Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses
Abstract
1. Introduction
2. Results
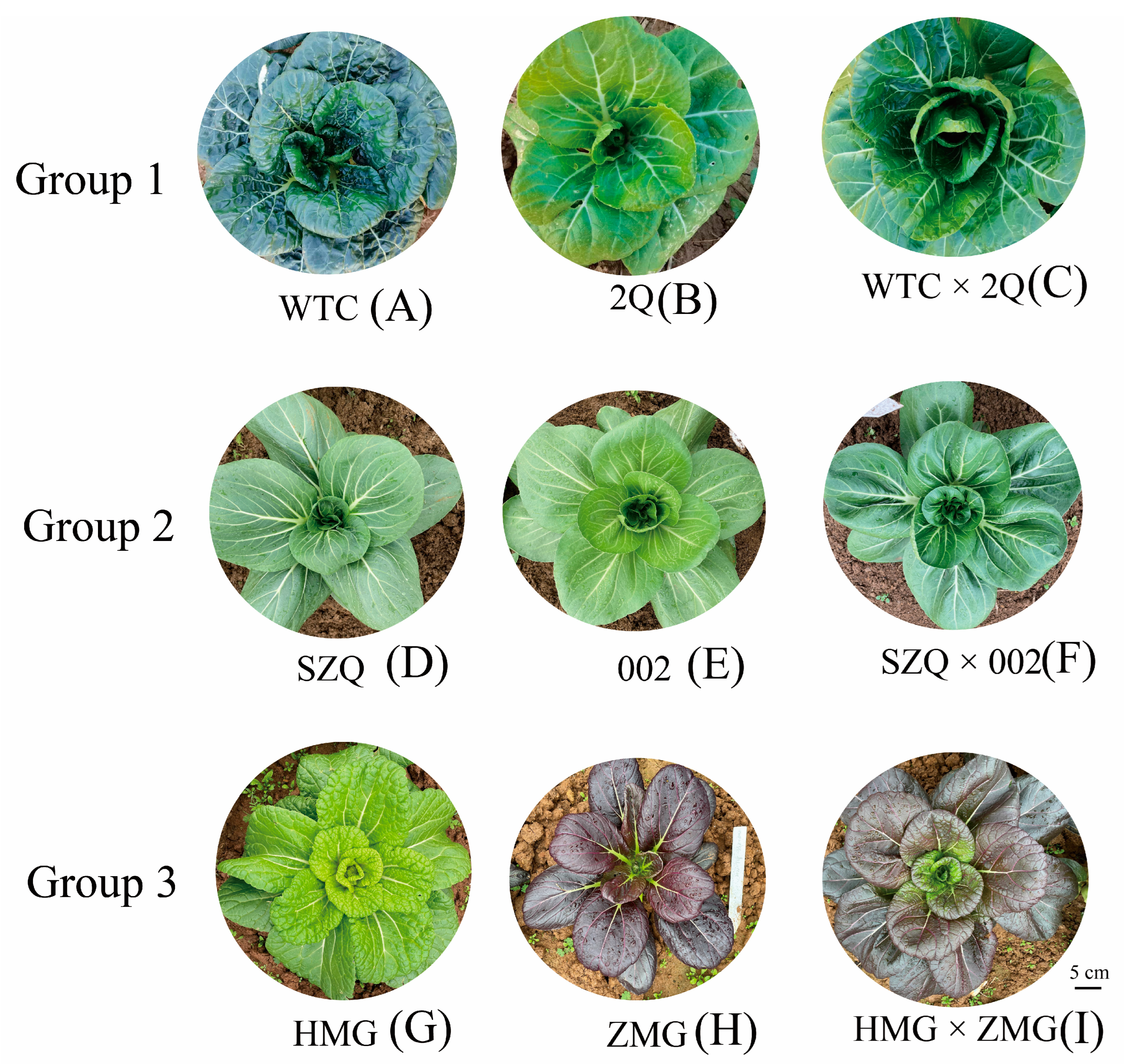
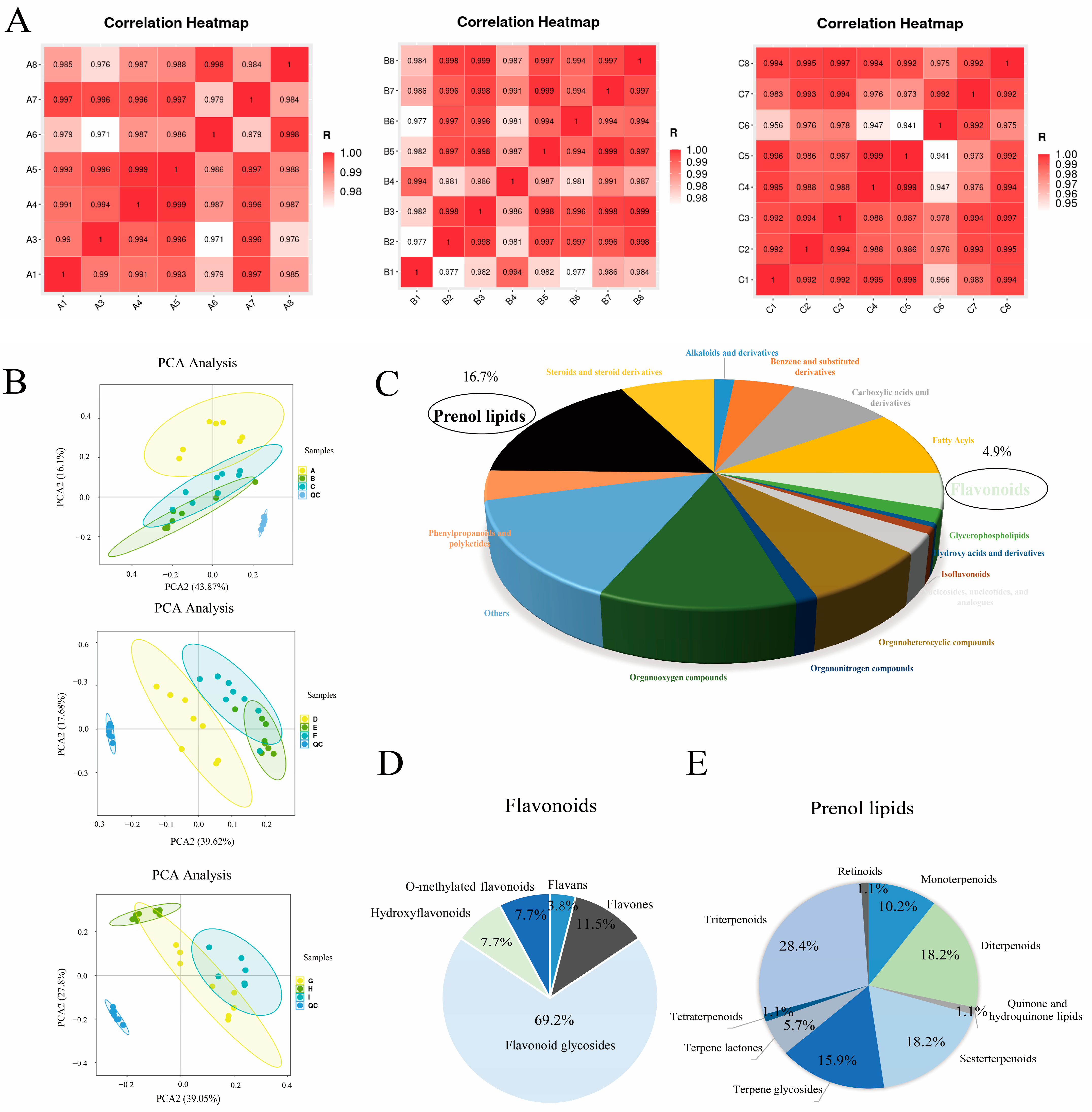
2.1. Analysis of Metabolites in Pak Choi Leaves
2.2. Transcriptome Analysis in Pak Choi Leaves
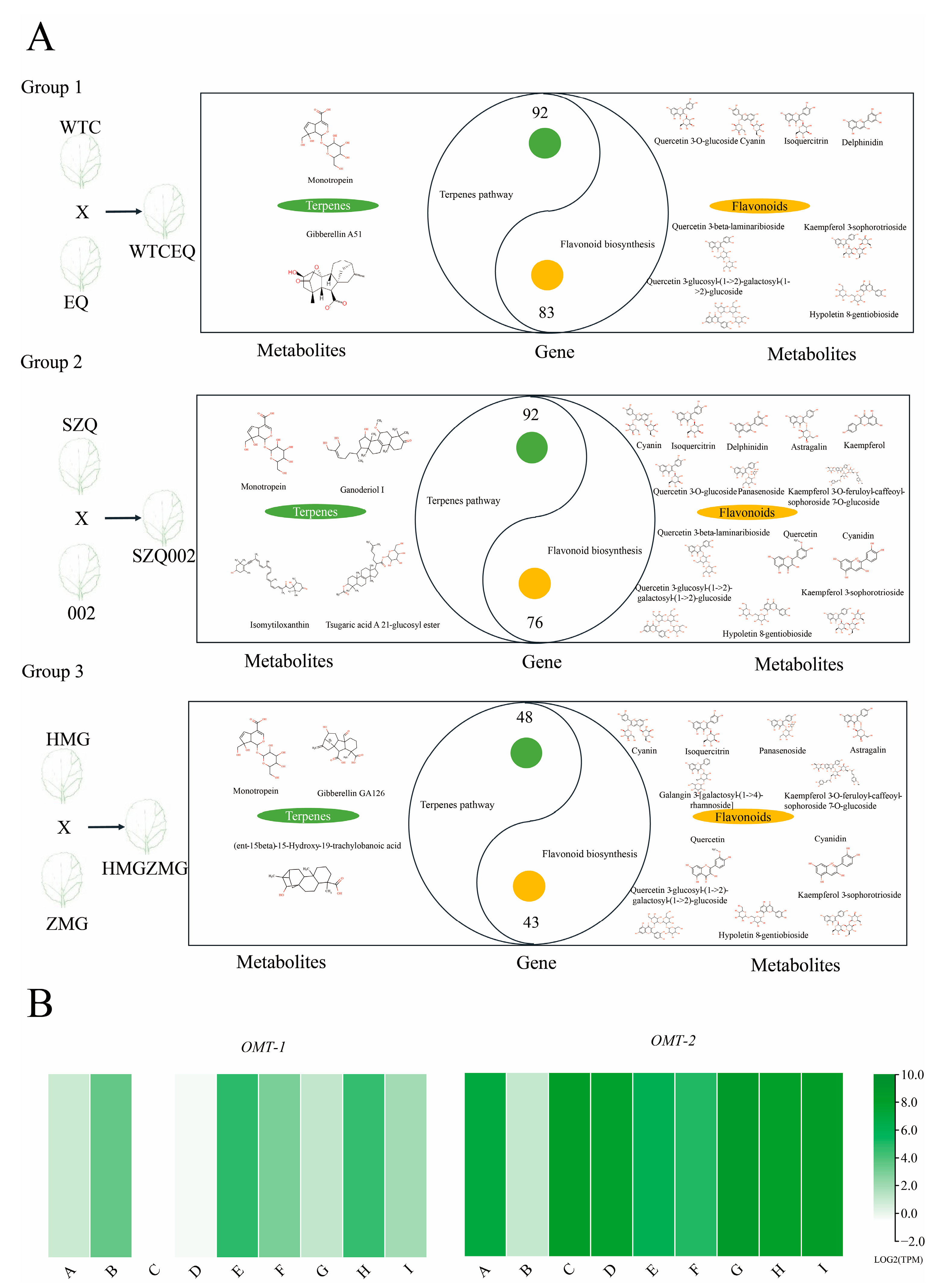
2.3. Association Analysis of Metabolic and Transcriptomic Data
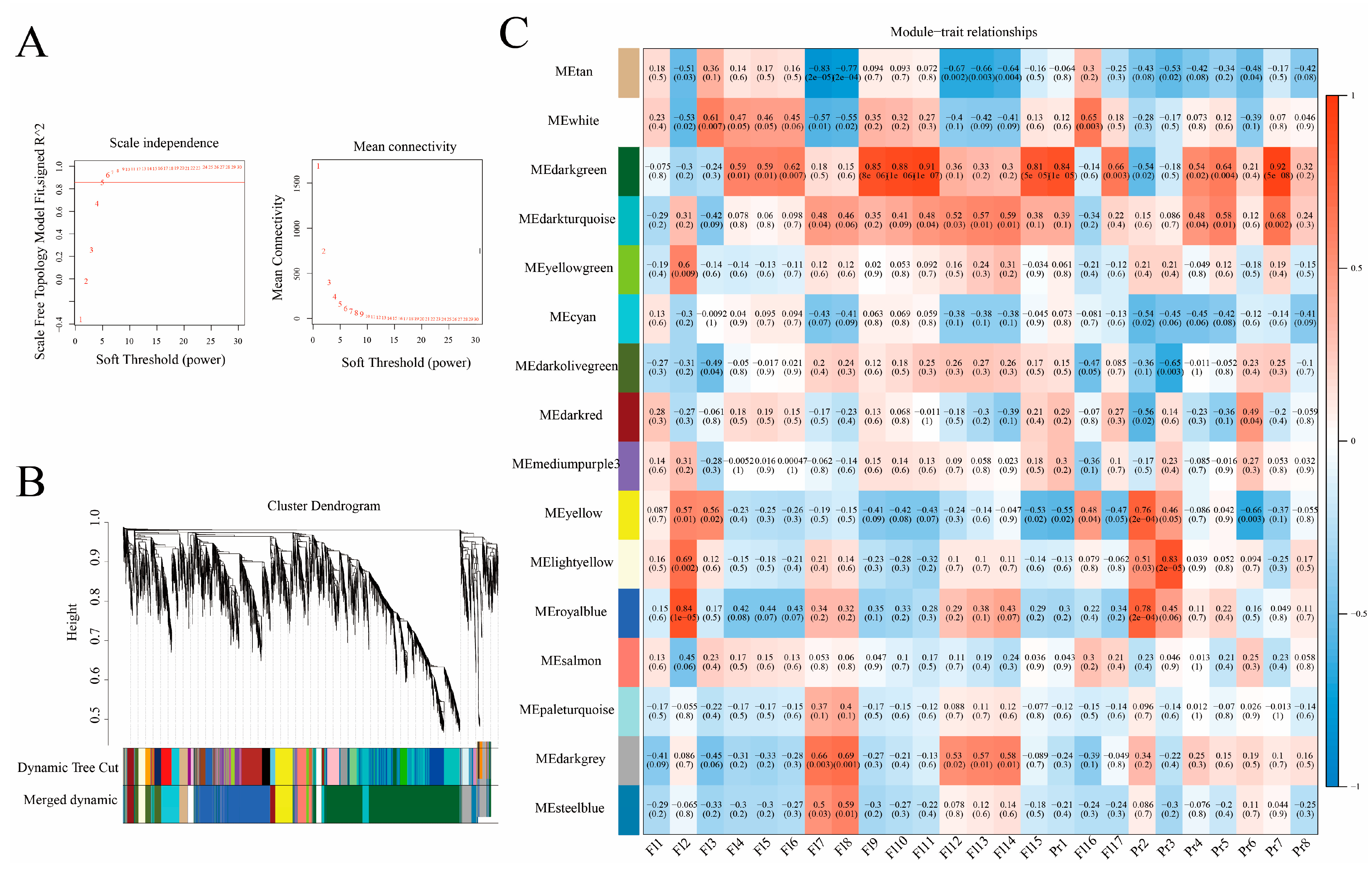
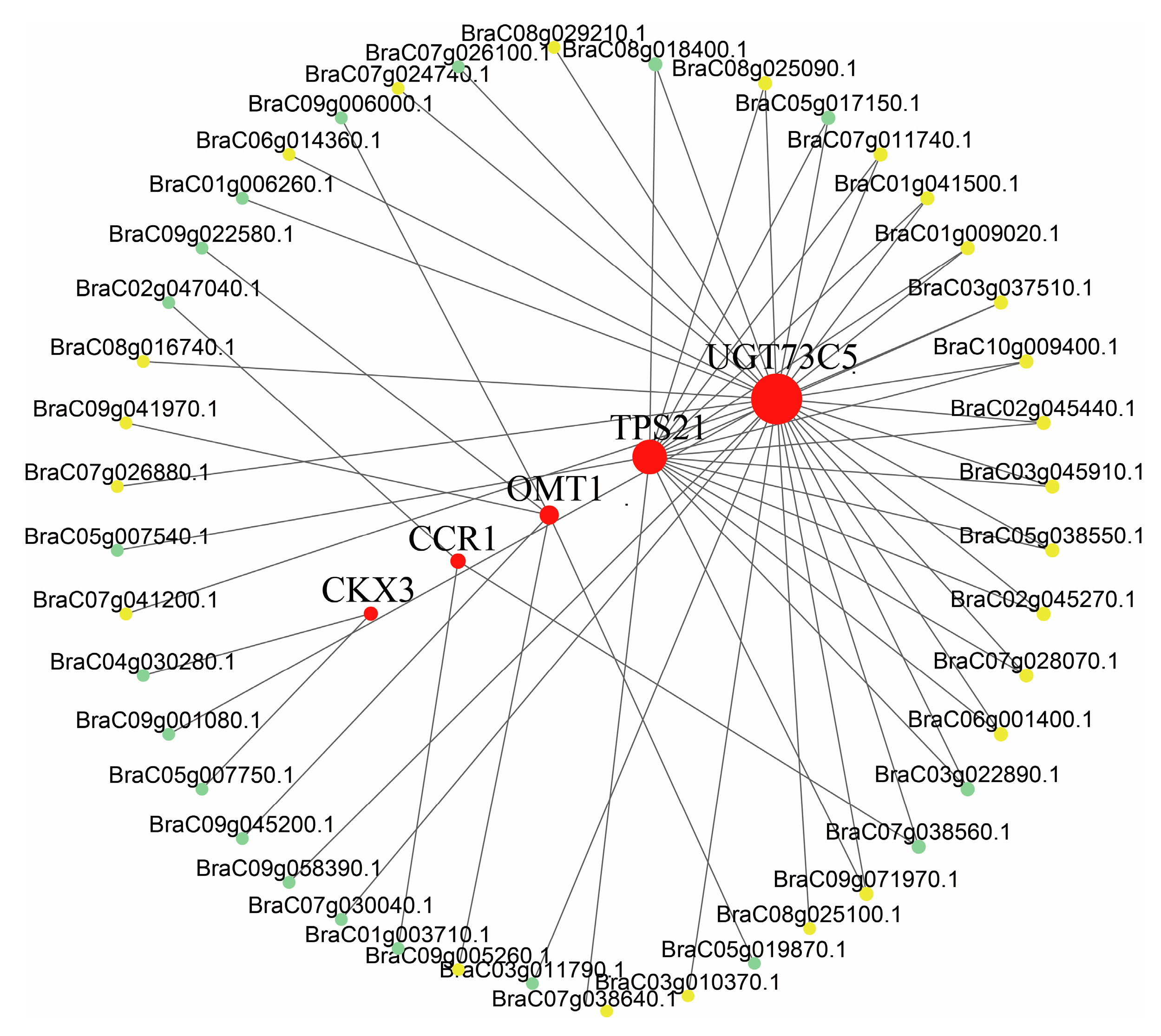
2.4. Co-Expression Analysis
2.5. Flavonoid and Terpenoid Regulation Networks in Pak Choi Leaves
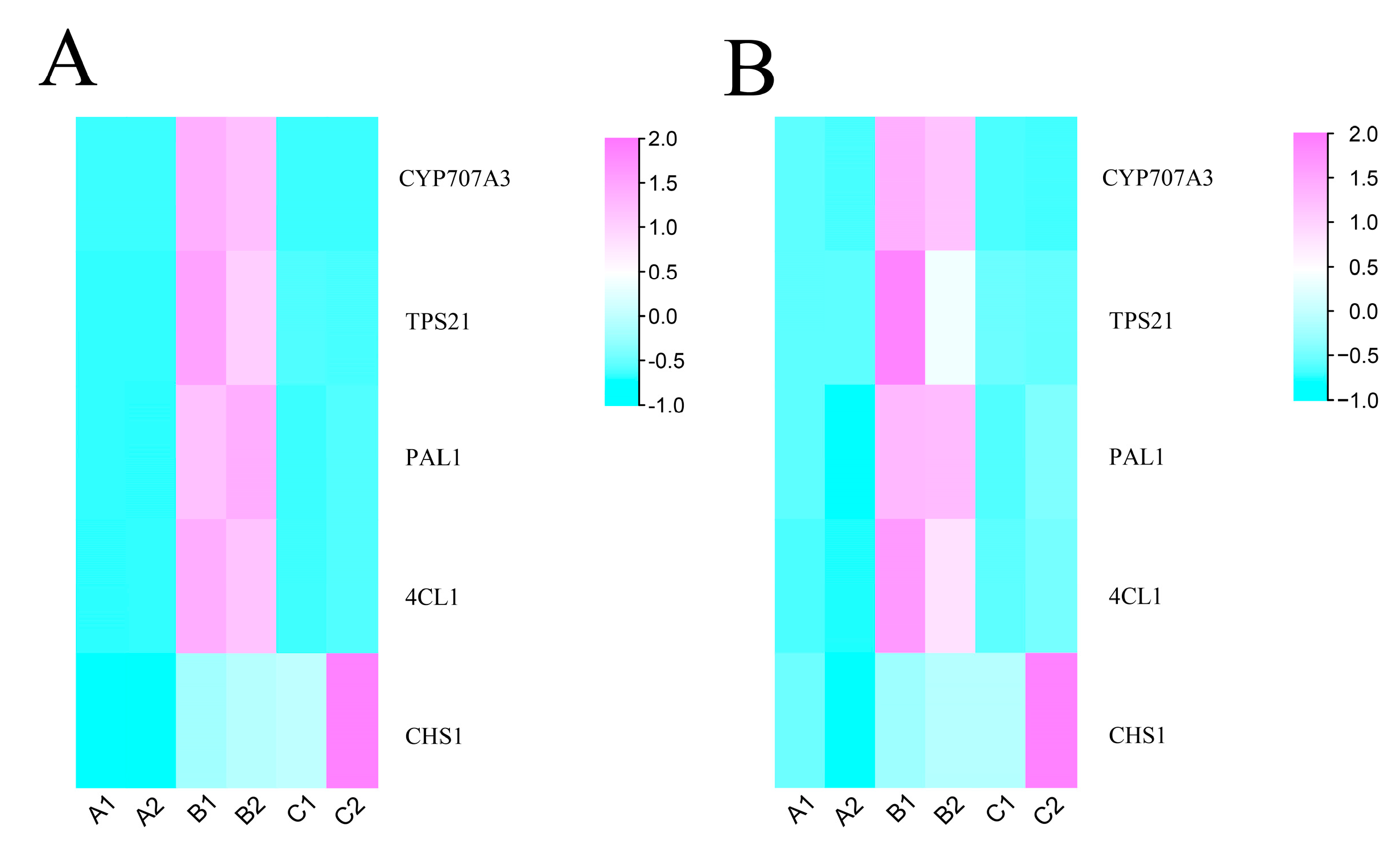
2.6. Validation of the Key DEGs Involved in Flavonoids and Terpenoids with qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Metabolite Extraction and Detection
4.3. Transcriptomic Analysis
4.4. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.5. Quantitative Real-Time PCR (qPCR) Expression Analyses
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.; Liao, X.; Yu, L.; Rao, S.; Chen, Q.; Zhu, Z.; Cong, X.; Zhang, W.; Ye, J.; Cheng, S.; et al. Combined metabolome and transcriptome analysis reveal the mechanism of selenate influence on the growth and quality of cabbage (Brassica oleracea var. capitata L.). Food Res. Int. 2022, 156, 111135. [Google Scholar] [CrossRef]
- Xie, J.; Wang, W.; Yang, T.; Zhang, Q.; Zhang, Z.; Zhang, X.; Li, N.; Zhi, L.; Ma, X.; Zhang, S.; et al. Large-scale genomic and transcriptomic profiles of rice hybrids reveal a core mechanism underlying heterosis. Genome Biol. 2022, 23, 264. [Google Scholar] [CrossRef]
- Thole, V.; Bassard, J.E.; Ramírez-González, R.; Trick, M.; Afshar, B.G.; Breitel, D.; Hill, L.; Foito, A.; Shepherd, L.; Freitag, S.; et al. RNA-seq, de novo transcriptome assembly and flavonoid gene analysis in 13 wild and cultivated berry fruit species with high content of phenolics. BMC Genom. 2019, 20, 995. [Google Scholar] [CrossRef]
- Feng, Z.; Sun, L.; Dong, M.; Fan, S.; Shi, K.; Qu, Y.; Zhu, L.; Shi, J.; Wang, W.; Liu, Y.; et al. Novel players in organogenesis and flavonoid biosynthesis in cucumber glandular trichomes. Plant Physiol. 2023, 192, 2723–2736. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, D.; Ma, F.N.; Yang, L.; Wu, B.; Xing, W.; Sun, P.; Chen, D.; Xu, B.; Song, S. Identification of key genes involved in flavonoid and terpenoid biosynthesis and the pathway of triterpenoid biosynthesis in Passiflora edulis. J. Integr. Agric. 2023, 22, 1412–1423. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, L.; Yang, J.; Hu, H.; Wei, G.; Cui, J.; Xu, J. Transcriptome and metabolome analyses reveal differences in terpenoid and flavonoid biosynthesis in Cryptomeria fortunei needles across different seasons. Front. Plant Sci. 2022, 13, 862746. [Google Scholar] [CrossRef] [PubMed]
- Apel, L.; Kammerer, D.; Stintzing, F.; Spring, O. Comparative metabolite profiling of triterpenoid saponins and flavonoids in flower color mutations of Primula veris L. Int. J. Mol. Sci. 2017, 18, 153. [Google Scholar] [CrossRef]
- Xue, T.; Zheng, X.; Chen, D.; Zhang, T.; Chen, Y.; Zhong, Q.; Chen, B.; Li, B. Metabolome and whole transcriptome analyses reveal the molecular mechanisms underlying terpenoids biosynthesis in Sapindus mukorossi fruits. Ind. Crops Prod. 2022, 181, 114810. [Google Scholar] [CrossRef]
- Khan, W.A.; Hu, H.; Cuin, T.A.; Hao, Y.; Ji, X.; Wang, J.; Hu, C. Untargeted metabolomics and comparative flavonoid analysis reveal the nutritional aspects of pak choi. Food Chem. 2022, 383, 132375. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, Y.; Guo, F.; Wang, G. Proteomic and metabolomic analyses reveal stage- and tissue-specific flavonoid accumulation in Ginkgo biloba. LWT 2022, 171, 114111. [Google Scholar] [CrossRef]
- Rao, S.; Gou, Y.; Yu, T.; Cong, X.; Gui, J.; Zhu, Z.; Zhang, W.; Liao, Y.; Ye, J.; Cheng, S.; et al. Effects of selenate on Se, flavonoid, and glucosinolate in broccoli florets by combined transcriptome and metabolome analyses. Food Res. Int. 2021, 146, 110463. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, H.; Lv, X.; Zheng, C.; Wu, Z.; Wang, N.; Wang, J.; Chen, H.; Wei, F. Profiling and spatial distribution of phenolic compounds in rapeseed by two-step extraction strategy and targeted metabolomics combined with chemometrics. Food Chem. 2023, 401, 134151. [Google Scholar] [CrossRef]
- Wang, P.; Gu, M.; Shao, S.; Chen, X.; Hou, B.; Ye, N.; Zhang, X. Changes in non-volatile and volatile metabolites associated with heterosis in tea plants (Camellia sinensis). J. Agric. Food Chem. 2022, 70, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Byrne, P.F.; McMullen, M.D.; Snook, M.E.; Musket, T.A.; Theuri, J.M.; Widstrom, N.W.; Wiseman, B.R.; Coe, E.H. Quantitative trait loci and metabolic pathways: Genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc. Natl. Acad. Sci. USA 1996, 93, 8820–8825. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Lee, Y.S.; Nguyen, V.B.; Giang, V.N.L.; Koo, H.J.; Park, H.-S.; Mohanan, P.; Song, Y.H.; Ryu, B.; Kang, K.B.; et al. Comparative transcriptome and metabolome analyses of four Panax species explore the dynamics of metabolite biosynthesis. J. Ginseng Res. 2022, 47, 44–53. [Google Scholar] [CrossRef]
- Qu, C.; Zhu, M.; Hu, R.; Niu, Y.; Chen, S.; Zhao, H.; Li, C.; Wang, Z.; Yin, N.; Sun, F.; et al. Comparative genomic analyses reveal the genetic basis of the yellow-seed trait in Brassica napus. Nat. Commun. 2023, 14, 5194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, C.; Chao, H.; Long, Y.; Wu, J.; Li, Z.; Ge, X.; Xia, H.; Yin, Y.; Batley, J.; et al. Integration of metabolome and transcriptome reveals flavonoid accumulation in the intergeneric hybrid between Brassica rapa and Raphanus sativus. Sci. Rep. 2019, 9, 18368. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yao, L.; Pecoraro, L.; Liu, C.; Wang, J.; Huang, L.; Gao, W. Cold stress regulates accumulation of flavonoids and terpenoids in plants by phytohormone, transcription process, functional enzyme, and epigenetics. Crit. Rev. Biotechnol. 2023, 43, 680–697. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y.; Gao, F.; Jin, W.; Li, S.; Kimani, S.; Yang, S.; Bao, T.; Gao, X.; Wang, L. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 4140–4158. [Google Scholar] [CrossRef]
- Shan, X.; Li, Y.; Yang, S.; Yang, Z.; Qiu, M.; Gao, R.; Han, T.; Meng, X.; Xu, Z.; Wang, L.; et al. The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytol. 2020, 228, 1864–1879. [Google Scholar] [CrossRef]
- Li, P.; Xia, E.; Fu, J.; Xu, Y.; Zhao, X.; Tong, W.; Tang, Q.; Tadege, M.; Fernie, A.R.; Zhao, J. Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants (Camellia sinensis). Plant J. 2022, 110, 1144–1165. [Google Scholar] [CrossRef]
- Singh, V.V.; Balbeer; Sharma, H.K.; Priyamedha; Ram, B.; Meena, H.S.; Rai, P.K. Heterosis and gene action studies for agro-physiological traits in Indian mustard (Brassica juncea L.). Vegetos 2022, 35, 803–809. [Google Scholar] [CrossRef]
- Yi, G.; Shin, H.; Park, H.R.; Park, J.E.; Ahn, J.H.; Lim, S.; Lee, J.G.; Lee, E.J.; Huh, J.H. Revealing biomass heterosis in the allodiploid ×Brassicoraphanus, a hybrid between Brassica rapa and Raphanus sativus, through integrated transcriptome and metabolites analysis. BMC Plant Biol. 2020, 20, 252. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lu, X.; Zhu, Y.; Pan, J.; Zhou, S.; Zhang, X.; Zhu, G.; Shang, Y.; Huang, S.; Zhang, C. The multi-omics basis of potato heterosis. J. Integr. Plant Biol. 2022, 64, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Arceo, M.; Gomez-Lopez, I.; Carr-Ugarte, H.; Eseberri, I.; Gonzalez, M.; Cano, M.P.; Portillo, M.P.; Gomez-Zorita, S. Anti-obesity effects of isorhamnetin and isorhamnetin conjugates. Int. J. Mol. Sci. 2022, 24, 299. [Google Scholar] [CrossRef]
- Wang, B.; Hou, M.; Shi, J.; Ku, L.; Song, W.; Li, C.; Ning, Q.; Li, X.; Li, C.; Zhao, B.; et al. De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nat. Genet. 2023, 55, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yue, M.; Liu, Y.; Zhang, N.; Lin, Y.; Zhang, Y.; Wang, Y.; Li, M.; Luo, Y.; Zhang, Y.; et al. A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × ananassa). Plant Biotechnol. J. 2023, 21, 1140–1158. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, B.; Yao, T.; Shen, C.; Wen, T.; Zhang, R.; Li, Y.; Le, Y.; Li, Z.; Zhang, X.; et al. Re enhances anthocyanin and proanthocyanidin accumulation to produce red foliated cotton and brown fiber. Plant Physiol. 2022, 189, 1466–1481. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zhao, C.; Wang, S.; Gao, N.; Wang, X.; Na, X.; Wang, X.; Bi, Y. PIF4-PAP1 interaction affects MYB-bHLH-WD40 complex formation and anthocyanin accumulation in Arabidopsis. J Plant Physiol. 2022, 268, 153558. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, X.; Gong, Q.; Cao, J.; Shen, W.; Yin, X.; Grierson, D.; Zhang, B.; Xu, C.; Li, X.; et al. Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 2021, 19, 671–688. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Zhou, L.; You, S.; Deng, H.; Chen, Y.; Alseekh, S.; Yuan, Y.; Fu, R.; Zhang, Z.; et al. Microtom metabolic network: Rewiring tomato metabolic regulatory network throughout the growth cycle. Mol. Plant. 2020, 13, 1203–1218. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021, 105, 167–181. [Google Scholar] [CrossRef]
- Mo, R.; Han, G.; Zhu, Z.; Essemine, J.; Dong, Z.; Li, Y.; Deng, W.; Qu, M.; Zhang, C.; Yu, C. The ethylene response factor ERF5 regulates anthocyanin biosynthesis in ‘Zijin’ mulberry fruits by interacting with MYBA and F3H Genes. Int. J. Mol. Sci. 2022, 23, 7615. [Google Scholar] [CrossRef]
- Wei, C.; Li, M.; Cao, X.; Jin, Z.; Zhang, C.; Xu, M.; Chen, K.; Zhang, B. Linalool synthesis related PpTPS1 and PpTPS3 are activated by transcription factor PpERF61 whose expression is associated with DNA methylation during peach fruit ripening. Plant Sci. 2022, 317, 111200. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, M.; Zhang, M.; Yang, M.; Dai, S.; Meng, Q.; Lv, W.; Zhuang, K. ETHYLENE-INSENSITIVE 3-LIKE 2 regulates beta-carotene and ascorbic acid accumulation in tomatoes during ripening. Plant Physiol. 2023, 192, 2067–2080. [Google Scholar] [CrossRef]
- Zhang, Y.; Ji, A.; Xu, Z.; Luo, H.; Song, J. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol. Biol. 2019, 100, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gao, M.; Zhao, Y.; Chen, Y.; Wu, L.; Yin, H.; Xiong, S.; Wang, S.; Wang, J.; Yang, Y.; et al. LcERF19, an AP2/ERF transcription factor from Litsea cubeba, positively regulates geranial and neral biosynthesis. Hortic. Res. 2022, 9, uhac093. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Han, T.; Bai, A.; Xu, H.; Wang, J.; Hou, X.; Li, Y. Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses. Int. J. Mol. Sci. 2024, 25, 3587. https://doi.org/10.3390/ijms25073587
Wang H, Han T, Bai A, Xu H, Wang J, Hou X, Li Y. Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses. International Journal of Molecular Sciences. 2024; 25(7):3587. https://doi.org/10.3390/ijms25073587
Chicago/Turabian StyleWang, Haibin, Tiantian Han, Aimei Bai, Huanhuan Xu, Jianjun Wang, Xilin Hou, and Ying Li. 2024. "Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses" International Journal of Molecular Sciences 25, no. 7: 3587. https://doi.org/10.3390/ijms25073587
APA StyleWang, H., Han, T., Bai, A., Xu, H., Wang, J., Hou, X., & Li, Y. (2024). Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses. International Journal of Molecular Sciences, 25(7), 3587. https://doi.org/10.3390/ijms25073587






