]
- Li, W.; Liu, W.; Mo, C.; Yi, M.; Gui, J. Two Novel lncRNAs Regulate Primordial Germ Cell Development in Zebrafish. Cells 2023, 12, 672. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, S.; Chen, X.; Xu, H.; Li, X. Expression and function of lncRNA MALAT-1 in the embryonic development of zebrafish. Gene 2019, 680, 65–71. [Google Scholar] [CrossRef]
- Pontier, D.B.; Gribnau, J. Xist regulation and function explored. Hum. Genet. 2011, 130, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes. Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Shi, M.; Ren, K.; Liu, Y.; Duan, Y.; Cheng, Y.; Zhang, W.; Xia, X.Q. Profiling the Spatial Expression Pattern and ceRNA Network of lncRNA, miRNA, and mRNA Associated with the Development of Intermuscular Bones in Zebrafish. Biology 2022, 12, 75. [Google Scholar] [CrossRef]
- Pauli, A.; Valen, E.; Lin, M.F.; Garber, M.; Vastenhouw, N.L.; Levin, J.Z.; Fan, L.; Sandelin, A.; Rinn, J.L.; Regev, A.; et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012, 22, 577–591. [Google Scholar] [CrossRef]
- Kaushik, K.; Leonard, V.E.; Kv, S.; Lalwani, M.K.; Jalali, S.; Patowary, A.; Joshi, A.; Scaria, V.; Sivasubbu, S. Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS ONE 2013, 8, e83616. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, X.; Xu, X.; Zhang, Y. Systematic identification and characterization of cardiac long intergenic noncoding RNAs in zebrafish. Sci. Rep. 2017, 7, 1250. [Google Scholar] [CrossRef]
- Schneider, C.; King, R.M.; Philipson, L. Genes specifically expressed at growth arrest of mammalian cells. Cell 1988, 54, 787–793. [Google Scholar] [CrossRef]
- Cabianca, D.S.; Casa, V.; Bodega, B.; Xynos, A.; Ginelli, E.; Tanaka, Y.; Gabellini, D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 2012, 149, 819–831. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Zhang, J.G.; Qin, R.H.; Dai, C.; Shi, P.; Yang, J.J.; Deng, Z.Y.; Shi, K.H. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology 2017, 386, 11–18. [Google Scholar] [CrossRef]
- Tu, Z.Q.; Li, R.J.; Mei, J.Z.; Li, X.H. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 4303–4309. [Google Scholar]
- Wang, K.; Li, J.; Xiong, G.; He, G.; Guan, X.; Yang, K.; Bai, Y. Negative regulation of lncRNA GAS5 by miR-196a inhibits esophageal squamous cell carcinoma growth. Biochem. Biophys. Res. Commun. 2018, 495, 1151–1157. [Google Scholar] [CrossRef]
- Li, W.; Zhao, W.; Lu, Z.; Zhang, W.; Yang, X. Long Noncoding RNA GAS5 Promotes Proliferation, Migration, and Invasion by Regulation of miR-301a in Esophageal Cancer. Oncol. Res. 2018, 26, 1285–1294. [Google Scholar] [CrossRef]
- Sun, M.; Jin, F.Y.; Xia, R.; Kong, R.; Li, J.H.; Xu, T.P.; Liu, Y.W.; Zhang, E.B.; Liu, X.H.; De, W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 2014, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ma, G.; Sun, S.; Xu, Y.; Wang, B. Polymorphism in the promoter region of lncRNA GAS5 is functionally associated with the risk of gastric cancer. Clin. Res. Hepatol. Gastroenterol. 2018, 42, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Vesterlund, L.; Jiao, H.; Unneberg, P.; Hovatta, O.; Kere, J. The zebrafish transcriptome during early development. BMC Dev. Biol. 2011, 11, 30. [Google Scholar] [CrossRef]
- Barbazuk, W.B.; Korf, I.; Kadavi, C.; Heyen, J.; Tate, S.; Wun, E.; Bedell, J.A.; McPherson, J.D.; Johnson, S.L. The syntenic relationship of the zebrafish and human genomes. Genome. Res. 2000, 10, 1351–1358. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Kalueff, A.V.; Stewart, A.M.; Gerlai, R. Zebrafish as an emerging model for studying complex brain disorders. Trends. Pharmacol. Sci. 2014, 35, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Bakkers, J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 2011, 91, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.I.; Currie, P.D. The zebrafish as a model for muscular dystrophy and congenital myopathy. Hum. Mol. Genet. 2003, 12, R265–R270. [Google Scholar] [CrossRef] [PubMed]
- Dubińska-Magiera, M.; Daczewska, M.; Lewicka, A.; Migocka-Patrzałek, M.; Niedbalska-Tarnowska, J.; Jagla, K. Zebrafish: A Model for the Study of Toxicants Affecting Muscle Development and Function. Int. J. Mol. Sci. 2016, 17, 1941. [Google Scholar] [CrossRef]
- Drummond, I.A. Kidney development and disease in the zebrafish. J. Am. Soc. Nephrol. 2005, 16, 299–304. [Google Scholar] [CrossRef]
- Santoriello, C.; Zon, L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Investig. 2012, 122, 2337–2343. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic. Acids. Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends. Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Kevin, C.W.; Howard, Y.C. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Li, K.; Blum, Y.; Verma, A.; Liu, Z.; Pramanik, K.; Leigh, N.R.; Chun, C.Z.; Samant, G.V.; Zhao, B.; Garnaas, M.K.; et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood 2010, 115, 133–139. [Google Scholar] [CrossRef]
- Chowdhury, T.A.; Koceja, C.; Eisa-Beygi, S.; Kleinstiver, B.P.; Kumar, S.N.; Lin, C.W.; Li, K.; Prabhudesai, S.; Joung, J.K.; Ramchandran, R. Temporal and Spatial Post-Transcriptional Regulation of Zebrafish tie1 mRNA by Long Noncoding RNA During Brain Vascular Assembly. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1562–1575. [Google Scholar] [CrossRef] [PubMed]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Kurian, L.; Aguirre, A.; Sancho-Martinez, I.; Benner, C.; Hishida, T.; Nguyen, T.B.; Reddy, P.; Nivet, E.; Krause, M.N.; Nelles, D.A.; et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 2015, 131, 1278–1290. [Google Scholar] [CrossRef]
- Sarangdhar, M.A.; Chaubey, D.; Bhatt, A.; Km, M.; Kumar, M.; Ranjan, S.; Pillai, B. A Novel Long Non-coding RNA, durga Modulates Dendrite Density and Expression of kalirin in Zebrafish. Front. Mol. Neurosci. 2017, 10, 95. [Google Scholar] [CrossRef]
- Lin, N.; Chang, K.Y.; Li, Z.; Gates, K.; Rana, Z.A.; Dang, J.; Zhang, D.; Han, T.; Yang, C.S.; Cunningham, T.J.; et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 2014, 53, 1005–1019. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, Y.; Gu, J.; Xie, S.; Xu, Y.; Zhu, G.; Wang, L.; Huang, J.; Ma, H.; Yao, J. Deep mRNA sequencing analysis to capture the transcriptome landscape of zebrafish embryos and larvae. PLoS ONE 2013, 8, e64058. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Coccia, E.M.; Cicala, C.; Charlesworth, A.; Ciccarelli, C.; Rossi, G.B.; Philipson, L.; Sorrentino, V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol. Cell Biol. 1992, 12, 3514–3521. [Google Scholar] [CrossRef]
- Goudarzi, M.; Berg, K.; Pieper, L.M.; Schier, A.F. Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility. Elife 2019, 8, e40815. [Google Scholar] [CrossRef]
- Gros, J.; Manceau, M.; Thomé, V.; Marcelle, C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 2005, 435, 954–958. [Google Scholar] [CrossRef]
- Asakura, A.; Rudnicki, M.A. Side population cells from diverse adult tissues are capable of in vitro hematopoietic differentiation. Exp. Hematol. 2002, 30, 1339–1345. [Google Scholar] [CrossRef]
- Chen, R.; Jiang, T.; She, Y.; Xie, S.; Zhou, S.; Li, C.; Ou, J.; Liu, Y. Comprehensive analysis of lncRNAs and mRNAs with associated co-expression and ceRNA networks in C2C12 myoblasts and myotubes. Gene 2018, 647, 164–173. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Sun, H.; Wang, H. Long non-coding RNAs in the regulation of skeletal myogenesis and muscle diseases. Cancer Lett. 2018, 417, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Chargé, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef] [PubMed]
- Gros, J.; Scaal, M.; Marcelle, C. A two-step mechanism for myotome formation in chick. Dev. Cell 2004, 6, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Mohammadabadi, M.; Bordbar, F.; Jensen, J.; Du, M.; Guo, W. Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals. Animals 2021, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Goulding, M.D.; Chalepakis, G.; Deutsch, U.; Erselius, J.R.; Gruss, P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991, 10, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Jostes, R.F.; Hui, T.E.; James, A.C.; Cross, F.T.; Schwartz, J.L.; Rotmensch, J.; Atcher, R.W.; Evans, H.H.; Mencl, J.; Bakale, G.; et al. In vitro exposure of mammalian cells to radon: Dosimetric considerations. Radiat. Res. 1991, 127, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Demignon, J.; Porteu, A.; Kahn, A.; Concordet, J.P.; Daegelen, D.; Maire, P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl. Acad. Sci. USA 1998, 95, 14220–14225. [Google Scholar] [CrossRef] [PubMed]
- Relaix, F.; Buckingham, M. From insect eye to vertebrate muscle: Redeployment of a regulatory network. Genes. Dev. 1999, 13, 3171–3178. [Google Scholar] [CrossRef] [PubMed]
- Hinits, Y.; Pan, L.; Walker, C.; Dowd, J.; Moens, C.B.; Hughes, S.M. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 2012, 369, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.; de Sena-Tomás, C.; George, V.; Werdich, A.A.; Kapur, S.; MacRae, C.A.; Targoff, K.L. Nkx genes establish second heart field cardiomyocyte progenitors at the arterial pole and pattern the venous pole through Isl1 repression. Development 2018, 145, dev161497. [Google Scholar] [CrossRef] [PubMed]
- Osborn, D.P.; Li, K.; Hinits, Y.; Hughes, S.M. Cdkn1c drives muscle differentiation through a positive feedback loop with Myod. Dev. Biol. 2011, 350, 464–475. [Google Scholar] [CrossRef]
- Ganassi, M.; Badodi, S.; Ortuste Quiroga, H.P.; Zammit, P.S.; Hinits, Y.; Hughes, S.M. Myogenin promotes myocyte fusion to balance fibre number and size. Nat. Commun. 2018, 9, 4232. [Google Scholar] [CrossRef]
- Lin, C.Y.; Lee, H.C.; Chen, H.C.; Hsieh, C.C.; Tsai, H.J. Normal function of Myf5 during gastrulation is required for pharyngeal arch cartilage development in zebrafish embryos. Zebrafish 2013, 10, 486–499. [Google Scholar] [CrossRef]
- Wang, Y.H.; Li, C.K.; Lee, G.H.; Tsay, H.J.; Tsai, H.J.; Chen, Y.H. Inactivation of zebrafish mrf4 leads to myofibril misalignment and motor axon growth disorganization. Dev. Dyn. 2008, 237, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of the Zebrafish (Danio Rerio); University of Oregon Press: Eugene, OR, USA, 1995; pp. 1–100. [Google Scholar]
- McCurley, A.T.; Callard, G.V. Characterization of housekeeping genes in zebrafish: Male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Li, M.; Sun, Y.; Sultan, Y.; Li, X. Systematic Identification of Long Noncoding RNAs during Three Key Organogenesis Stages in Zebrafish. Int. J. Mol. Sci. 2024, 25, 3440. https://doi.org/10.3390/ijms25063440
Zhou C, Li M, Sun Y, Sultan Y, Li X. Systematic Identification of Long Noncoding RNAs during Three Key Organogenesis Stages in Zebrafish. International Journal of Molecular Sciences. 2024; 25(6):3440. https://doi.org/10.3390/ijms25063440
Chicago/Turabian StyleZhou, Chune, Mengting Li, Yaoyi Sun, Yousef Sultan, and Xiaoyu Li. 2024. "Systematic Identification of Long Noncoding RNAs during Three Key Organogenesis Stages in Zebrafish" International Journal of Molecular Sciences 25, no. 6: 3440. https://doi.org/10.3390/ijms25063440





