Genome-Wide Identification and Expression Pattern Analysis of BAHD Acyltransferase Family in Taxus mairei
Abstract
:1. Introduction
2. Results
2.1. Identification of TwBAHD Family
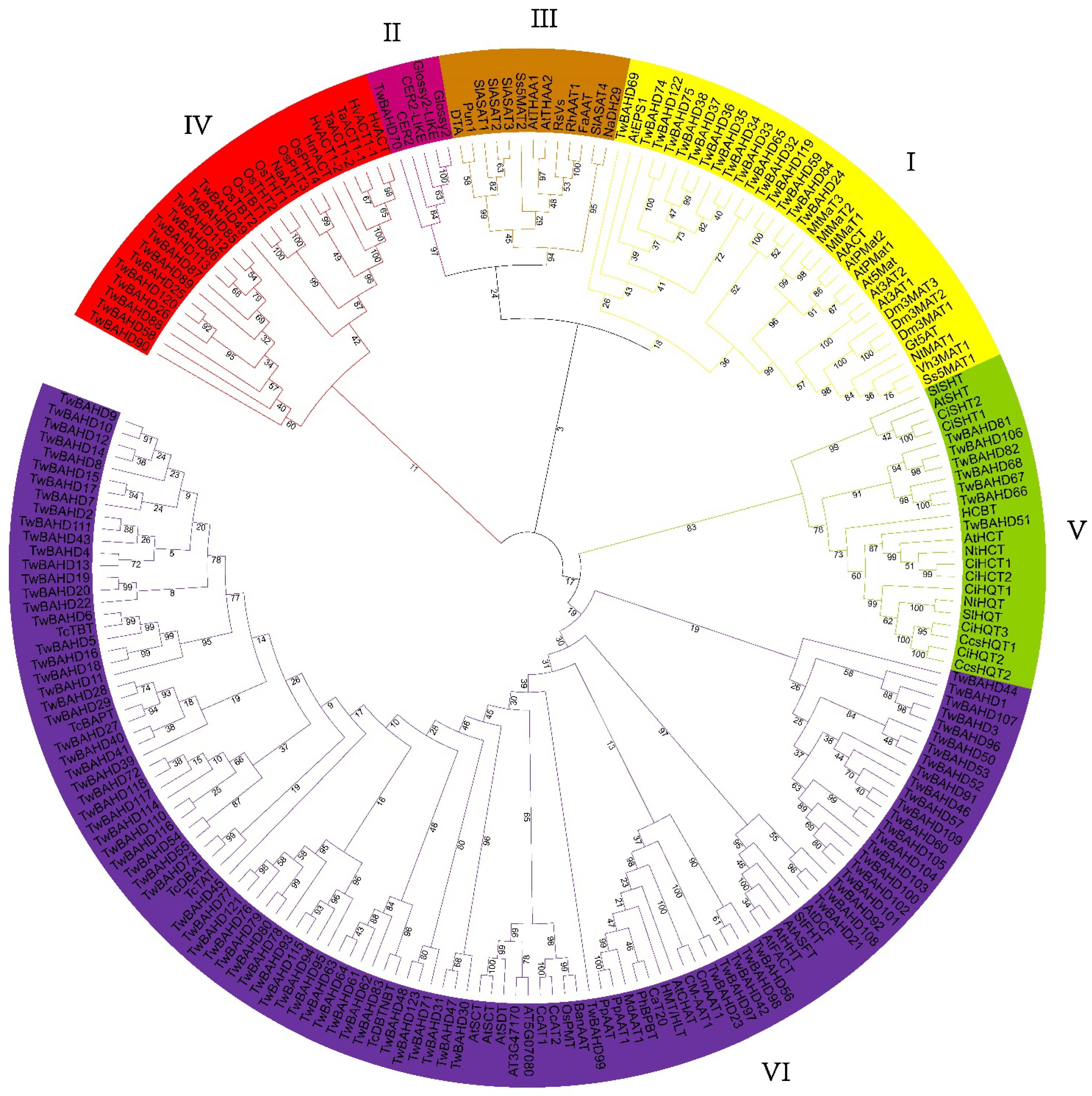
2.2. Phylogenetic Analysis of TwBAHD Family
2.3. Conserved Gene Structure and Protein Motif Analysis of TwBAHD Family
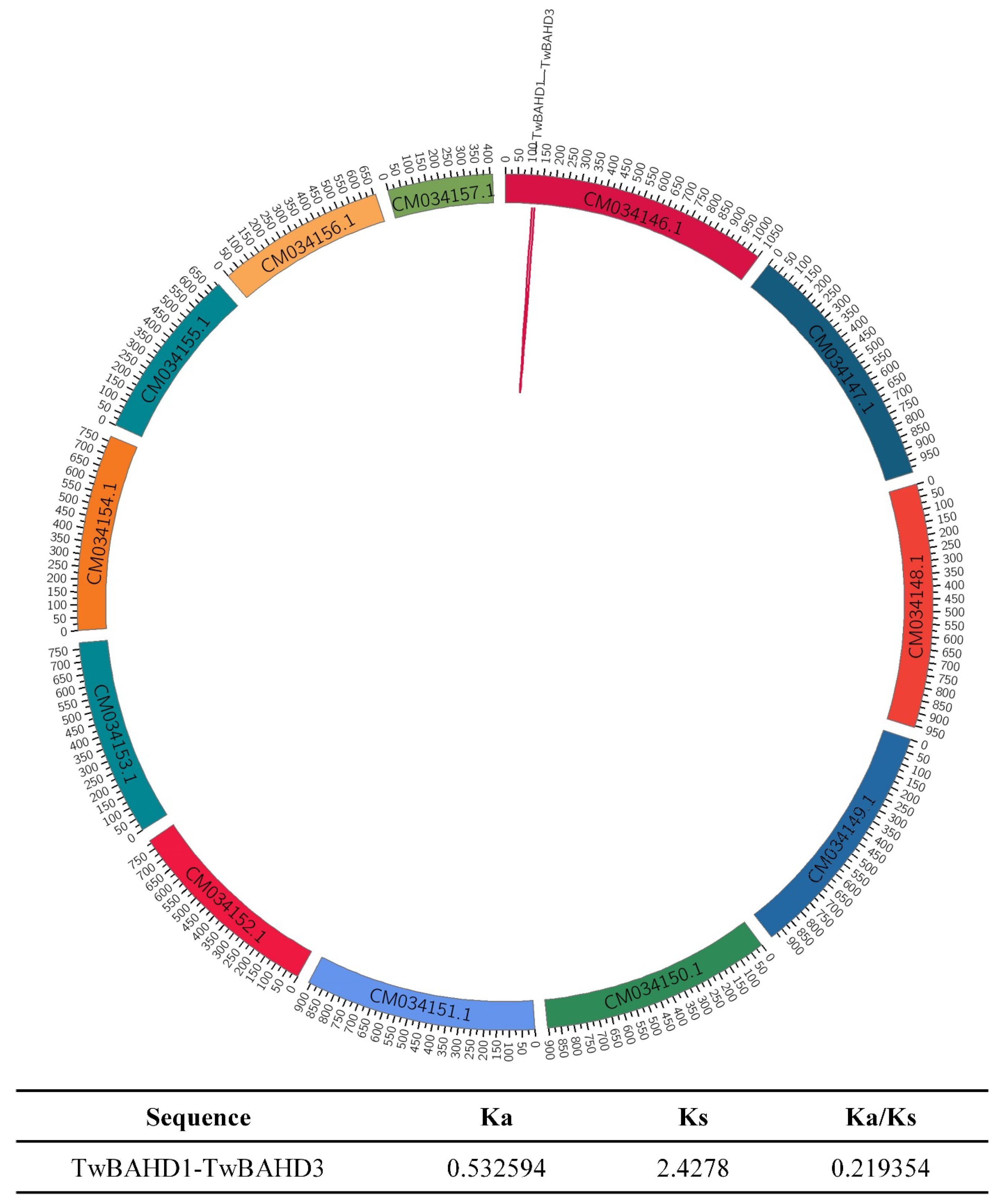
2.4. Chromosomal Location and Duplication Events of TwBAHDs
2.5. Cis-Acting Element Analysis in the Promoters of TwBAHD Genes
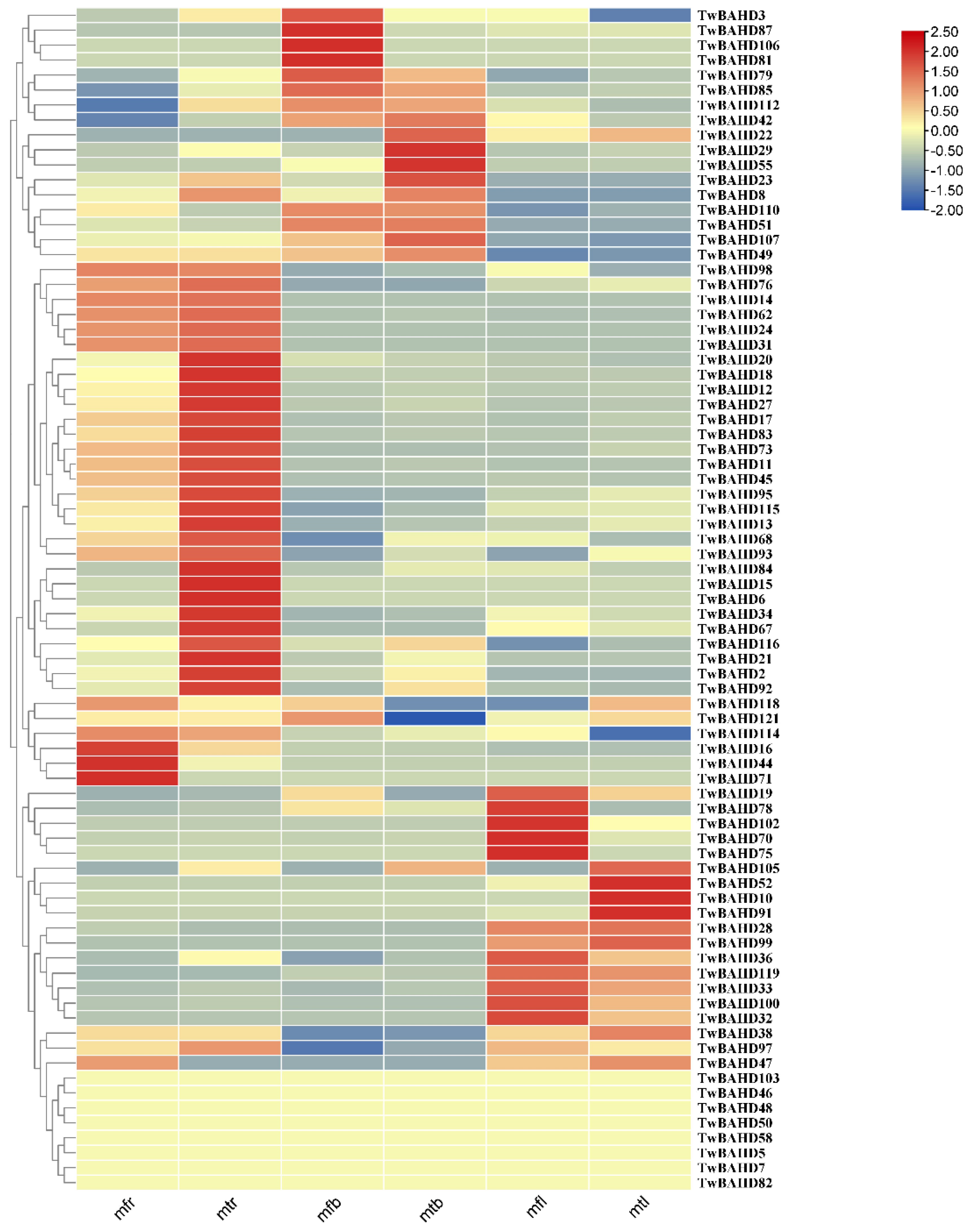
2.6. Expression Profile of TwBAHD Genes by RNA-seq
2.7. Gene Expression Analysis of TwBAHDs by qRT-PCR
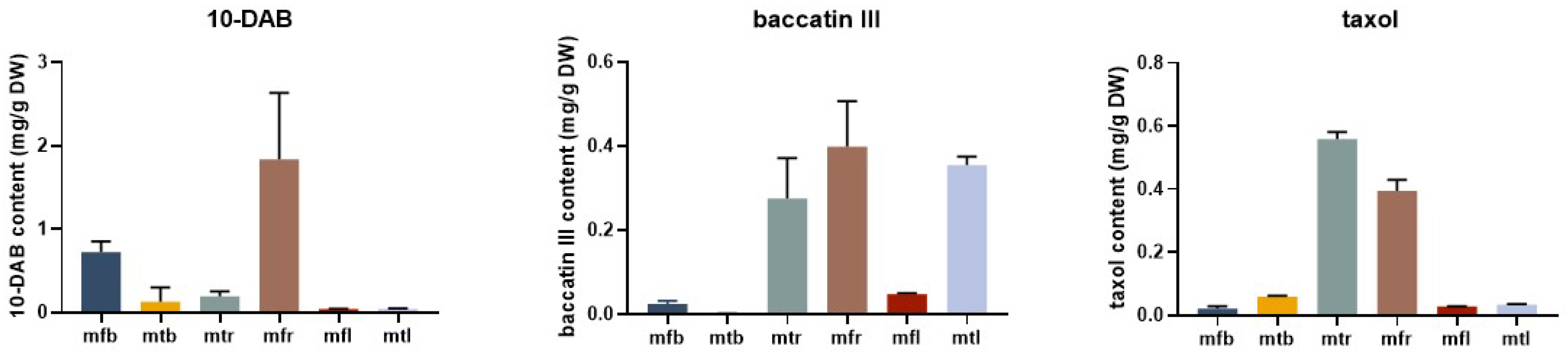
2.8. Content of Taxanes in Different Tissues of T. mairei
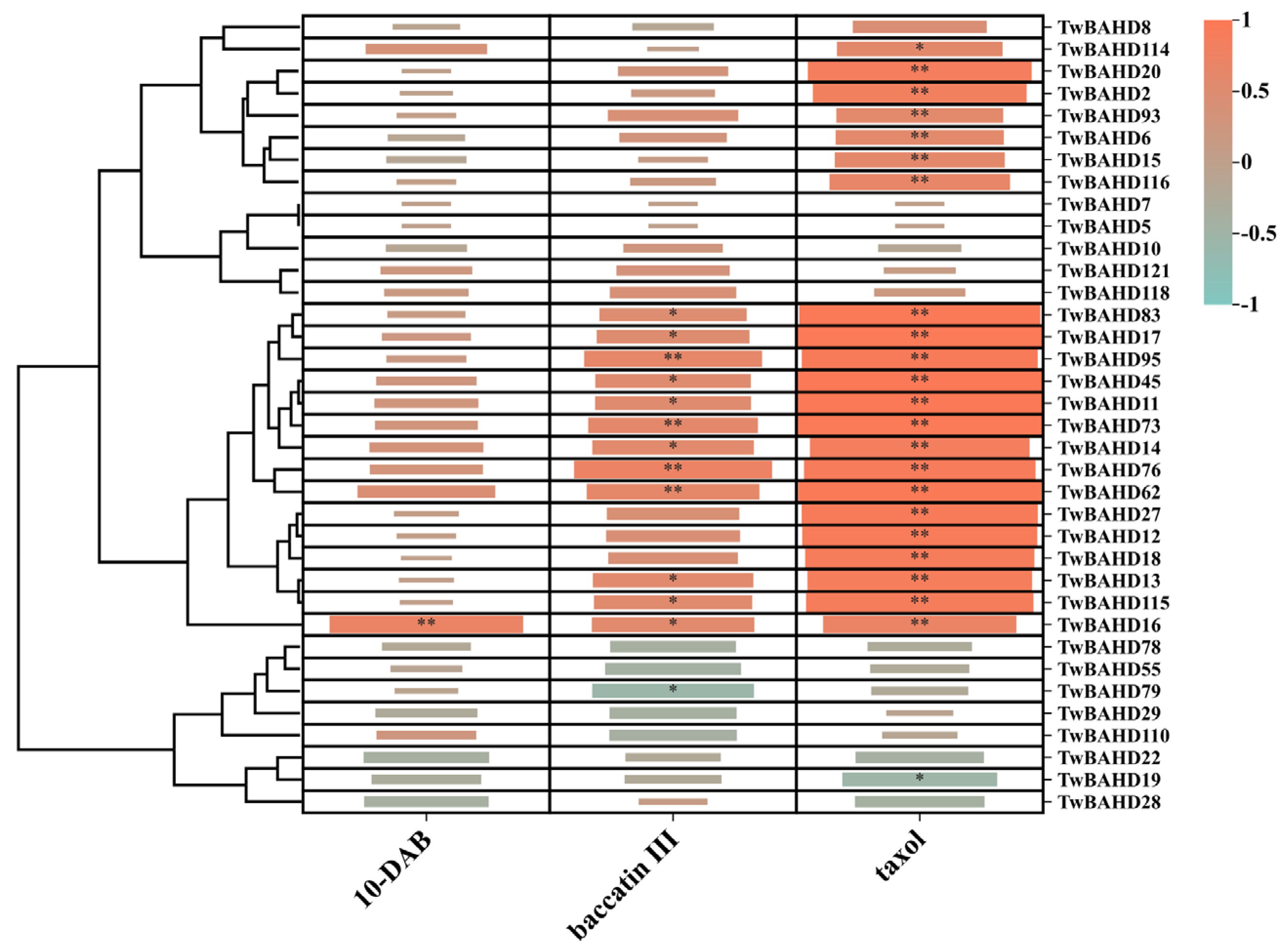
2.9. Correlation Analysis between TwBAHD Gene Expression and Taxane Levels
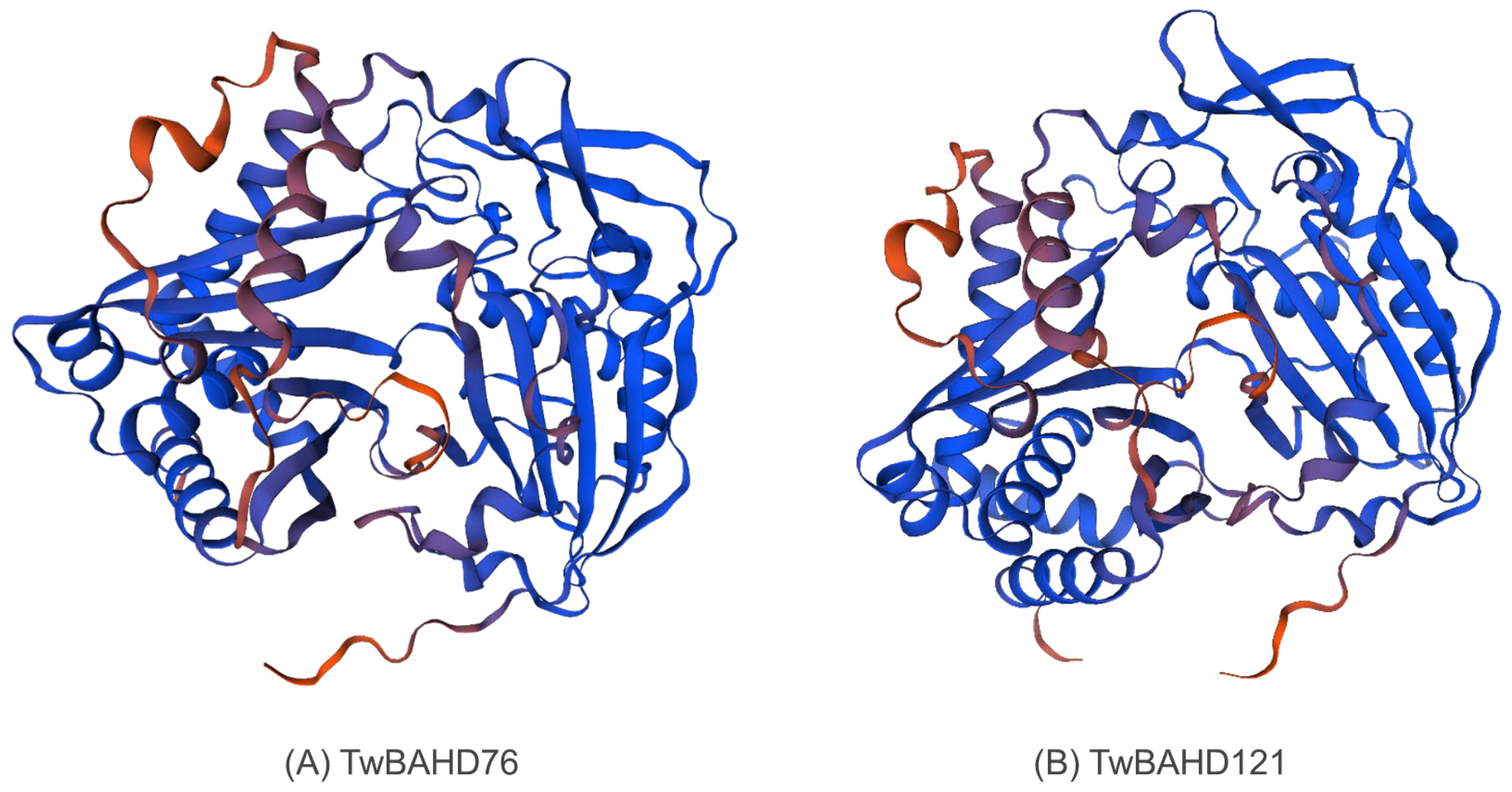
2.10. 3D Modeling of TwBAHD76 and TwBAHD121
2.11. Subcellular Localization of TwBAHD76 and TwBAHD121 Genes
3. Discussions
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of BAHD Family Members in Taxus mairei
4.3. Phylogenetic Analysis
4.4. Analysis of Gene Structures and Conserved Motifs
4.5. Chromosomal Distribution and Synteny Analysis
4.6. Analysis of Promoter Cis-Regulatory Element
4.7. Expression Patterns of the TwBAHD Gene Family Based on RNA-Seq Databases
4.8. RNA Extraction and qRT-PCR Analysis
4.9. Taxanes’ Determination and Correlation Analysis
4.10. Subcellular Localization of the TwBAHD Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riffi, O.; reda Kachmar, M.; M’Hamdi, Z.; Fliou, J.; Chakir, S.; Amechrouq, A. Study of the chemical composition and evaluation of the antioxidant and antimicrobial activity of Taxus baccata L. Arab. J. Chem. 2023, 16, 105334. [Google Scholar] [CrossRef]
- Alqahtani, F.Y.; Aleanizy, F.S.; El Tahir, E.; Alkahtani, H.M.; AlQuadeib, B.T. Paclitaxel. Profiles DrugSubst. Excip. Relat. Methodol. 2019, 44, 205–238. [Google Scholar] [CrossRef]
- Croteau, R.; Ketchum, R.E.; Long, R.M.; Kaspera, R.; Wildung, M.R. Taxol biosynthesis and molecular genetics. Phytochem. Rev. Proc. Phytochem. Soc. Eur. 2006, 5, 75–97. [Google Scholar] [CrossRef]
- Bocci, G.; Di Paolo, A.; Danesi, R. The pharmacological bases of the antiangiogenic activity of paclitaxel. Angiogenesis 2013, 16, 481–492. [Google Scholar] [CrossRef]
- Sabzehzari, M.; Zeinali, M.; Naghavi, M.R. Alternative sources and metabolic engineering of Taxol: Advances and future perspectives. Biotechnol. Adv. 2020, 43, 107569. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, L.; Zhuang, W.; Zhang, F.; Shu, X.; Wang, N.; Wang, Z. Recent Research Progress in Taxol Biosynthetic Pathway and Acylation Reactions Mediated by Taxus Acyltransferases. Molecules 2021, 26, 2855. [Google Scholar] [CrossRef]
- Walker, K.; Croteau, R. Taxol biosynthesis: Molecular cloning of a benzoyl-CoA:taxane 2alpha-O-benzoyltransferase cDNA from taxus and functional expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 13591–13596. [Google Scholar] [CrossRef]
- Walker, K.; Fujisaki, S.; Long, R.; Croteau, R. Molecular cloning and heterologous expression of the C-13 phenylpropanoid side chain-CoA acyltransferase that functions in Taxol biosynthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 12715–12720. [Google Scholar] [CrossRef] [PubMed]
- Walker, K.; Long, R.; Croteau, R. The final acylation step in taxol biosynthesis: Cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc. Natl. Acad. Sci. USA 2002, 99, 9166–9171. [Google Scholar] [CrossRef]
- Lin, S.L.; Wei, T.; Lin, J.F.; Guo, L.Q.; Wu, G.P.; Wei, J.B.; Huang, J.J.; Ouyang, P.L. Bio-production of Baccatin III, an Important Precursor of Paclitaxel by a Cost-Effective Approach. Mol. Biotechnol. 2018, 60, 492–505. [Google Scholar] [CrossRef]
- Jiménez-Barbero, J.; Amat-Guerri, F.; Snyder, J.P. The solid state, solution and tubulin-bound conformations of agents that promote microtubule stabilization. Curr. Med. Chem. Anti-Cancer Agents 2002, 2, 91–122. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Vogt, T. Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 2001, 213, 164–174. [Google Scholar] [CrossRef]
- Sah, B.; Subban, K.; Jayabaskaran, C. Biochemical insights into the recombinant 10-deacetylbaccatin III-10-β-O-acetyltransferase enzyme from the Taxol-producing endophytic fungus Lasiodiplodia theobromae. FEMS Microbiol. Lett. 2019, 366, fnz072. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, J.C. Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 2006, 9, 331–340. [Google Scholar] [CrossRef]
- Sharma, S.; Khare, P.; Kumar, A.; Chunduri, V.; Kumar, A.; Kapoor, P.; Mangal, P.; Kondepudi, K.K.; Bishnoi, M.; Garg, M. Anthocyanin-Biofortified Colored Wheat Prevents High Fat Diet-Induced Alterations in Mice: Nutrigenomics Studies. Mol. Nutr. Food Res. 2020, 64, e1900999. [Google Scholar] [CrossRef]
- St-Pierre, B.; De Luca, V. Chapter Nine—Evolution of Acyltransferase Genes: Origin and Diversification of the BAHD Superfamily of Acyltransferases Involved in Secondary Metabolism. In Recent Advances in Phytochemistry; Romeo, J.T., Ibrahim, R., Varin, L., De Luca, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2000; Volume 34, pp. 285–315. [Google Scholar]
- Dudareva, N.; D’Auria, J.C.; Nam, K.H.; Raguso, R.A.; Pichersky, E. Acetyl-CoA:benzylalcohol acetyltransferase—An enzyme involved in floral scent production in Clarkia breweri. Plant J. Cell Mol. Biol. 1998, 14, 297–304. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, B.; Laflamme, P.; Alarco, A.M.; De Luca, V. The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J. Cell Mol. Biol. 1998, 14, 703–713. [Google Scholar] [CrossRef]
- Fujiwara, H.; Tanaka, Y.; Yonekura-Sakakibara, K.; Fukuchi-Mizutani, M.; Nakao, M.; Fukui, Y.; Yamaguchi, M.; Ashikari, T.; Kusumi, T. cDNA cloning, gene expression and subcellular localization of anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J. Cell Mol. Biol. 1998, 16, 421–431. [Google Scholar] [CrossRef]
- Wang, L.; Chen, K.; Zhang, M.; Ye, M.; Qiao, X. Catalytic function, mechanism, and application of plant acyltransferases. Crit. Rev. Biotechnol. 2022, 42, 125–144. [Google Scholar] [CrossRef]
- Fayad, M.A.; Charles, S.; Shelvy, S.; Sheeja, T.E.; Sangeetha, K.; Angadi, U.B.; Tandon, G.; Iquebal, M.A.; Jaiswal, S.; Kumar, D. Whole genome based identification of BAHD acyltransferase gene involved in piperine biosynthetic pathway in black pepper. J. Biomol. Struct. Dyn. 2024. [Google Scholar] [CrossRef]
- Yu, X.H.; Gou, J.Y.; Liu, C.J. BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: Bioinformatics and gene expression. Plant Mol. Biol. 2009, 70, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wang, Z.; Zhuang, W.; Wang, T.; Xie, Y. Family characteristics, phylogenetic reconstruction, and potential applications of the plant BAHD acyltransferase family. Front. Plant Sci. 2023, 14, 1218914. [Google Scholar] [CrossRef]
- Xia, Y.; Nikolau, B.J.; Schnable, P.S. Developmental and hormonal regulation of the arabidopsis CER2 gene that codes for a nuclear-localized protein required for the normal accumulation of cuticular waxes. Plant Physiol. 1997, 115, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Yamane, M.; Takenoya, M.; Yajima, S.; Sue, M. Crystal structure of barley agmatine coumaroyltransferase, an N-acyltransferase from the BAHD superfamily. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2020, 76, 590–596. [Google Scholar] [CrossRef]
- Yamane, M.; Takenoya, M.; Yajima, S.; Sue, M. Molecular and structural characterization of agmatine coumaroyltransferase in Triticeae, the key regulator of hydroxycinnamic acid amide accumulation. Phytochemistry 2021, 189, 112825. [Google Scholar] [CrossRef] [PubMed]
- Kage, U.; Karre, S.; Kushalappa, A.C.; McCartney, C. Identification and characterization of a fusarium head blight resistance gene TaACT in wheat QTL-2DL. Plant Biotechnol. J. 2017, 15, 447–457. [Google Scholar] [CrossRef]
- Burhenne, K.; Kristensen, B.K.; Rasmussen, S.K. A new class of N-hydroxycinnamoyltransferases. Purification, cloning, and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J. Biol. Chem. 2003, 278, 13919–13927. [Google Scholar] [CrossRef]
- Moglia, A.; Acquadro, A.; Eljounaidi, K.; Milani, A.M.; Cagliero, C.; Rubiolo, P.; Genre, A.; Cankar, K.; Beekwilder, J.; Comino, C. Genome-Wide Identification of BAHD Acyltransferases and In vivo Characterization of HQT-like Enzymes Involved in Caffeoylquinic Acid Synthesis in Globe Artichoke. Front. Plant Sci. 2016, 7, 1424. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Gou, J.; Liao, Q.; Li, Y.; Zhou, Q.; Bi, G.; Li, C.; Du, R.; Wang, X.; Sun, T.; et al. The Taxus genome provides insights into paclitaxel biosynthesis. Nat. Plants 2021, 7, 1026–1036. [Google Scholar] [CrossRef]
- Bartley, L.E.; Peck, M.L.; Kim, S.R.; Ebert, B.; Manisseri, C.; Chiniquy, D.M.; Sykes, R.; Gao, L.; Rautengarten, C.; Vega-Sánchez, M.E.; et al. Overexpression of a BAHD acyltransferase, OsAt10, alters rice cell wall hydroxycinnamic acid content and saccharification. Plant Physiol. 2013, 161, 1615–1633. [Google Scholar] [CrossRef]
- Kusano, H.; Li, H.; Minami, H.; Kato, Y.; Tabata, H.; Yazaki, K. Evolutionary Developments in Plant Specialized Metabolism, Exemplified by Two Transferase Families. Front. Plant Sci. 2019, 10, 794. [Google Scholar] [CrossRef]
- Kuang, X.; Sun, S.; Wei, J.; Li, Y.; Sun, C. Iso-Seq analysis of the Taxus cuspidata transcriptome reveals the complexity of Taxol biosynthesis. BMC Plant Biol. 2019, 19, 210. [Google Scholar] [CrossRef] [PubMed]
- Moghe, G.; Kruse, L.H.; Petersen, M.; Scossa, F.; Fernie, A.R.; Gaquerel, E.; D’Auria, J.C. BAHD Company: The Ever-Expanding Roles of the BAHD Acyltransferase Gene Family in Plants. Annu. Rev. Plant Biol. 2023, 74, 165–194. [Google Scholar] [CrossRef]
- Peng, M.; Gao, Y.; Chen, W.; Wang, W.; Shen, S.; Shi, J.; Wang, C.; Zhang, Y.; Zou, L.; Wang, S.; et al. Evolutionarily Distinct BAHD N-Acyltransferases Are Responsible for Natural Variation of Aromatic Amine Conjugates in Rice. Plant Cell 2016, 28, 1533–1550. [Google Scholar] [CrossRef] [PubMed]
- de Vries, L.; MacKay, H.A.; Smith, R.A.; Mottiar, Y.; Karlen, S.D.; Unda, F.; Muirragui, E.; Bingman, C.; Vander Meulen, K.; Beebe, E.T.; et al. pHBMT1, a BAHD-family monolignol acyltransferase, mediates lignin acylation in poplar. Plant Physiol. 2022, 188, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Ullah, C.; Chen, Y.H.; Ortega, M.A.; Tsai, C.J. The diversity of salicylic acid biosynthesis and defense signaling in plants: Knowledge gaps and future opportunities. Curr. Opin. Plant Biol. 2023, 72, 102349. [Google Scholar] [CrossRef] [PubMed]
- Kriegshauser, L.; Knosp, S.; Grienenberger, E.; Tatsumi, K.; Gütle, D.D.; Sørensen, I.; Herrgott, L.; Zumsteg, J.; Rose, J.K.C.; Reski, R.; et al. Function of the HYDROXYCINNAMOYL-CoA:SHIKIMATE HYDROXYCINNAMOYL TRANSFERASE is evolutionarily conserved in embryophytes. Plant Cell 2021, 33, 1472–1491. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.P.; Liu, H.; Gao, S.; Cheng, A.X. Functional Characterization of a Hydroxyacid/Alcohol Hydroxycinnamoyl Transferase Produced by the Liverwort Marchantia emarginata. Molecules 2017, 22, 1854. [Google Scholar] [CrossRef]
- Abdullah; Faraji, S.; Heidari, P.; Poczai, P. The BAHD Gene Family in Cacao (Theobroma cacao, Malvaceae): Genome-Wide Identification and Expression Analysis. Front. Ecol. Evol. 2021, 9, 707708. [Google Scholar] [CrossRef]
- Liu, L.; Xu, H.; Zhang, W.; Xing, J.; Zhu, M.; Zhang, Y.; Wang, Y. Genome-Wide Analysis of the BAHD Family in Welsh Onion and CER2-LIKEs Involved in Wax Metabolism. Genes 2023, 14, 1286. [Google Scholar] [CrossRef]
- Walker, K.; Schoendorf, A.; Croteau, R. Molecular cloning of a taxa-4(20),11(12)-dien-5alpha-ol-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Arch. Biochem. Biophys. 2000, 374, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Biol. Sci. 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, R.; Jiang, S.Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Tuominen, L.K.; Johnson, V.E.; Tsai, C.J. Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genom. 2011, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Souleyre, E.J.; Greenwood, D.R.; Friel, E.N.; Karunairetnam, S.; Newcomb, R.D. An alcohol acyl transferase from apple (cv. Royal Gala), MpAAT1, produces esters involved in apple fruit flavor. FEBS J. 2005, 272, 3132–3144. [Google Scholar] [CrossRef]
- Li, D.; Xu, Y.; Xu, G.; Gu, L.; Li, D.; Shu, H. Molecular cloning and expression of a gene encoding alcohol acyltransferase (MdAAT2) from apple (cv. Golden Delicious). Phytochemistry 2006, 67, 658–667. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Manríquez, D.; Flores, F.B.; Regad, F.; Bouzayen, M.; Latché, A.; Pech, J.C. Functional characterization of a melon alcohol acyl-transferase gene family involved in the biosynthesis of ester volatiles. Identification of the crucial role of a threonine residue for enzyme activity *. Plant Mol. Biol. 2005, 59, 345–362. [Google Scholar] [CrossRef]
- Boatright, J.; Negre, F.; Chen, X.; Kish, C.M.; Wood, B.; Peel, G.; Orlova, I.; Gang, D.; Rhodes, D.; Dudareva, N. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 2004, 135, 1993–2011. [Google Scholar] [CrossRef]
- Xu, Y.; Tie, W.; Yan, Y.; Xu, B.; Liu, J.; Li, M.; Yang, J.; Zeng, J.; Hu, W.; Jin, Z. Identification and expression of the BAHD family during development, ripening, and stress response in banana. Mol. Biol. Rep. 2021, 48, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Nakayama, T.; Nishino, T. Proposed mechanism and functional amino acid residues of malonyl-CoA:anthocyanin 5-O-glucoside-6‴-O-malonyltransferase from flowers of Salvia splendens, a member of the versatile plant acyltransferase family. Biochemistry 2003, 42, 1764–1771. [Google Scholar] [CrossRef]
- Bayer, A.; Ma, X.; Stöckigt, J. Acetyltransfer in natural product biosynthesis--functional cloning and molecular analysis of vinorine synthase. Bioorg. Med. Chem. 2004, 12, 2787–2795. [Google Scholar] [CrossRef]
- Unno, H.; Ichimaida, F.; Suzuki, H.; Takahashi, S.; Tanaka, Y.; Saito, A.; Nishino, T.; Kusunoki, M.; Nakayama, T. Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J. Biol. Chem. 2007, 282, 15812–15822. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Koepke, J.; Panjikar, S.; Fritzsch, G.; Stöckigt, J. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily *. J. Biol. Chem. 2005, 280, 13576–13583. [Google Scholar] [CrossRef]
- Zhang, W.; Li, J.; Dong, Y.; Huang, Y.; Qi, Y.; Bai, H.; Li, H.; Shi, L. Genome-wide identification and expression of BAHD acyltransferase gene family shed novel insights into the regulation of linalyl acetate and lavandulyl acetate in lavender. J. Plant Physiol. 2024, 292, 154143. [Google Scholar] [CrossRef]
- Zhuang, W.; Shu, X.; Lu, X.; Wang, T.; Zhang, F.; Wang, N.; Wang, Z. Genome-wide analysis and expression profiles of PdeMYB transcription factors in colored-leaf poplar (Populus deltoids). BMC Plant Biol. 2021, 21, 432. [Google Scholar] [CrossRef] [PubMed]
- Murayama, K.; Kato-Murayama, M.; Sato, T.; Hosaka, T.; Ishiguro, K.; Mizuno, T.; Kitao, K.; Honma, T.; Yokoyama, S.; Tanaka, Y.; et al. Anthocyanin 5,3′-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin. Phytochemistry 2021, 186, 112727. [Google Scholar] [CrossRef]
- Zhang, J.F.; Gong, S.; Guo, Z.G. Effects of different elicitors on 10-deacetylbaccatin III-10-O-acetyltransferase activity and cytochrome P450 monooxygenase content in suspension cultures of Taxus cuspidata cells. Cell Biol. Int. Rep. 2010, 18, e00009. [Google Scholar] [CrossRef]
- Baebler, S.; Camloh, M.; Kovac, M.; Ravnikar, M.; Zel, J. Jasmonic acid stimulates taxane production in cell suspension culture of yew (Taxus x media). Planta Med. 2002, 68, 475–476. [Google Scholar] [CrossRef]
- Hao, D.C.; Ge, G.; Xiao, P.; Zhang, Y.; Yang, L. The first insight into the tissue specific taxus transcriptome via Illumina second generation sequencing. PLoS ONE 2011, 6, e21220. [Google Scholar] [CrossRef] [PubMed]
- Ashihara, H.; Ludwig, I.A.; Crozier, A. Biosynthesis of Purine Alkaloids. In Plant Nucleotide Metabolism—Biosynthesis, Degradation, and Alkaloid Formation; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2020; pp. 231–258. [Google Scholar] [CrossRef]
- Nevarez, D.M.; Mengistu, Y.A.; Nawarathne, I.N.; Walker, K.D. An N-aroyltransferase of the BAHD superfamily has broad aroyl CoA specificity in vitro with analogues of N-dearoylpaclitaxel. J. Am. Chem. Soc. 2009, 131, 5994–6002. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Viner, C.; Ishak, C.A.; Johnson, J.; Walker, N.J.; Shi, H.; Sjöberg-Herrera, M.K.; Shen, S.Y.; Lardo, S.M.; Adams, D.J.; Ferguson-Smith, A.C.; et al. Modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet. Genome Biol. 2024, 25, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.T.; Kong, Y.Z.; Wang, Q.; Sun, Y.H.; Gong, D.P.; Lv, J.; Liu, G.S. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan Hered. 2015, 37, 91–97. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kęska, K.; Szcześniak, M.W.; Makałowska, I.; Czernicka, M. Long-Term Waterlogging as Factor Contributing to Hypoxia Stress Tolerance Enhancement in Cucumber: Comparative Transcriptome Analysis of Waterlogging Sensitive and Tolerant Accessions. Genes 2021, 12, 189. [Google Scholar] [CrossRef]
- Li, L.; Chen, Y.; Ma, Y.; Wang, Z.; Wang, T.; Xie, Y. Optimization of Taxol Extraction Process Using Response Surface Methodology and Investigation of Temporal and Spatial Distribution of Taxol in Taxus mairei. Molecules 2021, 26, 5485. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, D.; Wang, Z.; Zhuang, W.; Zhang, F.; Xie, Y.; Wang, T. Genome-Wide Identification and Expression Pattern Analysis of BAHD Acyltransferase Family in Taxus mairei. Int. J. Mol. Sci. 2024, 25, 3777. https://doi.org/10.3390/ijms25073777
Xu D, Wang Z, Zhuang W, Zhang F, Xie Y, Wang T. Genome-Wide Identification and Expression Pattern Analysis of BAHD Acyltransferase Family in Taxus mairei. International Journal of Molecular Sciences. 2024; 25(7):3777. https://doi.org/10.3390/ijms25073777
Chicago/Turabian StyleXu, Donghuan, Zhong Wang, Weibing Zhuang, Fan Zhang, Yinfeng Xie, and Tao Wang. 2024. "Genome-Wide Identification and Expression Pattern Analysis of BAHD Acyltransferase Family in Taxus mairei" International Journal of Molecular Sciences 25, no. 7: 3777. https://doi.org/10.3390/ijms25073777
APA StyleXu, D., Wang, Z., Zhuang, W., Zhang, F., Xie, Y., & Wang, T. (2024). Genome-Wide Identification and Expression Pattern Analysis of BAHD Acyltransferase Family in Taxus mairei. International Journal of Molecular Sciences, 25(7), 3777. https://doi.org/10.3390/ijms25073777




