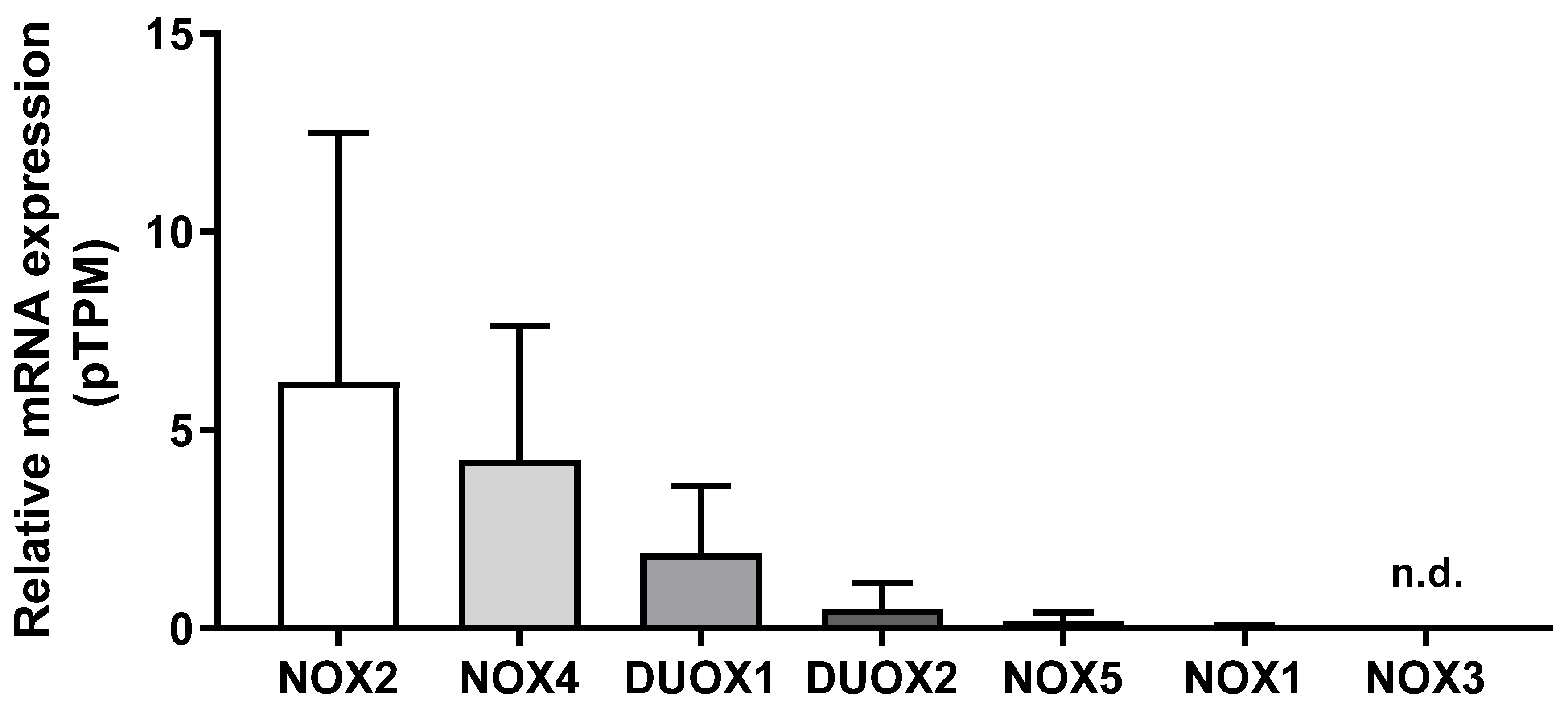Abstract
NADPH oxidase enzymes (NOX) are involved in all stages of carcinogenesis, but their expression levels and prognostic value in breast cancer (BC) remain unclear. Thus, we aimed to assess the expression and prognostic value of NOX enzymes in BC samples using online databases. For this, mRNA expression from 290 normal breast tissue samples and 1904 BC samples obtained from studies on cBioPortal, Kaplan–Meier Plotter, and The Human Protein Atlas were analyzed. We found higher levels of NOX2, NOX4, and Dual oxidase 1 (DUOX1) in normal breast tissue. NOX1, NOX2, and NOX4 exhibited higher expression in BC, except for the basal subtype, where NOX4 expression was lower. DUOX1 mRNA levels were lower in all BC subtypes. NOX2, NOX4, and NOX5 mRNA levels increased with tumor progression stages, while NOX1 and DUOX1 expression decreased in more advanced stages. Moreover, patients with low expression of NOX1, NOX4, and DUOX1 had lower survival rates than those with high expression of these enzymes. In conclusion, our data suggest an overexpression of NOX enzymes in breast cancer, with certain isoforms showing a positive correlation with tumor progression.
1. Introduction
Breast cancer (BC) is the second most commonly diagnosed cancer and the most frequent malignancy in women worldwide. In 2018, an estimated 2.1 million new diagnoses and approximately 626,679 BC-related deaths have occurred [1]. Due to the inherent molecular and clinical heterogeneity, BC is associated with diverse risk factors, etiologies, treatment effectiveness, and prognosis [2,3,4,5]. Most studies profiling gene expression signatures have classified BC into five major molecular subtypes: luminal A (estrogen receptor (ER)-positive, progesterone receptor (PR)-positive, human epidermal growth factor receptor 2 (HER2)-negative), luminal B (ER-positive, PR-positive, HER2-positive), HER2-enriched (ER-negative, PR- negative, HER2-positive), basal-like or triple-negative (ER-negative, PR-negative, HER2-negative), and normal breast-like tumors (unclassified) [6,7]. More recently, the claudin-low subtype has been identified and characterized as negative for ER, PR, and HER2 expression, but with low expression of Ki67 and high expression of epithelial–mesenchymal transition (EMT)-related genes, which distinguishes this subtype from the basal-like [8].
The mammalian nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (NOX) are transmembrane proteins that carry electrons across biological membranes, reducing molecular oxygen (O2) to superoxide anion (O2•−) or hydrogen peroxide (H2O2). The NOX enzymes family contains seven members, which are NOX1-5 and Dual oxidases 1 and 2 (DUOX1-2). All isoforms exhibit six highly conserved transmembrane domains, one NADPH binding site in the C-terminal region, one FAD binding site, and two histidine-linked heme groups in the transmembrane domains III and IV. Additionally, NOX5 and DUOX 1-2 show an intracellular calcium-binding site that is closely related to their activation. Unlike all other sources of reactive oxygen species (ROS), NOX enzymes generate ROS as their main function [9].
Accumulated evidence in recent decades highlights the role of NOX-derived ROS in virtually all steps of carcinogenesis (initiation, promotion, and progression) of different organs/tissues. Furthermore, NOX enzymes have been suggested as a prospective target in novel therapeutic approaches [10,11]. Regarding BC, some studies have shown that NOX might be important players in this disease, although most of them were conducted under in vitro conditions utilizing cell lines, and the expression levels and prognostic value of NOX in patients remain elusive [12,13,14]. Thus, the aims of this study were first to assess the expression and prognostic value of individual NOX family members in the different BC molecular subtypes using online databases.
2. Results
2.1. Expression of NOX Family Genes in the Breast Tissue
We first compared the mRNA levels of NOX family members in 290 samples of normal breast tissue using the HPA database. As shown in Figure 1, NOX2 mRNA levels were the most expressed, which was followed by NOX4, DUOX1, DUOX2, NOX5, and NOX1. NOX3 mRNA levels were undetectable. Therefore, we did not evaluate NOX3 in the next analysis.
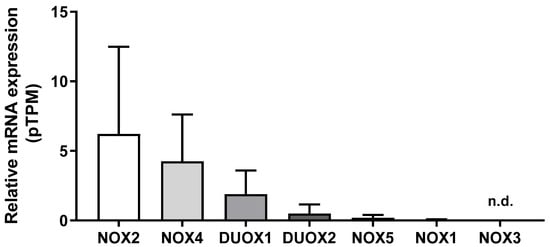
Figure 1.
NADPH oxidases mRNA levels in the breast. Differences among NOX family mRNA levels in normal breast tissue. Data were obtained from the online platform Human Protein Atlas (https://www.proteinatlas.org, accessed on 2 July 2022). Results are represented as the mean protein-coding transcripts per million (pTPM) ± standard deviation (N = 290; n.d.: not detectable).
2.2. Comparison of NOX mRNA Levels in Non-Tumoral and Breast Tumor Tissues
We next compared NOX mRNA levels between non-tumoral and breast tumor tissues. No data concerning DUOX2 expression were available, so this isoform was not analyzed. Due to breast cancer heterogeneity, we divided BC samples according to the five major molecular subtypes: luminal A, luminal B, HER2, basal, and claudin-low.
As demonstrated in Table 1, we observed that NOX1 mRNA levels were higher in the HER2 and basal subtypes when compared to the normal group. Similarly, the mRNA levels for NOX2 showed an increase in the HER2 and basal subtypes, in addition to claudin-low. Concerning NOX4 mRNA levels, they were revealed to be high in the luminal A, HER2, and claudin-low subtypes in comparison to normal tissue, while in the basal subtype, NOX4 mRNA levels were decreased. There were no statistically significant differences in NOX5 expression in any of the tumor subtypes when compared to the normal group. In relation to DUOX1, we detected a decrease in the mRNA levels in the luminal A, luminal B, HER2, and claudin-low subtypes when compared to the control.

Table 1.
Comparison of NOX1, NOX2, NOX4, and DUOX1 mRNA levels in non-tumoral and different subtypes of breast tumor tissues.
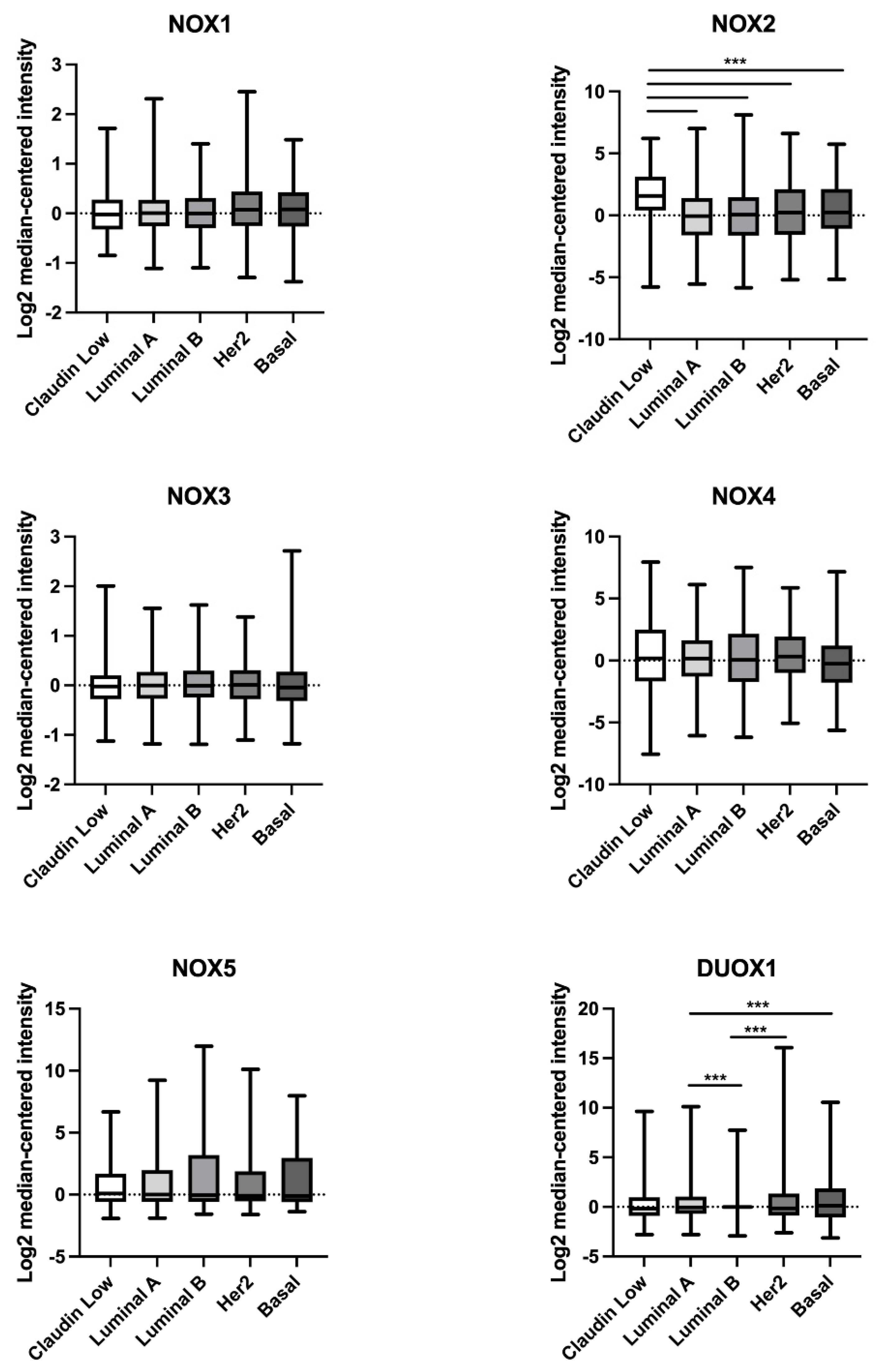
As demonstrated in Figure 2, NOX2 mRNA levels were higher in the claudin-low BC subtype in comparison to the luminal A, luminal B, Her2, and basal subtypes. Furthermore, DUOX1 mRNA levels were lower in the luminal B BC subtype when compared to luminal A and Her2, and in luminal A in comparison to the basal subtype.
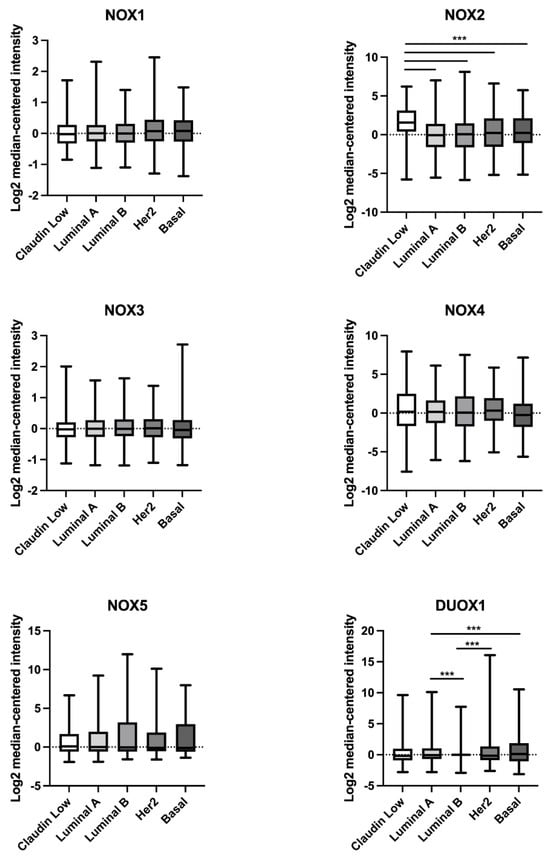
Figure 2.
Comparison of NOX1, NOX2, NOX3, NOX4, NOX5, and DUOX1 mRNA levels in different subtypes of breast tumor tissues. Data were obtained from the cBioPortal online platform (http://www.cbioportal.org/, accessed on 3 July 2022) (N = 1904). Claudin-low (N = 199); luminal A (N = 679); luminal B (N = 461); HER2 (N = 220); basal (N = 199). *** p < 0.001.
2.3. mRNA Expression of NOX Genes at Different Stages of Breast Tumor Development
Another parameter we have taken into consideration is BC staging. The American Joint Committee on Cancer (AJCC) TNM staging system takes into account three primary factors: tumor size (T), regional lymph node status (N), and distant metastases (M). These factors are used to determine an overall stage, which ranges from 0 to 4. Stage 0 is referred to as “carcinoma in situ”, indicating that the cancer remains confined to the primary layer of cells where it originated and has not spread. In this context, stage 1 BC is anatomically defined as a tumor smaller than 2 cm in size and shows no involvement of lymph nodes. On the other hand, stage 4 represents a state of advanced disease with distant metastases [4,15].
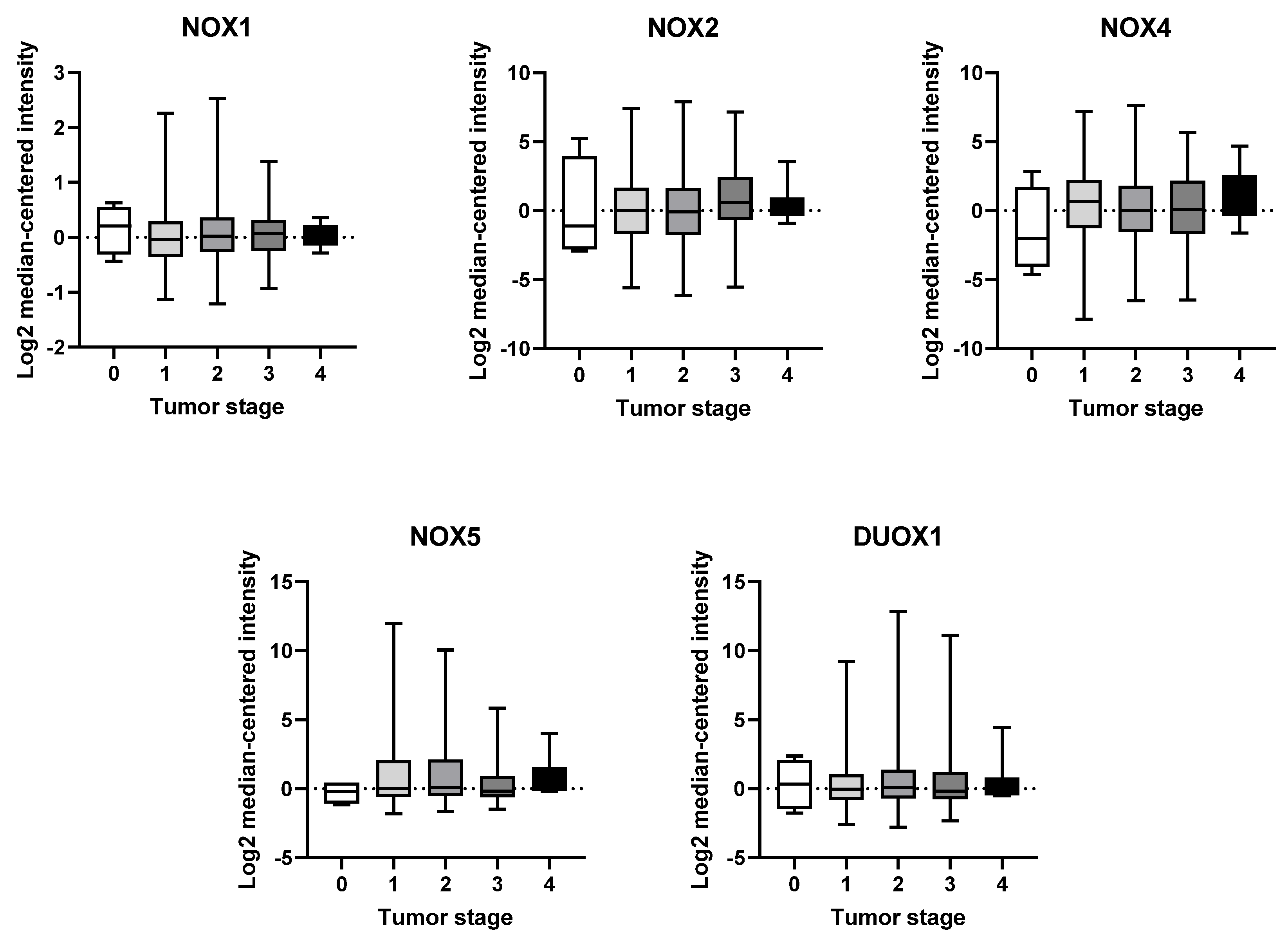
Based on our analysis, we have not observed any significant variations in the expression of any NOX isoform across the different tumor stages (Figure 3).
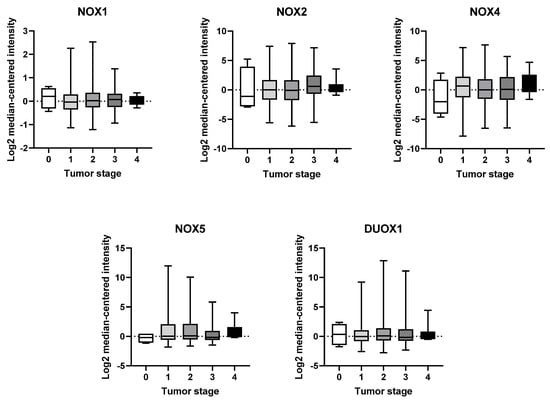
Figure 3.
mRNA expression of NOX family at different stages of breast tumor. NOX1, NOX2, NOX4, NOX5, and DUOX1 were evaluated across tumor staging of breast tissues. Data were obtained from the cBioPortal online platform (http://www.cbioportal.org/, accessed on 3 July 2022) (N = 1904).
2.4. Survival Analysis of NOX Members in BC Patients
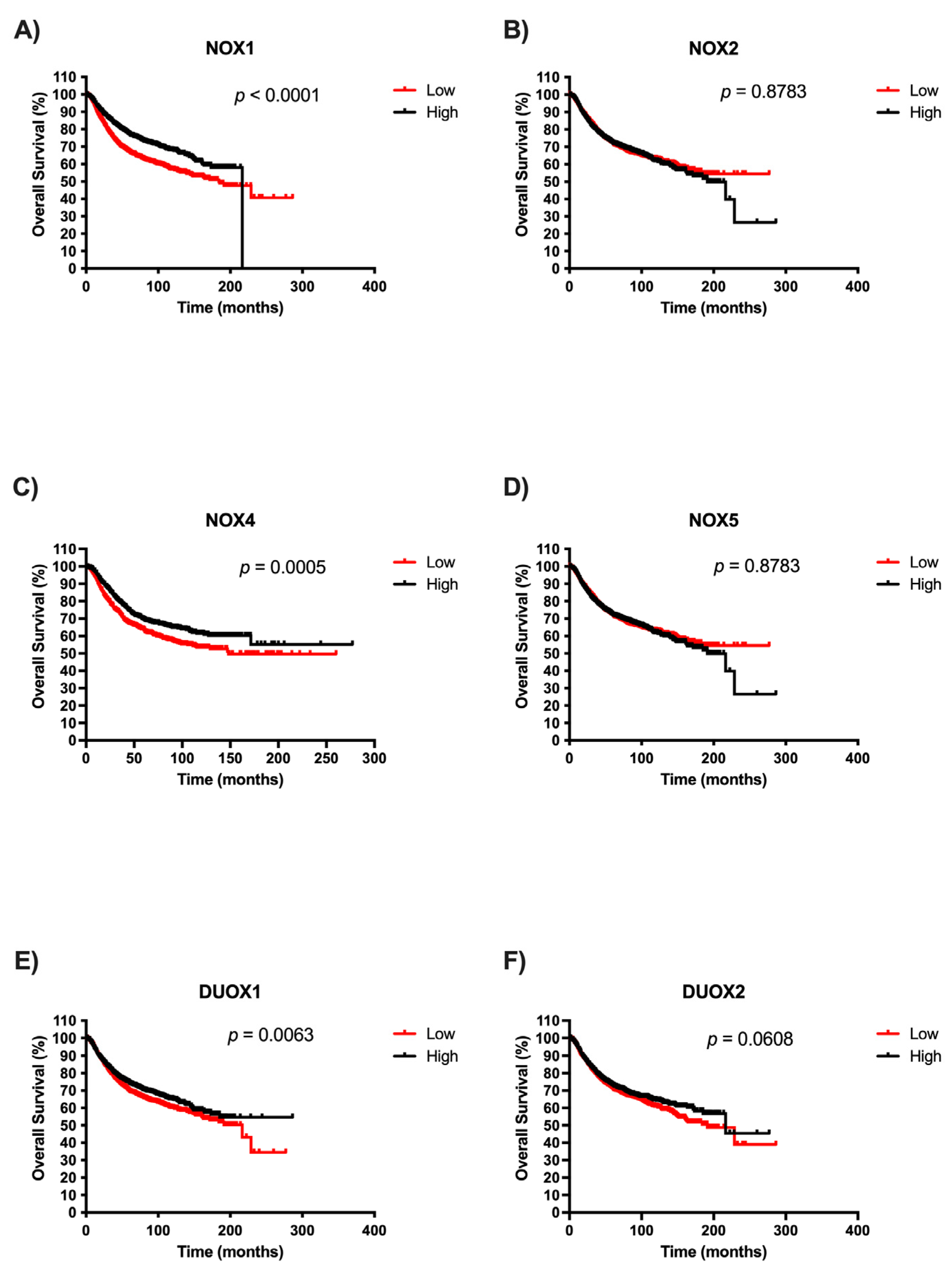
Afterwards, we examined the potential prognostic values of NOX isoforms in BC. The survival curves are demonstrated in Figure 4. It is important to highlight that there were no patient survival data related to high NOX1 expression after 200 months. We observed that patients with low expression of NOX1, NOX4, and DUOX1 exhibited significantly unfavorable prognoses in BC when compared with patients with high expression of those NOXs (Figure 4A, Figure 4C, and Figure 4E, respectively). Moreover, NOX2, NOX5, and DUOX2 expression were not associated with overall survival in BC (Figure 4B, Figure 4D, and Figure 4F, respectively).
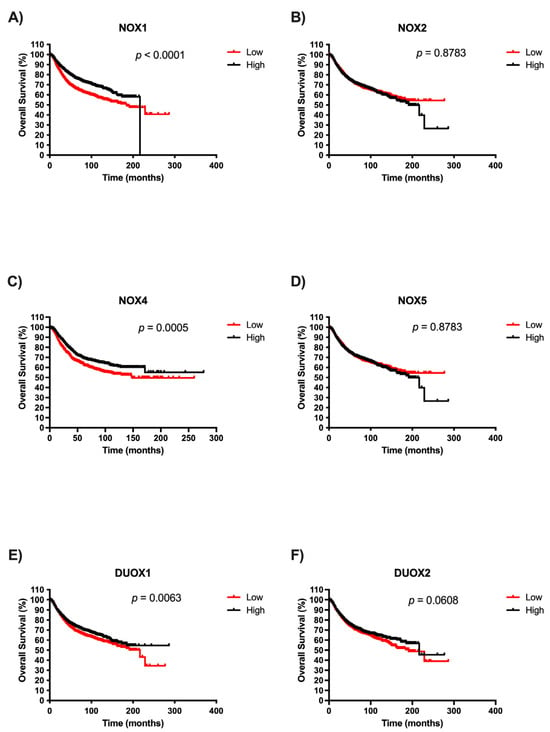
Figure 4.
Survival analysis according to NOX family gene expression in BC patients. Comparison between the survival of BC patients with high or low expression of NOX1 (A), NOX2 (B), NOX4 (C), NOX5 (D), DUOX1 (E), and DUOX2 (F) in tumor breast tissues using Kaplan–Meier plotter analysis.
2.5. NOX Gene Expression in Relation to Estrogen Receptor Status in Breast Cancer Samples
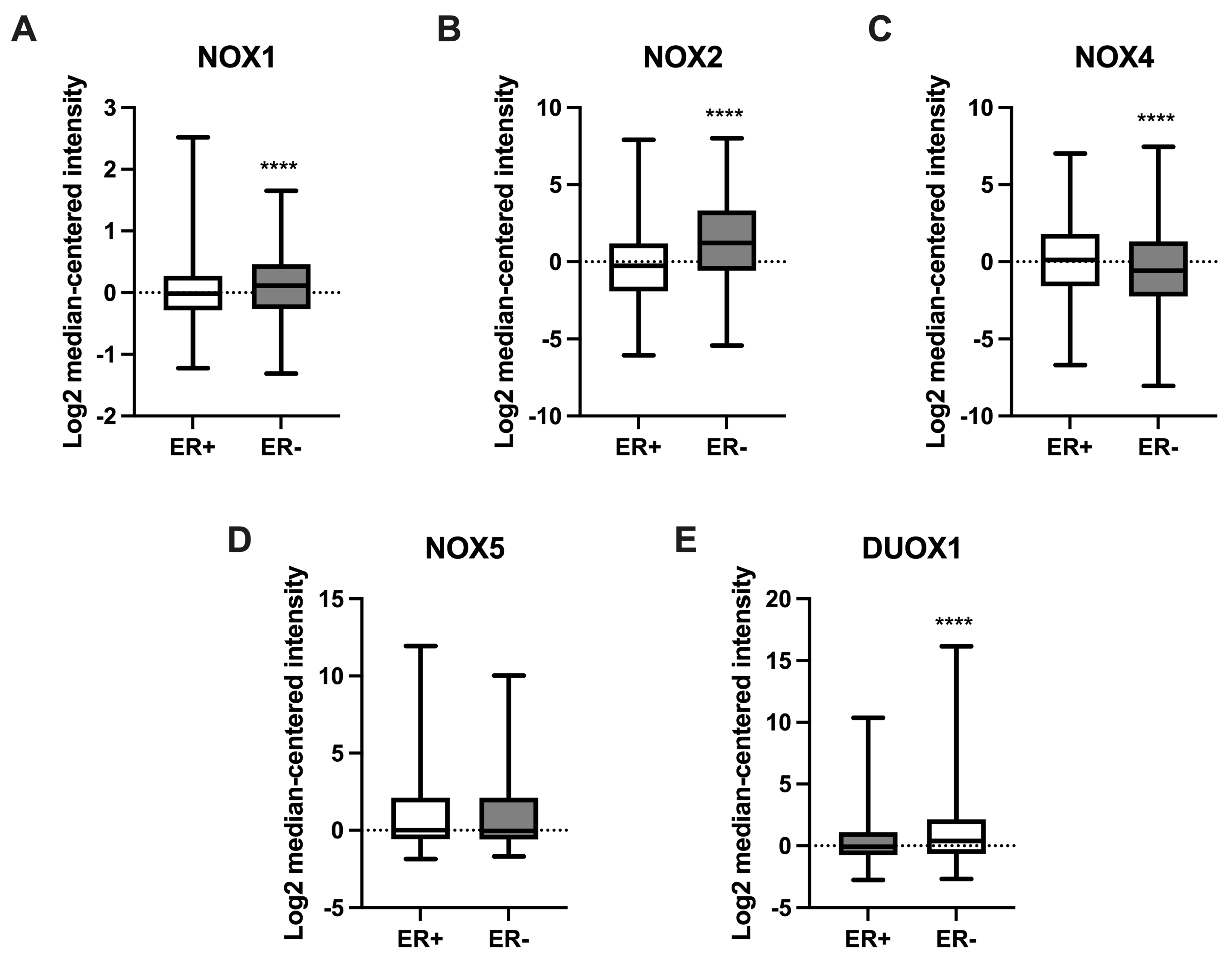
We further compared NOX mRNA levels between BC tissues that were positive or negative for the presence of estrogen receptors (ERs). As shown in Figure 5, NOX1 (Figure 5A), NOX2 (Figure 5B), and DUOX1 (Figure 5E) were higher in tissues that were ER-negative when compared to the ER-positive group. On the other hand, NOX4 expression (Figure 5C) was lower in ER-negative tissues in comparison to those with ER-positive status.
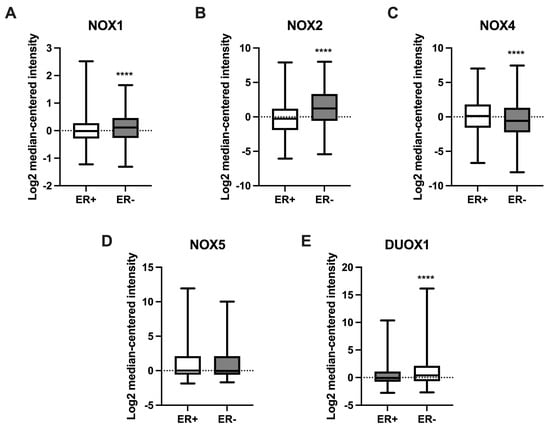
Figure 5.
Expression of NOX family genes in relation to estrogen receptor (ER) status in BC samples. mRNA levels of NOX1 (A), NOX2 (B), NOX4 (C), NOX5 (D), and DUOX1 (E) in relation to the presence (ER+) or absence (ER-) of ER in tumor breast tissues. Data were obtained from the cBioPortal online platform (http://www.cbioportal.org/, accessed on 3 July 2022). N = 1904. **** p < 0.0001.
3. Discussion
Here, we show that NOX2 was the most expressed NOX isoform in normal tissue. NOX2 was originally the first identified NOX. It is highly expressed in leukocytes, where it has an essential role in phagocytic oxidative burst and host defense [11]. NOX2 expression was followed by NOX4, the only constitutively active isoform, which has been linked to physiological functions such as differentiation and memory formation [16,17]. Next, we observed DUOX1 and DUOX2 expression, respectively. Although DUOX2 plays an imperative role in hormone synthesis in the thyroid gland, DUOX1 has a diverse tissue distribution, but not a well-established physiological function [18,19]. It seems to be implicated in differentiation, wound responses, and host defense (reviewed in [20]). However, the role of DUOX1 and 2 in mammary tissue physiology is not known. Subsequently, we noticed NOX5, an isoform whose expression is typically found in the endothelium, spermatocytes, and lymphoid organs [21], and, finally, NOX1, which was mainly expressed in the colon and associated with gut immune modulation [22]. NOX3 mRNA levels were undetectable in our analysis, which might be due in part to its restricted distribution, found in high levels, for instance, in the inner ear, participating in ontogenesis [23]. To the best of our knowledge, no other study has evaluated the expression levels of NOX enzymes in normal human breast tissue.
Subsequently, we evaluated the expression levels of each NOX isoform by comparing the normal tissue to the different BC molecular subtypes. In a time in which personalized medicine is moving toward even more individualized treatment, the identification of new biomarkers among the subtypes should facilitate management planning. We found that the luminal A subtype had higher levels of NOX4 mRNA and lower levels of DUOX1. This subtype is low-grade, associated with a better prognosis, and tends to grow slowly [24]. In the luminal B subtype, which is known to grow slightly faster than luminal A cancers and to show worse recurrence-free survival at 5 years and 10 years [25], we observed a significant decrease in DUOX1 expression. In line with these findings, the HER2 subtype presented higher expression levels of NOX1, NOX2, and NOX4, but lower levels of DUOX1. The HER2 BC subtype was characterized as growing faster than luminal cancers and having a worse prognosis. Interestingly, these three subtypes exhibited low levels of DUOX1 expression. Data from our group demonstrated that DUOX1 was downregulated in BC cell lines and tissues. Moreover, non-tumor breast cells silenced for DUOX1 had a higher proliferation rate, as well as less adherence and migratory capacity when compared to control cells, suggesting that DUOX1 may play a tumor-suppressive role [26], which can be disrupted in luminal A, B, and HER2 BC subtypes according to our analysis.
Furthermore, in basal-like, also called triple-negative and considered the most aggressive and resistant BC subtype, NOX1 and NOX2 expression were significantly higher, in contrast to NOX4, which was diminished when compared to non-tumoral tissue. Some studies concerning NOX enzymes and BC have shown an upregulation of NOX1 in tumor tissues compared to normal tissues [27,28]. Desouki et al. (2005) demonstrated that NOX1 is highly expressed in breast tumors (86%). The authors also showed crosstalk between the mitochondria and NOX1 controlling redox signaling, and suggested that a disruption of this pathway could be involved in breast tumorigenesis [27]. Another study has proposed that the activation of the G protein-coupled receptor (GPCR) leukotriene receptor BLT2 promotes cell survival through the induction of NOX1- and NOX4-derived ROS in an ERK- and AKT-dependent mechanism in BC cell lines [29]. In addition, activation of the RAS-Erk1/2-NOX1 pathway was observed in a population of BC stem-like cells exposed to low concentrations of combined carcinogens, with an important function in the maintenance of increased cell proliferation [30].
NOX2 and NOX4 expression levels were higher in the claudin-low BC subtype, which is marked by aggressiveness, resistance to treatment, and poor prognosis [2]. Moreover, we observed higher NOX2 levels in three other BC subtypes (HER2, basal, and claudin-low). NOX2 has been related to angiogenesis, immune suppression, metastasis, and pro-survival signals in tissues such as the lung, as well as in leukemia and melanoma cells [31,32,33,34,35]. NOX2 was found in lipid rafts (LRs) of BC cell lines, whereas the disruption of these structures resulted in downregulation of the enzyme [36]. Mukawera et al. demonstrated that IKKε overexpression in BC cells was dependent on NOX2, leading to increased proliferation. NOX4 expression was also upregulated in three BC subtypes (luminal A, HER2, and claudin-low). Overexpression of NOX4 in non-tumor breast cells has been associated with increased senescence, cellular transformation, and resistance to apoptosis, suggesting an oncogenic role of this enzyme. The same authors also showed that NOX4 was overexpressed in various BC cell lines and primary breast tumors [12]. Additionally, another group has suggested NOX4 as a key player in TGFβ-induced epithelial-to-mesenchymal transition and migration of non-tumor and metastatic human breast cells [13]. In conformity, a subsequent study reported the efficiency of attenuating metastasis in BC cells by targeting NOX4 [37]. Our findings, in conjunction with those exposed in the literature, reinforce the great relevance of NOX4 in BC initiation and development.
Although, herein, we observed no changes in NOX5 expression levels among the BC subtypes, previous reports have identified NOX5 upregulation in BC, but they analyzed a limited number of samples with no molecular subtype distinction [14,38]. More recently, leptin has emerged as a potential activator of NOX5-derived ROS production in human epithelial mammary cells, linking obesity-associated hyperleptinemia with mammary tumorigenesis [39].
Surprisingly, in the present study, we did not identify significant alterations in NOX expression over tumor stages in our analysis. However, we verified the presence of increased expression levels of NOX1, NOX2, and DUOX1 in ER- BC tissues, and significantly low levels of NOX4 in ER+. ER is one of the main prognostic biomarkers in BC. The lack of the receptor is associated with a more aggressive profile of the tumor [40]. There are few studies correlating NOX expression and tumor progression. Liu and colleagues revealed a greater expression of NOX1 according to tumor progression in lung cancer [41]. Moreover, high expression of NOX4 and NOX5 were correlated to poorer overall survival in patients with end-stage gastric cancer [42].
Some studies have proposed that the expression level of NOX enzymes can influence the overall survival of patients. Individuals with hepatocarcinoma seem to have better prognoses when high expression levels of NOX4 mRNA are found, while patients with high levels of NOX2 present poor prognoses [43]. Analysis of gastric tumors has correlated NOX2 overexpression with longer patient survival, in contrast with NOX4 overexpression, which is associated with poorer overall patient survival [44]. It has also been reported that the high expression of DUOXs is related to reduced mortality in thyroid cancer [45]. In our analysis, patients with lower NOX1, NOX4, and DUOX1 expression exhibited shorter survival compared to patients with higher levels. Although we have observed high levels of expression of these proteins in tumor tissues, the survival data suggest that the loss of these NOX may occur in more undifferentiated cells and, consequently, more aggressive tumors, which would explain the lower survival rates of patients.
In conclusion, our data shed light on the role of NOX isoforms in BC. We highlight here that NOX2 is the most expressed NOX isoform in normal breast tissue, whereas, in conjunction with NOX4, it displays high levels in different BC subtypes. In the face of the plurality of BC molecular and histological characteristics, taking NOX expression into account could add significant value to the design and management of new therapies. However, many studies are still needed to elucidate the role of each NOX member in the different types of cancer and their implications.
4. Materials and Methods
4.1. Gene Expression Profiles in Breast Tissue
To analyze the expression of NOX family genes in normal breast tissue, we used the database The Human Protein Atlas (HPA) (https://www.proteinatlas.org, accessed on 2 July 2022). Individual genes (NOX1, CYBB NOX2, NOX3, NOX4, NOX5, DUOX1, and DUOX2) were evaluated. All quantitative transcriptome data (RNA-Seq) were retrieved from the Cancer Genome Atlas (TCGA) portal. A total of 290 normal breast samples were analyzed. The data were downloaded and plotted using the Graphpad Prism 9 software (GraphPad Software, Inc., San Diego, CA, USA).
4.2. Analysis of Gene Expression in Breast Tumor Tissue
The evaluation of NOX mRNA levels in BC patients was achieved through the online analysis tool cBioPortal (http://www.cbioportal.org/, accessed on 3 July 2022). The expression of NOX mRNA levels in human BC and normal tissues was obtained using the METABRIC study [3,45], containing 1904 samples from patients who had their breast tissue samples collected. We compared NOX mRNA levels between the different subtypes of BC and the non-tumoral tissue, as well as the influence of estrogen receptor status.
4.3. Analysis of Patient Survival According to NOX Expression
The significance of the prognosis in BC patients with different levels of NOX isoforms was performed using the Kaplan–Meier database (http://kmplot.com/analysis/, accessed on 15 August 2022). The BC database was established using gene expression data and survival information from 876 patients. Individual genes (NOX1, CYBB NOX2, NOX4, NOX5, DUOX1, and DUOX2) were evaluated on the platform. The patient cases were divided into two groups according to the median expression of the gene (high vs. low expression). The survival of patients in both groups was then analyzed using the Kaplan–Meier survival graph. A p-value less than 0.05 was considered statistically significant. The data were downloaded as text, which was replotted.
4.4. Statistical Analysis
Normality was verified using the D’Agostino and Pearson test. The data were analyzed using ANOVA, except in the results with only 2 groups, in which the t test was applied. Log2 median-centered intensity was calculated by subtracting the median from the values of both groups. The results were expressed as the mean ± standard deviation.
Author Contributions
Conceptualization, R.S.F. and A.d.V.e.S.; methodology, A.d.V.e.S., C.C.d.F., L.M.P. and P.H.M.T.; formal analysis, A.d.V.e.S., C.C.d.F., L.M.P. and P.H.M.T.; data curation, A.d.V.e.S., C.C.d.F., P.H.M.T., A.C.F.F. and R.S.F.; writing—original draft preparation, A.d.V.e.S., C.C.d.F. and R.S.F.; writing—review and editing, L.M.P., P.H.M.T., A.C.F.F. and R.S.F.; supervision, R.S.F.; project administration, R.S.F.; funding acquisition, R.S.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The APC was funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Turkoz, F.P.; Solak, M.; Petekkaya, I.; Keskin, O.; Kertmen, N.; Sarici, F.; Arik, Z.; Babacan, T.; Ozisik, Y.; Altundag, K. Association between common risk factors and molecular subtypes in breast cancer patients. Breast 2013, 22, 344–350. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA—J. Am. Med. Assoc. 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Blows, F.M.; Driver, K.E.; Schmidt, M.K.; Broeks, A.; van Leeuwen, F.E.; Wesseling, J.; Cheang, M.C.; Gelmon, K.; Nielsen, T.O.; Blomqvist, C.; et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010, 7, e1000279. [Google Scholar] [CrossRef]
- Cheang, M.C.U.; Martin, M.; Nielsen, T.O.; Prat, A.; Voduc, D.; Rodriguez-Lescure, A.; Ruiz, A.; Chia, S.; Shepherd, L.; Ruiz-Borrego, M.; et al. Defining Breast Cancer Intrinsic Subtypes by Quantitative Receptor Expression. Oncologist 2015, 20, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, G.K.; Zhao, X.; Band, H.; Band, V. Histological, molecular and functional subtypes of breast cancers. Cancer Biol. Ther. 2010, 10, 955–960. [Google Scholar] [CrossRef]
- Drummond, G.R.; Selemidis, S.; Griendling, K.K.; Sobey, C.G. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat. Rev. Drug Discov. 2011, 10, 453–471. [Google Scholar] [CrossRef] [PubMed]
- Parascandolo, A.; Laukkanen, M.O. Carcinogenesis and reactive oxygen species signaling: Interaction of the NADPH oxidase NOX1-5 and superoxide dismutase 1-3 signal transduction pathways. Antioxid. Redox Signal. 2019, 30, 443–486. [Google Scholar] [CrossRef] [PubMed]
- Block, K.; Gorin, Y. Aiding and abetting roles of NOX oxidases in cellular transformation. Nat. Rev. Cancer 2012, 12, 627–637. [Google Scholar] [CrossRef]
- Graham, K.A.; Kulawiec, M.; Owens, K.M.; Li, X.; Desouki, M.M.; Chandra, D.; Singh, K.K. NADPH oxidase 4 is an oncoprotein localized to mitochondria. Cancer Biol. Ther. 2010, 10, 223–231. [Google Scholar] [CrossRef]
- Boudreau, H.E.; Casterline, B.W.; Rada, B.; Korzeniowska, A.; Leto, T.L. Nox4 involvement in TGF-beta and SMAD3-driven induction of the epithelial-to-mesenchymal transition and migration of breast epithelial cells. Free Radic. Biol. Med. 2012, 53, 1489–1499. [Google Scholar] [CrossRef]
- Dho, S.H.; Kim, J.Y.; Lee, K.P.; Kwon, E.S.; Lim, J.C.; Kim, C.J.; Jeong, D.; Kwon, K.S. STAT5A-mediated NOX5-L expression promotes the proliferation and metastasis of breast cancer cells. Exp. Cell Res. 2017, 351, 51–58. [Google Scholar] [CrossRef]
- Giuliano, A.E.; Connolly, J.L.; Edge, S.B.; Mittendorf, E.A.; Rugo, H.S.; Solin, L.J.; Weaver, D.L.; Winchester, D.J.; Hortobagyi, G.N. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 290–303. [Google Scholar] [CrossRef]
- Schröder, K.; Wandzioch, K.; Helmcke, I.; Brandes, R.P. Nox4 acts as a switch between differentiation and proliferation in preadipocytes. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 239–245. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Ago, T.; Kuroda, J.; Wakisaka, Y.; Tachibana, M.; Komori, M.; Shibahara, T.; Nakashima, H.; Nakashima, K.; Kitazono, T. Nox4 Promotes Neural Stem/Precursor Cell Proliferation and Neurogenesis in the Hippocampus and Restores Memory Function Following Trimethyltin-Induced Injury. Neuroscience 2019, 398, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.R.; Marden, C.C.; Ward-Bailey, P.; Gagnon, L.H.; Bronson, R.T.; Donahue, L.R. Congenital hypothyroidism, dwarfism, and hearing impairment caused by a missense mutation in the mouse dual oxidase 2 gene, Duox2. Mol. Endocrinol. 2007, 21, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Grasberger, H. Defects of thyroidal hydrogen peroxide generation in congenital hypothyroidism. Mol. Cell. Endocrinol. 2010, 322, 99–106. [Google Scholar] [CrossRef]
- De Faria, C.C.; Fortunato, R.S. The role of dual oxidases in physiology and cancer. Genet. Mol. Biol. 2020, 43, e20190096. [Google Scholar] [CrossRef] [PubMed]
- Jagnandan, D.; Church, J.E.; Banfi, B.; Stuehr, D.J.; Marrero, M.B.; Fulton, D.J.R. Novel mechanism of activation of NADPH oxidase 5: Calcium sensitization via phosphorylation. J. Biol. Chem. 2007, 282, 6494–6507. [Google Scholar] [CrossRef]
- Schwerd, T.; Bryant, R.V.; Pandey, S.; Capitani, M.; Meran, L.; Cazier, J.B.; Jung, J.; Mondal, K.; Parkes, M.; Mathew, C.G.; et al. NOX1 loss-of-function genetic variants in patients with inflammatory bowel disease. Mucosal Immunol. 2018, 11, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Paffenholz, R.; Bergstrom, R.A.; Pasutto, F.; Wabnitz, P.; Munroe, R.J.; Jagla, W.; Heinzmann, U.; Marquardt, A.; Bareiss, A.; Laufs, J.; et al. Vestibular defects in head-tilt mice result from mutations in Nox3, encoding an NADPH oxidase. Genes Dev. 2004, 18, 486–491. [Google Scholar] [CrossRef]
- Dumitrescu, R.G. Interplay between genetic and epigenetic changes in breast cancer subtypes. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2018; pp. 19–34. [Google Scholar] [CrossRef]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24, S26–S35. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, R.S.; Gomes, L.R.; Munford, V.; Pessoa, C.F.; Quinet, A.; Hecht, F.; Kajitani, G.S.; Milito, C.B.; Carvalho, D.P.; Menck, C.F.M. DUOX1 silencing in mammary cell alters the response to genotoxic stress. Oxidative Med. Cell. Longev. 2018, 2018, 3570526. [Google Scholar] [CrossRef]
- Desouki, M.M.; Kulawiec, M.; Bansal, S.; Das, G.; Singh, K.K. Cross talk between mitochondria and superoxide generating NADPH oxidase in breast and ovarian tumors. Cancer Biol. Ther. 2005, 4, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- Pervin, S.; Tran, L.; Urman, R.; Braga, M.; Parveen, M.; Li, S.A.; Chaudhuri, G.; Singh, R. Oxidative stress specifically downregulates survivin to promote breast tumour formation. Br. J. Cancer 2013, 108, 848–858. [Google Scholar] [CrossRef]
- Choi, J.A.; Lee, J.W.; Kim, H.; Kim, E.Y.; Seo, J.M.; Ko, J.; Kim, J.H. Pro-survival of estrogen receptor-negative breast cancer cells is regulated by a BLT2-reactive oxygen species-linked signaling pathway. Carcinogenesis 2009, 31, 543–551. [Google Scholar] [CrossRef]
- Pluchino, L.A.; Wang, H.-C.R. Chronic Exposure to Combined Carcinogens Enhances Breast Cell Carcinogenesis with Mesenchymal and Stem-Like Cell Properties. PLoS ONE 2014, 9, e108698. [Google Scholar] [CrossRef] [PubMed]
- Ushio-Fukai, M.; Nakamura, Y. Reactive oxygen species and angiogenesis: NADPH oxidase as target for cancer therapy. Cancer Lett. 2008, 266, 37–52. [Google Scholar] [CrossRef]
- Kusmartsev, S.; Nefedova, Y.; Yoder, D.; Gabrilovich, D.I. Antigen-Specific Inhibition of CD8 + T Cell Response by Immature Myeloid Cells in Cancer Is Mediated by Reactive Oxygen Species. J. Immunol. 2004, 172, 989–999. [Google Scholar] [CrossRef]
- Van Der Weyden, L.; Arends, M.J.; Campbell, A.D.; Bald, T.; Wardle-Jones, H.; Griggs, N.; Velasco-Herrera, M.D.C.; Tüting, T.; Sansom, O.J.; Karp, N.A.; et al. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature 2017, 541, 233–236. [Google Scholar] [CrossRef]
- Maraldi, T.; Prata, C.; Sega, F.V.D.; Caliceti, C.; Zambonin, L.; Fiorentini, D.; Hakim, G. NAD(P)H oxidase isoform Nox2 plays a prosurvival role in human leukaemia cells. Free Radic. Res. 2009, 43, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Brar, S.S.; Kennedy, T.P.; Sturrock, A.B.; Huecksteadt, T.P.; Quinn, M.T.; Whorton, A.R.; Hoidal, J.R. An NAD(P)H oxidase regulates growth and transcription in melanoma cells. Am. J. Physiol. Cell Physiol. 2002, 282, C1212–C1224. [Google Scholar] [CrossRef] [PubMed]
- Malla, R.R.; Raghu, H.; Rao, J.S. Regulation of NADPH oxidase (Nox2) by lipid rafts in breast carcinoma cells. Int. J. Oncol. 2010, 37, 1483–1493. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, Z.; Hu, X. Inhibiting cancer metastasis via targeting NAPDH oxidase 4. Biochem. Pharmacol. 2013, 86, 253–266. [Google Scholar] [CrossRef]
- Juhasz, A.; Ge, Y.; Markel, S.; Chiu, A.; Matsumoto, L.; van Balgooy, J.; Roy, K.; Doroshow, J.H. Expression of NADPH oxidase homologues and accessory genes in human cancer cell lines, tumours and adjacent normal tissues. Free Radic. Res. 2009, 43, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Mahbouli, S.; Der Vartanian, A.; Ortega, S.; Rougé, S.; Vasson, M.P.; Rossary, A. Leptin induces ROS via NOX5 in healthy and neoplastic mammary epithelial cells. Oncol. Rep. 2017, 38, 3254–3264. [Google Scholar] [CrossRef] [PubMed]
- Fragomeni, S.M.; Sciallis, A.; Jeruss, J.S. Molecular Subtypes and Local-Regional Control of Breast Cancer. Surg. Oncol. Clin. N. Am. 2018, 27, 95–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pei, C.; Yan, S.; Liu, G.; Liu, G.; Chen, W.; Cui, Y.; Liu, Y. NADPH oxidase 1-dependent ROS is crucial for TLR4 signaling to promote tumor metastasis of non-small cell lung cancer. Tumor Biol. 2015, 36, 1493–1502. [Google Scholar] [CrossRef]
- You, X.; Ma, M.; Hou, G.; Hu, Y.; Shi, X. Gene expression and prognosis of NOX family members in gastric cancer. OncoTargets Ther. 2018, 11, 3065–3074. [Google Scholar] [CrossRef] [PubMed]
- Eun, H.S.; Cho, S.Y.; Joo, J.S.; Kang, S.H.; Moon, H.S.; Lee, E.S.; Kim, S.H.; Lee, B.S. Gene expression of NOX family members and their clinical significance in hepatocellular carcinoma. Sci. Rep. 2017, 7, 11060. [Google Scholar] [CrossRef] [PubMed]
- Pulcrano, M.; Boukheris, H.; Talbot, M.; Caillou, B.; Dupuy, C.; Virion, A.; De Vathaire, F.; Schlumberger, M. Poorly differentiated follicular thyroid carcinoma: Prognostic factors and relevance of histological classification. Thyroid 2007, 17, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Chin, S.F.; Rueda, O.M.; Vollan, H.K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.J.; et al. The somatic mutation profiles of 2433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).