Genome-Wide Identification and Expression Profiling of the ABF Transcription Factor Family in Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Results
2.1. Identification of Members of the Wheat ABF TF Family
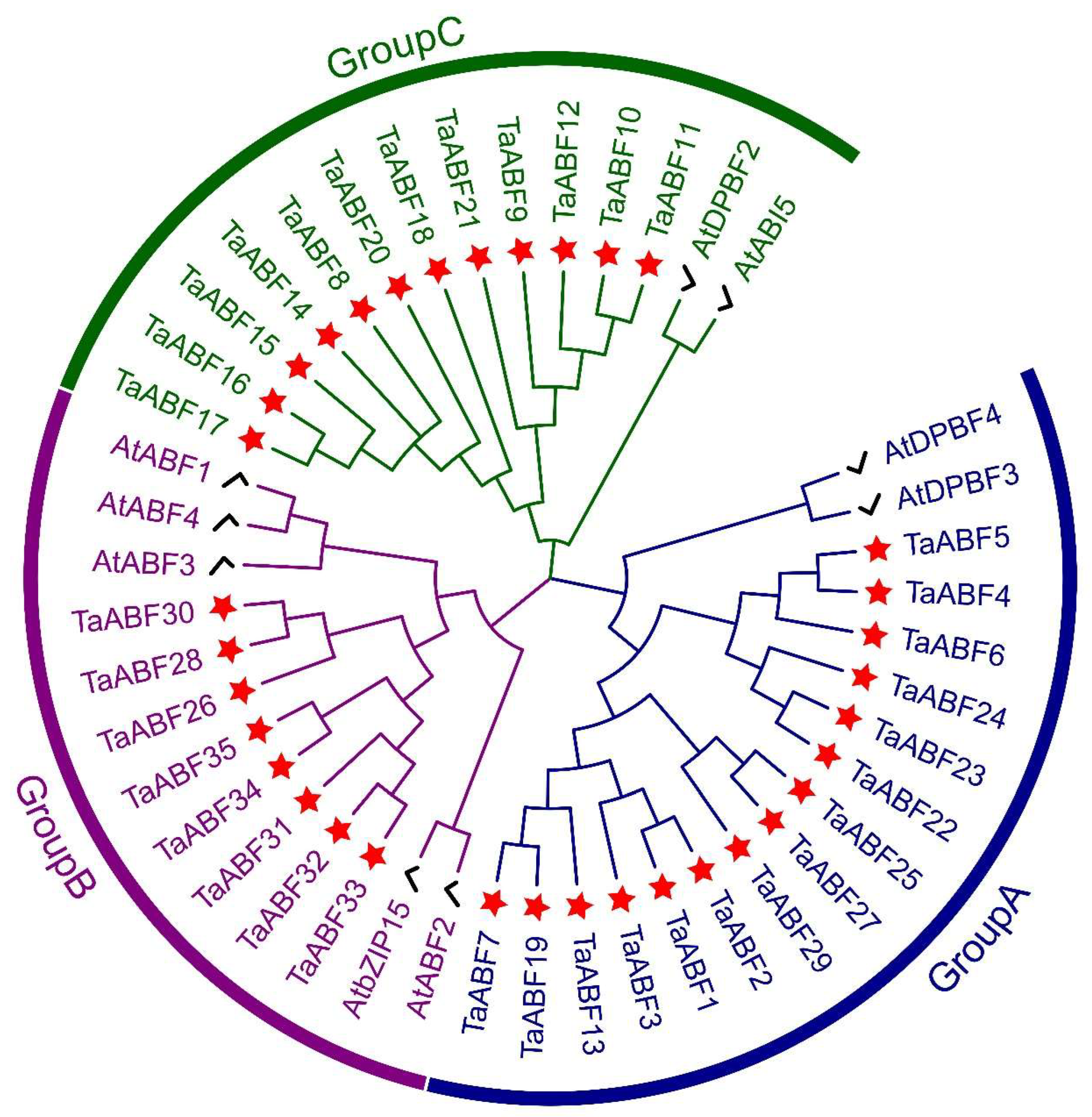
2.2. Phylogenetic Analyses and Classification of Wheat ABF TF
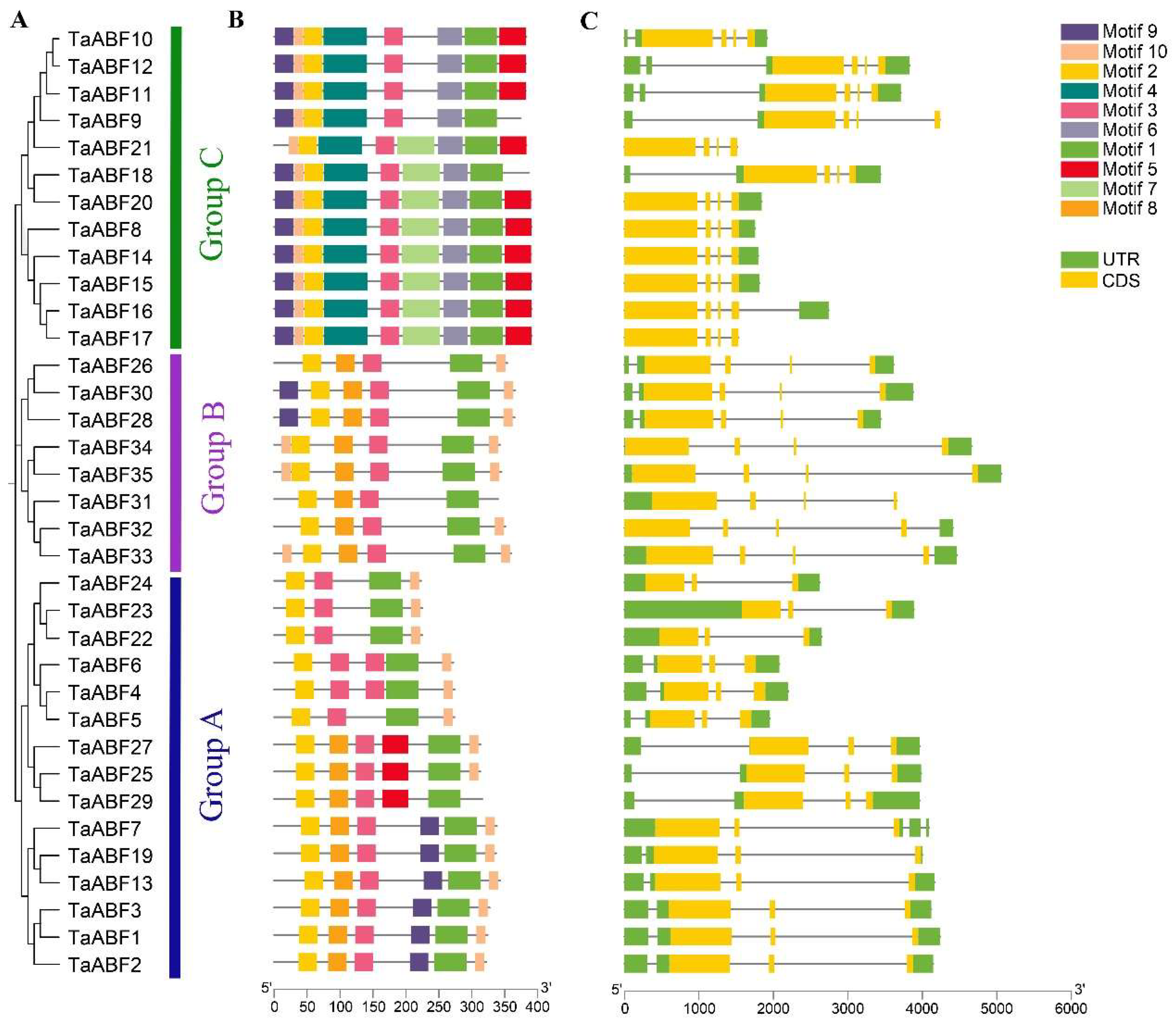
2.3. Conserved Motif and Gene Structure Analyses of Wheat TF
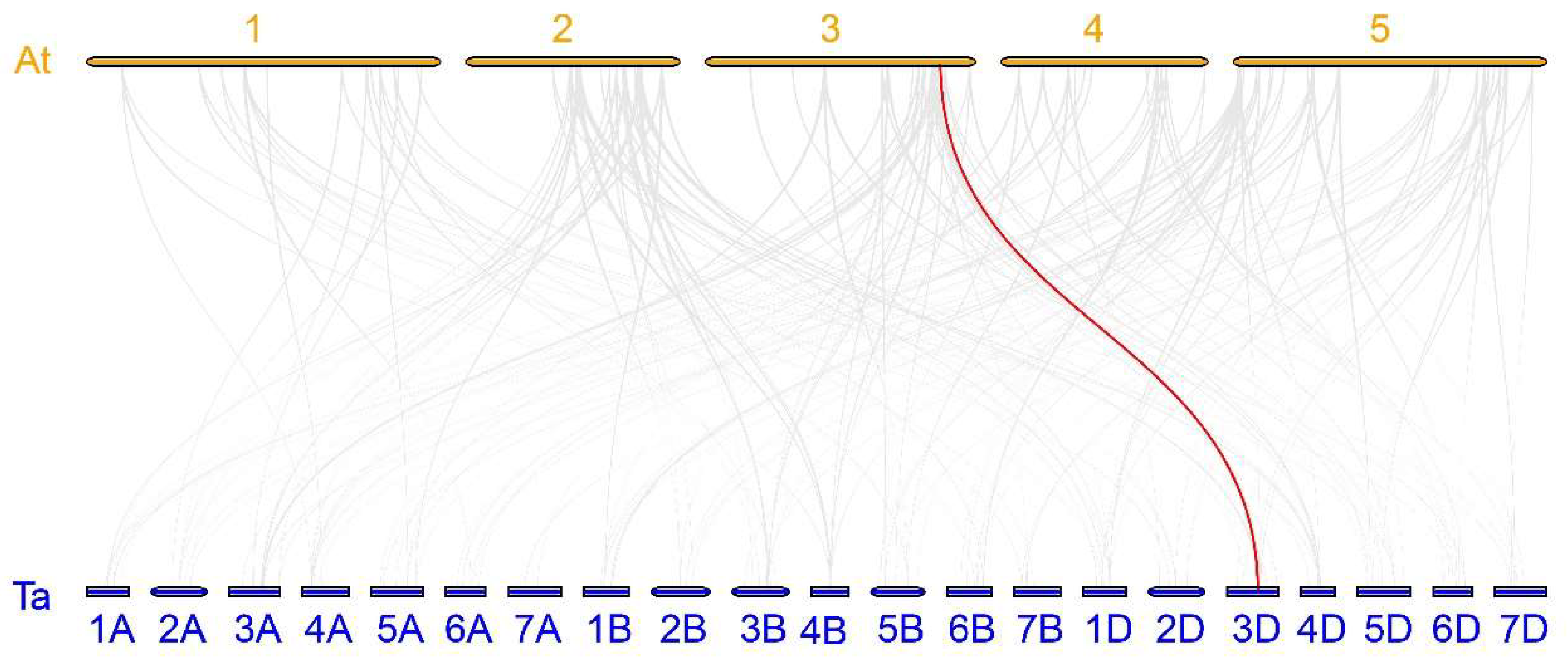
2.4. Expansion Pattern Analyses of TaABF Gene
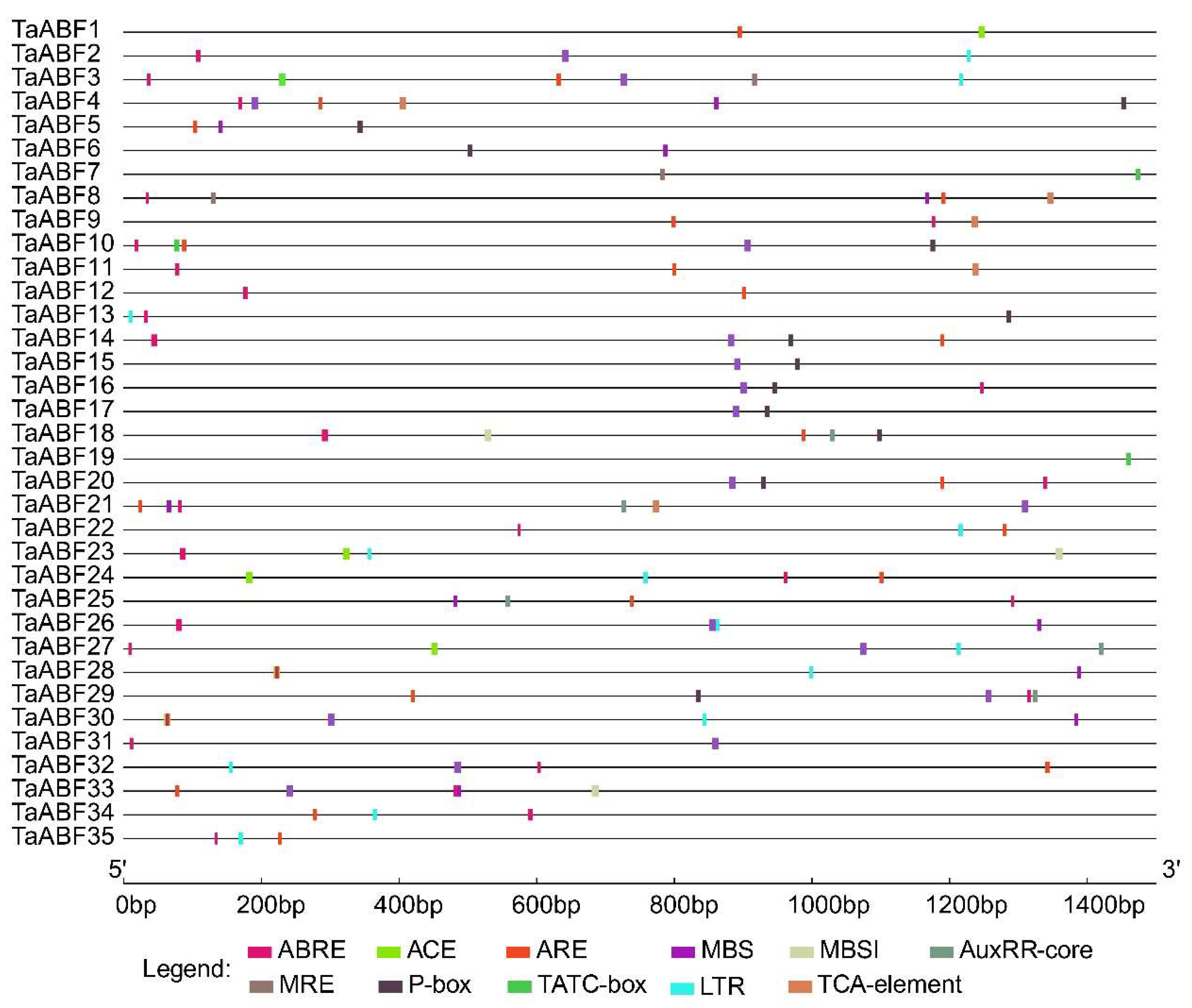
2.5. Analyses of Cis-Acting Regulatory Element Analyses in TaABF Genes
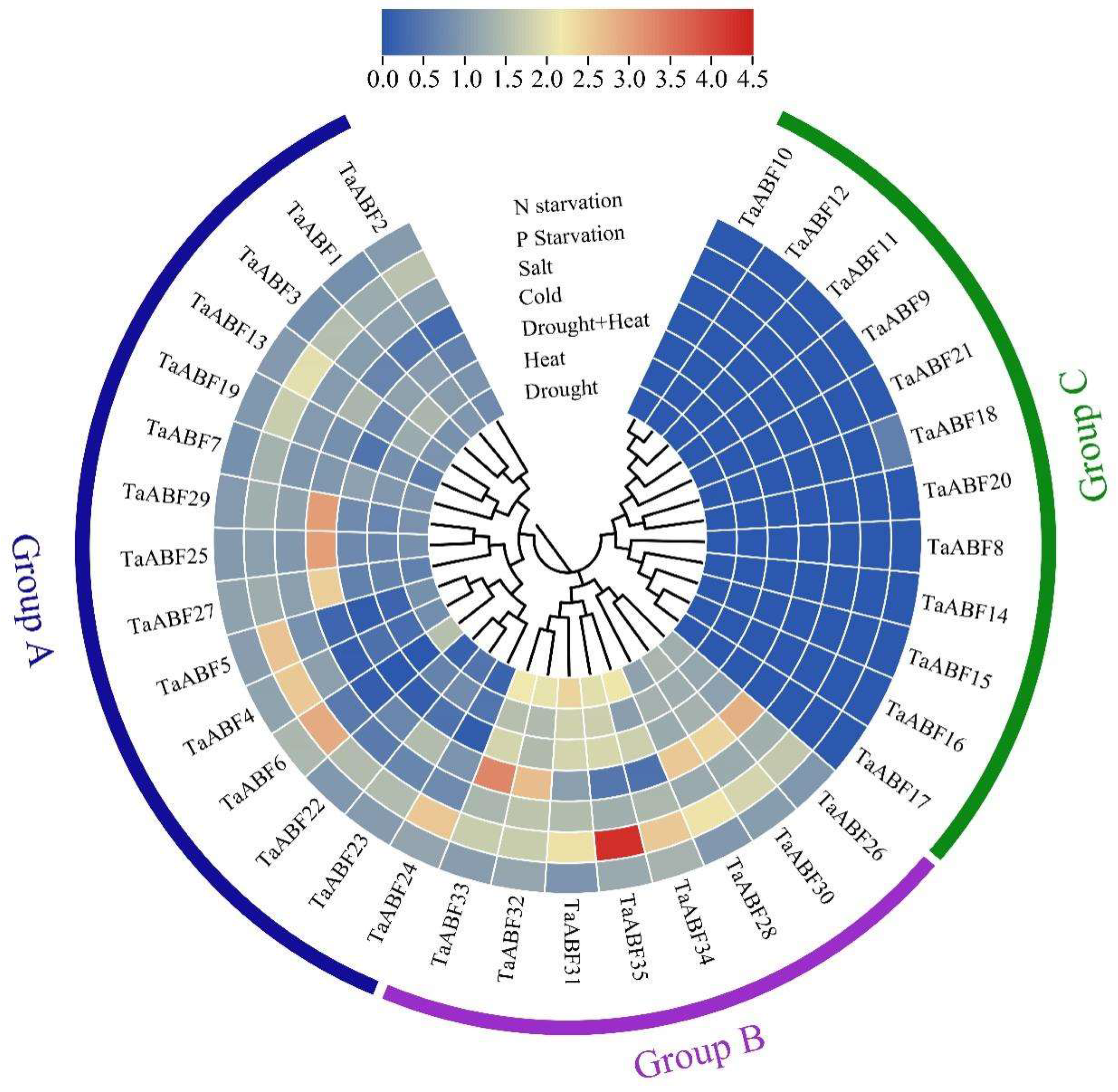
2.6. Analyses of TaABF Gene Expression in Response to Spatial and Abiotic Stressors
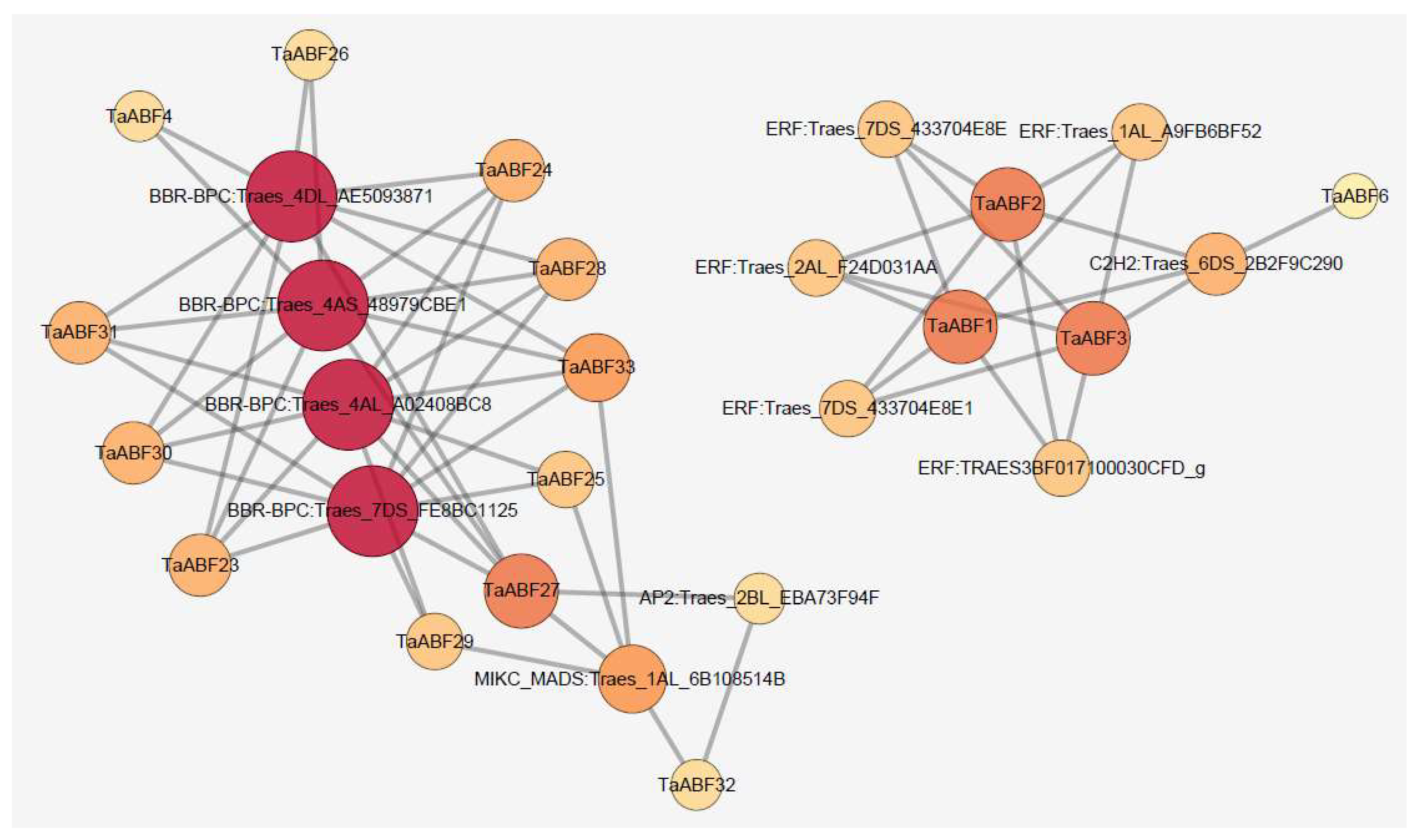
2.7. TaABF Gene Regulatory Networks
3. Discussion
4. Materials and Methods
4.1. Wheat TaABF Family Member Identification
4.2. Phylogenetic Tree, Gene Structure, Conserved Motif, and Cis-Acting Element Analyses
4.3. Chromosomal Localization and Gene Duplication Analyses
4.4. TaABF Gene Expression Profiling
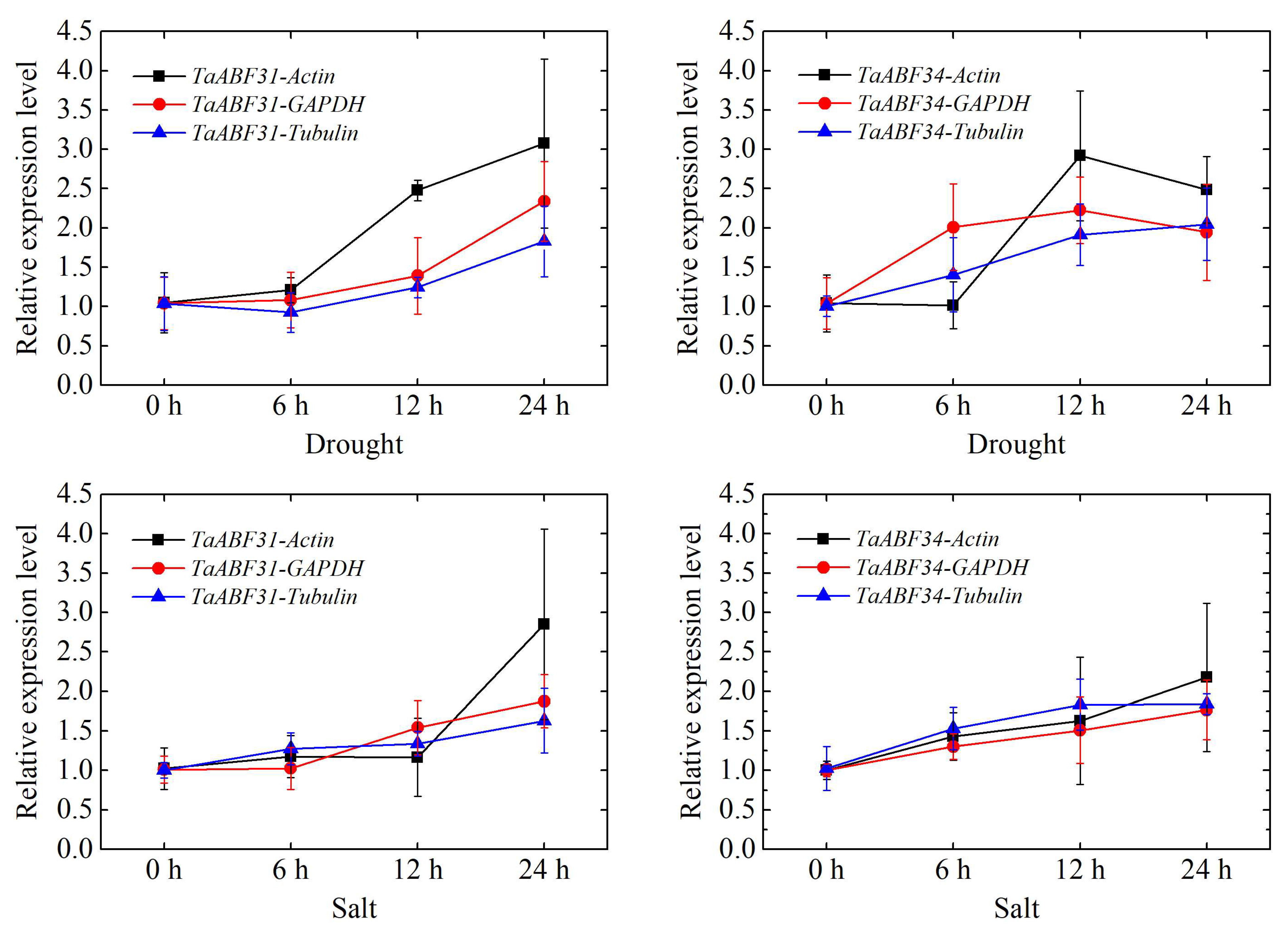
4.5. Verification of TaABF Gene Responses to Salt and Drought Stress
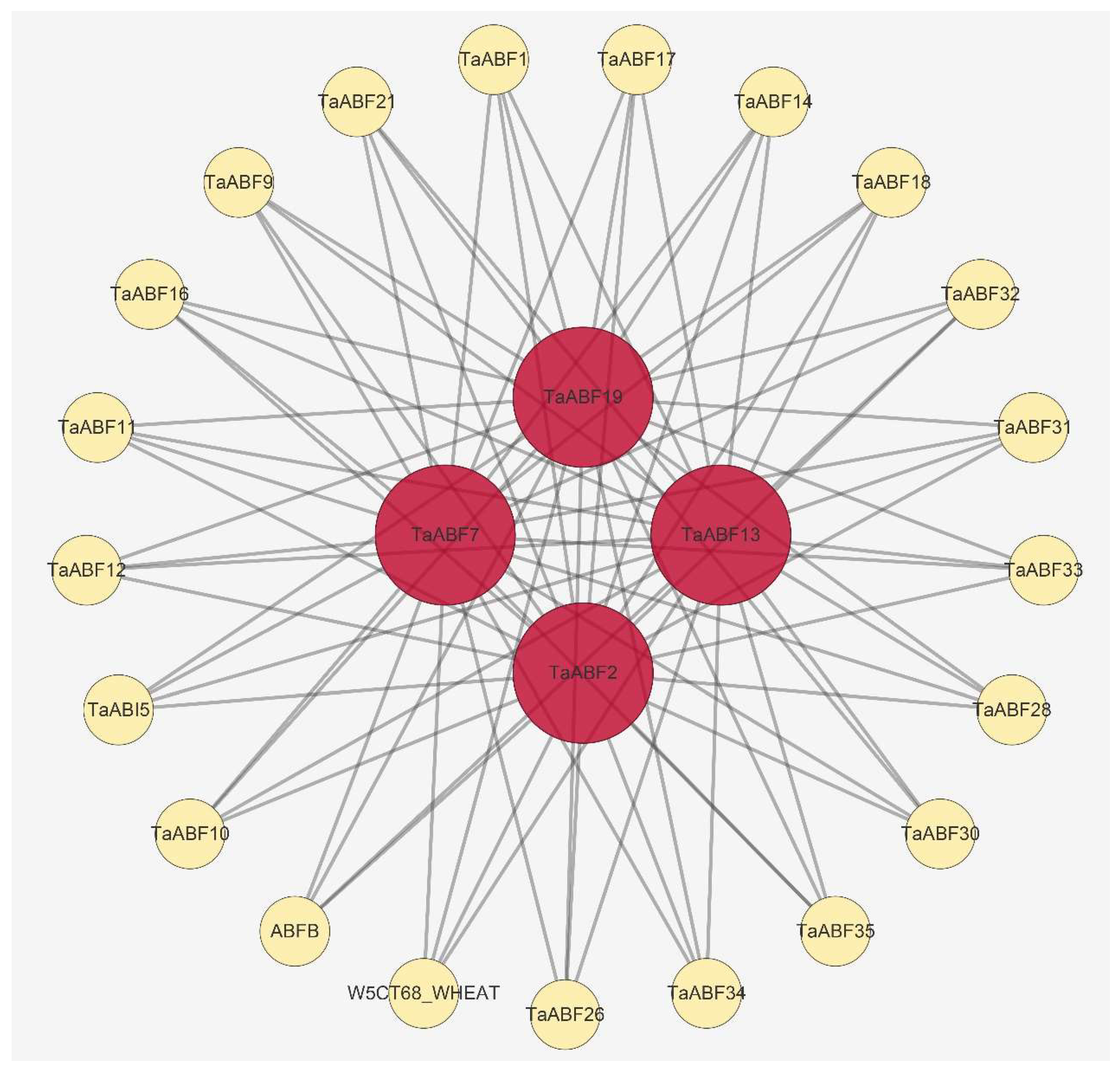
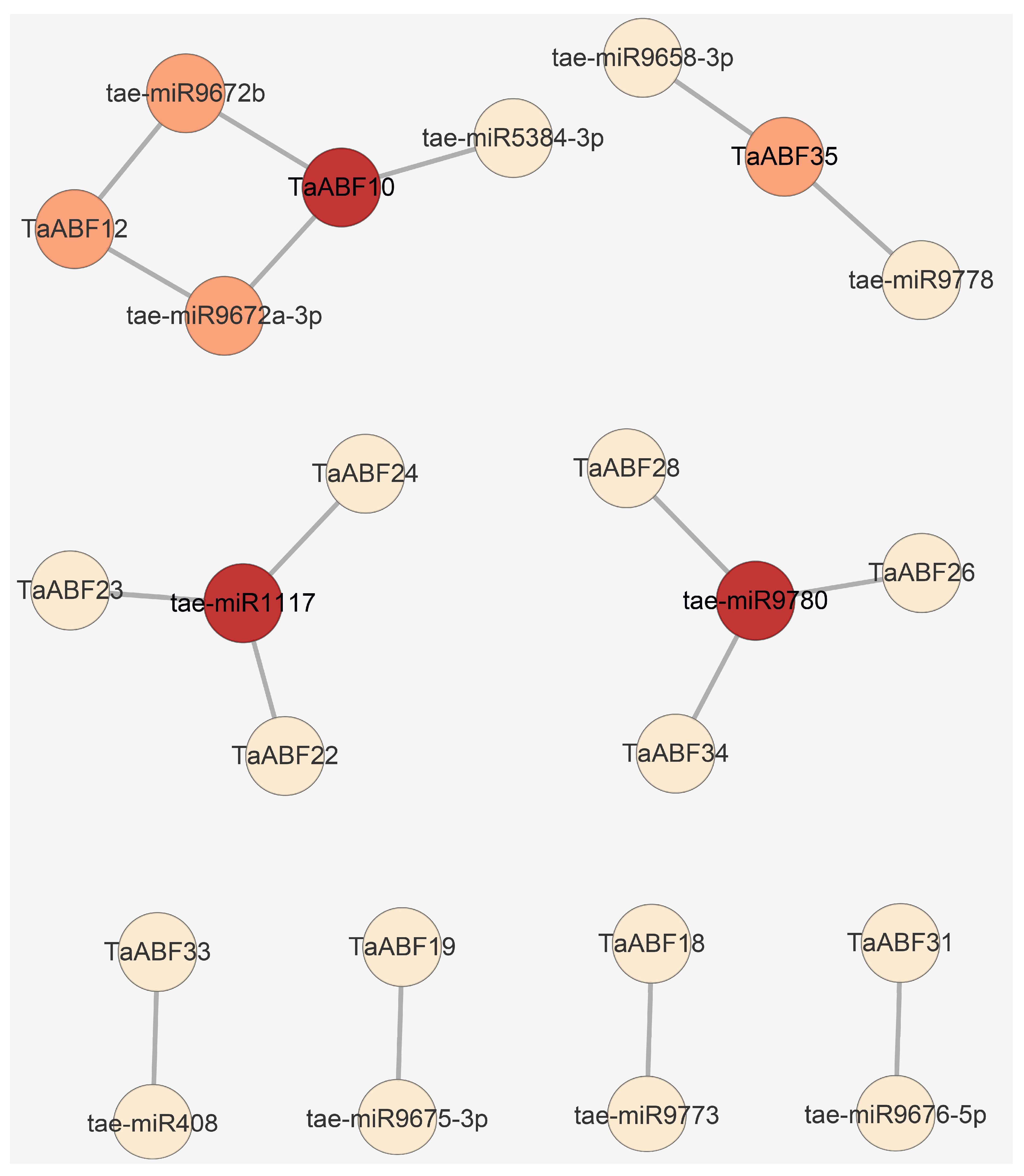
4.6. Protein–Protein Interaction (PPI), miRNA-TaABFs Interaction, and TF Regulatory Network Predictive Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hossain, A.; Skalicky, M.; Brestic, M.; Maitra, S.; Ashraful Alam, M.; Syed, M.A.; Hossain, J.; Sarkar, S.; Saha, S.; Bhadra, P.; et al. Consequences and Mitigation Strategies of Abiotic Stresses in Wheat (Triticum aestivum L.) under the Changing Climate. Agronomy 2021, 11, 241. [Google Scholar] [CrossRef]
- Balfourier, F.; Bouchet, S.; Robert, S.; De Oliveira, R.; Rimbert, H.; Kitt, J.; Choulet, F.; Appels, R.; Feuillet, C.; Keller, B.; et al. Worldwide Phylogeography and History of Wheat Genetic Diversity. Sci. Adv. 2019, 5, eaav0536. [Google Scholar] [CrossRef]
- Shewry, P.R.; Hey, S.J. The Contribution of Wheat to Human Diet and Health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Shen, J.; Yuan, L.; Jiang, R.; Chen, X.; Davies, W.J.; Zhang, F. Improving Crop Productivity and Resource Use Efficiency to Ensure Food Security and Environmental Quality in China. J. Exp. Bot. 2012, 63, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Hossain, M.A. Wheat Production in Changing Environments; Spinger: Dordrecht, The Netherlands, 2019; ISBN 9789811368837. [Google Scholar]
- Rejeb, I.; Pastor, V.; Mauch-Mani, B. Plant Responses to Simultaneous Biotic and Abiotic Stress: Molecular Mechanisms. Plants 2014, 3, 458–475. [Google Scholar] [CrossRef]
- Baloglu, M.C.; Inal, B.; Kavas, M.; Unver, T. Diverse Expression Pattern of Wheat Transcription Factors against Abiotic Stresses in Wheat Species. Gene 2014, 550, 117–122. [Google Scholar] [CrossRef]
- He, Q.; Cai, H.; Bai, M.; Zhang, M.; Chen, F.; Huang, Y.; Priyadarshani, S.V.G.N.; Chai, M.; Liu, L.; Liu, Y.; et al. A Soybean bZIP Transcription Factor GmbZIP19 Confers Multiple Biotic and Abiotic Stress Responses in Plant. Int. J. Mol. Sci. 2020, 21, 4701. [Google Scholar] [CrossRef] [PubMed]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The Leucine Zipper: A Hypothetical Structure Common to a New Class of DNA Binding Proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Hurst, H.C. Transcription Factors. 1: bZIP Proteins. Protein Profile 1994, 1, 123–168. [Google Scholar]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP Transcription Factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Liu, X.; Chu, Z. Genome-Wide Evolutionary Characterization and Analysis of bZIP Transcription Factors and Their Expression Profiles in Response to Multiple Abiotic Stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 227. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP Transcription Factor Family—An Update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Fassler, J.; Landsman, D.; Acharya, A.; Moll, J.R.; Bonovich, M.; Vinson, C. B-ZIP Proteins Encoded by the Drosophila Genome: Evaluation of Potential Dimerization Partners. Genome Res. 2002, 12, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Foster, R.; Chua, N.H. Plant bZIP Protein DNA Binding Specificity. J. Mol. Biol. 1993, 230, 1131–1144. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Qian, Y.; Jiang, M.; Xu, J.; Yang, J.; Zhang, T.; Gou, L.; Pi, E. Regulation Mechanisms of Plant Basic Leucine Zippers to Various Abiotic Stresses. Front. Plant Sci. 2020, 11, 1258. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.X.; Qin, B.; Fang, Q.; Zhang, W.J.; Zhang, Z.Y.; Liu, Y.C.; Li, W.J.; Du, C.; Liu, X.; Zhang, Y.; et al. Genome-Wide Identification, Phylogeny and Expression Analysis of the bZIP Gene Family in Alfalfa (Medicago sativa). Biotechnol. Biotechnol. Equip. 2021, 35, 905–916. [Google Scholar] [CrossRef]
- Pourabed, E.; Ghane Golmohamadi, F.; Soleymani Monfared, P.; Razavi, S.M.; Shobbar, Z.S. Basic Leucine Zipper Family in Barley: Genome-Wide Characterization of Members and Expression Analysis. Mol. Biotechnol. 2015, 57, 12–26. [Google Scholar] [CrossRef]
- Feng, R.J.; Ren, M.Y.; Lu, L.F.; Peng, M.; Guan, X.; Zhou, D.B.; Zhang, M.Y.; Qi, D.F.; Li, K.; Tang, W.; et al. Involvement of Abscisic Acid-Responsive Element-Binding Factors in Cassava (Manihot esculenta) Dehydration Stress Response. Sci. Rep. 2019, 9, 12661. [Google Scholar] [CrossRef]
- Jin, Z.; Xu, W.; Liu, A. Genomic Surveys and Expression Analysis of bZIP Gene Family in Castor Bean (Ricinus communis L.). Planta 2014, 239, 299–312. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and Expression Analyses of Soybean Basic Leucine Zipper Transcription Factor Family. BMC Genom. 2018, 19, 159. [Google Scholar] [CrossRef]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-Wide Analysis of the bZIP Transcription Factors in Cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-Wide Analysis and Expression Profile of the bZIP Transcription Factor Gene Family in Grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cheng, K.; Wan, L.; Yan, L.; Jiang, H.; Liu, S.; Lei, Y.; Liao, B. Genome-Wide Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Six Legume Genomes. BMC Genom. 2015, 16, 1053. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-Wide Analysis of bZIP-Encoding Genes in Maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.; Wu, Z.; Cheng, Z.; Zhang, S.; Liu, H.; Huang, Q. Genome-Wide Identification, Evolutionary Patterns, and Expression Analysis of bZIP Gene Family in Olive (Olea europaea L.). Genes 2020, 11, 510. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic Survey and Gene Expression Analysis of the Basic Leucine Zipper Transcription Factor Family in Rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S.-Y. Genome-Wide Expansion and Expression Divergence of the Basic Leucine Zipper Transcription Factors in Higher Plants with an Emphasis on Sorghum. J. Integr. Plant Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y. The Role of ABF Family bZIP Class Transcription Factors in Stress Response. Physiol. Plant. 2006, 126, 519–527. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 Are Master Transcription Factors That Cooperatively Regulate ABRE-Dependent ABA Signaling Involved in Drought Stress Tolerance and Require ABA for Full Activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Fiallos-Salguero, M.S.; Li, J.; Li, Y.; Xu, J.; Fang, P.; Wang, Y.; Zhang, L.; Tao, A. Identification of AREB/ABF Gene Family Involved in the Response of ABA under Salt and Drought Stresses in Jute (Corchorus olitorius L.). Plants 2023, 12, 1161. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal Role of the AREB/ABF-SnRK2 Pathway in ABRE-mediated Transcription in Response to Osmotic Stress in Plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Zhao, B.Y.; Hu, Y.F.; Li, J.; Yao, X.; Liu, K. BnaABF2, a bZIP Transcription Factor from Rapeseed (Brassica napus L.), Enhances Drought and Salt Tolerance in Transgenic Arabidopsis. Bot. Stud. 2016, 57, 12. [Google Scholar] [CrossRef]
- Muñiz García, M.N.; Giammaria, V.; Grandellis, C.; Téllez-Iñón, M.T.; Ulloa, R.M.; Capiati, D.A. Characterization of StABF1, a Stress-Responsive bZIP Transcription Factor from Solanum tuberosum L. That Is Phosphorylated by StCDPK2 in Vitro. Planta 2012, 235, 761–778. [Google Scholar] [CrossRef]
- Huang, X.S.; Liu, J.H.; Chen, X.J. Overexpression of PtrABF Gene, a bZIP Transcription Factor Isolated from Poncirus Trifoliata, Enhances Dehydration and Drought Tolerance in Tobacco via Scavenging ROS and Modulating Expression of Stress-Responsive Genes. BMC Plant Biol. 2010, 10, 230. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, L.; Xin, M.; Xing, H.; Zhang, Y.; Wu, J.; Xu, D.; Wang, Y.; Shang, Y.; Wei, L.; et al. Overexpression of GhABF3 Increases Cotton (Gossypium hirsutum L.) Tolerance to Salt and Drought. BMC Plant Biol. 2022, 22, 313. [Google Scholar] [CrossRef]
- Liu, J.; Chu, J.; Ma, C.; Jiang, Y.; Ma, Y.; Xiong, J.; Cheng, Z.-M. Overexpression of an ABA-Dependent Grapevine bZIP Transcription Factor, VvABF2, Enhances Osmotic Stress in Arabidopsis. Plant Cell Rep. 2019, 38, 587–596. [Google Scholar] [CrossRef]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.-Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF Transcription Factors Function Predominantly in Gene Expression Downstream of SNRK2 Kinases in Abscisic Acid Signalling in Response to Osmotic Stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef]
- Hossain, M.A.; Cho, J.-I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-Binding bZIP Transcription Factor OsABF2 Is a Positive Regulator of Abiotic Stress and ABA Signaling in Rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef]
- Kerr, T.C.C.; Abdel-Mageed, H.; Aleman, L.; Lee, J.; Payton, P.; Cryer, D.; Allen, R.D. Ectopic Expression of Two AREB/ABF Orthologs Increases Drought Tolerance in Cotton (Gossypium hirsutum). Plant Cell Environ. 2018, 41, 898–907. [Google Scholar] [CrossRef]
- Li, F.; Mei, F.; Zhang, Y.; Li, S.; Kang, Z.; Mao, H. Genome-Wide Analysis of the AREB/ABF Gene Lineage in Land Plants and Functional Analysis of TaABF3 in Arabidopsis. BMC Plant Biol. 2020, 20, 558. [Google Scholar] [CrossRef]
- Bai, J.; Song, M.J.; Gao, J.; Li, G. Whole Genome Duplication and Dispersed Duplication Characterize the Evolution of the Plant PINOID Gene Family across Plant Species. Gene 2022, 829, 146494. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Gao, R.; Luo, Y.; Yun, F.; Wu, X.; Wang, P.; Liao, W. Genome-Wide Identification, Expression Profile, and Alternative Splicing Analysis of CAMTA Family Genes in Cucumber (Cucumis sativus L.). Agronomy 2021, 11, 1827. [Google Scholar] [CrossRef]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a Positive Regulator of ABA Response and Drought Tolerance in Rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef]
- Wang, X.; Guo, C.; Peng, J.; Li, C.; Wan, F.; Zhang, S.; Zhou, Y.; Yan, Y.; Qi, L.; Sun, K.; et al. ABRE—BINDING FACTORS Play a Role in the Feedback Regulation of ABA Signaling by Mediating Rapid ABA Induction of ABA Co-receptor Genes. New Phytol. 2019, 221, 341–355. [Google Scholar] [CrossRef]
- Wang, J.; Li, Q.; Mao, X.; Li, A.; Jing, R. Wheat Transcription Factor TaAREB3 Participates in Drought and Freezing Tolerances in Arabidopsis. Int. J. Biol. Sci. 2016, 12, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and Functional Diversification of MIRNA Genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Jian, C.; Lv, J.; Yan, Y.; Chi, Q.; Li, Z.; Wang, Q.; Zhang, J.; Liu, X.; Zhao, H. Identification and Characterization of microRNAs in the Flag Leaf and Developing Seed of Wheat (Triticum aestivum L.). BMC Genom. 2014, 15, 289. [Google Scholar] [CrossRef]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective Small RNA Destruction by the Expression of a Short Tandem Target Mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef]
- Pegler, J.; Oultram, J.; Grof, C.; Eamens, A. Profiling the Abiotic Stress Responsive microRNA Landscape of Arabidopsis thaliana. Plants 2019, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Hai, B.; Guo, J.; Li, Y.; Zhang, L. Characterization of Wheat miRNAs and Their Target Genes Responsive to Cadmium Stress. Plant Physiol. Biochem. 2016, 101, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.; Gupta, O.P.; Pandey, D.M.; Sharma, I.; Sharma, P. Identification of New Stress-Induced microRNA and Their Targets in Wheat Using Computational Approach. Plant Signal. Behav. 2013, 8, e23932. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Duhan, J.S.; Jaiswal, S.; Angadi, U.B.; Sharma, R.; Raghav, N.; Gupta, O.P.; Sheoran, S.; Sharma, P.; Singh, R.; et al. RNA-Seq Analysis of Developing Grains of Wheat to Intrigue Into the Complex Molecular Mechanism of the Heat Stress Response. Front. Plant Sci. 2022, 13, 904392. [Google Scholar] [CrossRef] [PubMed]
- Fileccia, V.; Ingraffia, R.; Amato, G.; Giambalvo, D.; Martinelli, F. Identification of microRNAS Differentially Regulated by Water Deficit in Relation to Mycorrhizal Treatment in Wheat. Mol. Biol. Rep. 2019, 46, 5163–5174. [Google Scholar] [CrossRef]
- Kneeshaw, S.; Keyani, R.; Delorme-Hinoux, V.; Imrie, L.; Loake, G.J.; Le Bihan, T.; Reichheld, J.P.; Spoel, S.H. Nucleoredoxin Guards against Oxidative Stress by Protecting Antioxidant Enzymes. Proc. Natl. Acad. Sci. USA 2017, 114, 8414–8419. [Google Scholar] [CrossRef]
- Li, J.; Jiao, Z.; He, R.; Sun, Y.; Xu, Q.; Zhang, J.; Jiang, Y.; Li, Q.; Niu, J. Gene Expression Profiles and microRNA Regulation Networks in Tiller Primordia, Stem Tips, and Young Spikes of Wheat Guomai 301. Genes 2019, 10, 686. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Kim, S.Y.; Hyeon, D.Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.H.; Kim, S.K.; Hong, J.C.; et al. The Arabidopsis NAC Transcription Factor ANAC096 Cooperates with bZIP-Type Transcription Factors in Dehydration and Osmotic Stress Responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef]
- Li, X.; Li, X.; Li, M.; Yan, Y.; Liu, X.; Li, L. Dual Function of NAC072 in ABF3-Mediated ABA-Responsive Gene Regulation in Arabidopsis. Front. Plant Sci. 2016, 7, 166111. [Google Scholar] [CrossRef]
- Liu, J.; Shu, D.; Tan, Z.; Ma, M.; Guo, N.; Gao, S.; Duan, G.; Kuai, B.; Hu, Y.; Li, S.; et al. The Arabidopsis IDD14 Transcription Factor Interacts with bZIP-type ABFS/AREBS and Cooperatively Regulates ABA-mediated Drought Tolerance. New Phytol. 2022, 236, 929–942. [Google Scholar] [CrossRef]
- Mu, Y.; Liu, Y.; Bai, L.; Li, S.; He, C.; Yan, Y.; Yu, X.; Li, Y. Cucumber CsBPCs Regulate the Expression of CsABI3 during Seed Germination. Front. Plant Sci. 2017, 8, 250645. [Google Scholar] [CrossRef] [PubMed]
- Gong, R.; Cao, H.; Zhang, J.; Xie, K.; Wang, D.; Yu, S. Divergent Functions of the GAGA-binding Transcription Factor Family in Rice. Plant J. 2018, 94, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Theune, M.L.; Bloss, U.; Brand, L.H.; Ladwig, F.; Wanke, D. Phylogenetic Analyses and GAGA-Motif Binding Studies of BBR/BPC Proteins Lend to Clues in GAGA-Motif Recognition and a Regulatory Role in Brassinosteroid Signaling. Front. Plant Sci. 2019, 10, 466. [Google Scholar] [CrossRef]
- Wu, J.; Mohamed, D.; Dowhanik, S.; Petrella, R.; Gregis, V.; Li, J.; Wu, L.; Gazzarrini, S. Spatiotemporal Restriction of FUSCA3 Expression by Class I BPCs Promotes Ovule Development and Coordinates Embryo and Endosperm Growth. Plant Cell 2020, 32, 1886–1904. [Google Scholar] [CrossRef] [PubMed]
- Sahu, A.; Singh, R.; Verma, P.K. Plant BBR/BPC Transcription Factors: Unlocking Multilayered Regulation in Development, Stress and Immunity. Planta 2023, 258, 31. [Google Scholar] [CrossRef] [PubMed]
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-Binding Factor Basic pentacysteine6 Recruits the polycomb-repressive complex1 Component like heterochromatin protein1 to GAGA DNA motifs. Plant Physiol. 2015, 168, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Liu, Y.; Yang, L.; He, H.; Huang, Y.; Fang, L.; Scheller, H.V.; Jiang, M.; Zhang, A. Cell Wall β-1,4-Galactan Regulated by the BPC1/BPC2-GALS1 Module Aggravates Salt Sensitivity in Arabidopsis thaliana. Mol. Plant 2021, 14, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, M.; Fang, L. BASIC PENTACYSTEINE2 Negatively Regulates Osmotic Stress Tolerance by Modulating LEA4-5 Expression in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 168, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, X.; Zhang, J.; Zhao, H.; Tan, S.; Xu, W.; Pan, J.; Yang, F.; Pi, E. ERF Subfamily Transcription Factors and Their Function in Plant Responses to Abiotic Stresses. Front. Plant Sci. 2022, 13, 1042084. [Google Scholar] [CrossRef]
- Park, S.I.; Kwon, H.J.; Cho, M.H.; Song, J.S.; Kim, B.G.; Baek, J.; Kim, S.L.; Ji, H.; Kwon, T.R.; Kim, K.H.; et al. The OsERF115/AP2EREBP110 Transcription Factor Is Involved in the Multiple Stress Tolerance to Heat and Drought in Rice Plants. Int. J. Mol. Sci. 2021, 22, 7181. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Wang, Y.; Chory, J. Two Interacting Ethylene Response Factors Regulate Heat Stress Response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of Rice Transcription Factor OsERF19 in Response to ABA and Salt Stress Responses. Plant Physiol. Biochem. 2021, 167, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhu, Z.; Zhang, L.; Fang, L.; Zhang, J.; Wang, Q.; Li, S.; Liang, Z.; Xin, H. Overexpression of Ethylene Response Factors VaERF080 and VaERF087 from Vitis amurensis Enhances Cold Tolerance in Arabidopsis. Sci. Hortic. 2019, 243, 320–326. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Yang, C.; Cheng, Y.; Cai, Z.; Nian, H.; Ma, Q. GsERF1 Enhances Arabidopsis thaliana Aluminum Tolerance through an Ethylene-Mediated Pathway. BMC Plant Biol. 2022, 22, 258. [Google Scholar] [CrossRef]
- Sun, Y.; Li, J.Q.; Yan, J.Y.; Yuan, J.J.; Li, G.X.; Wu, Y.R.; Xu, J.M.; Huang, R.F.; Harberd, N.P.; Ding, Z.J.; et al. Ethylene Promotes Seed Iron Storage during Arabidopsis Seed Maturation via ERF95 Transcription Factor. J. Integr. Plant Biol. 2020, 62, 1193–1212. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A Parallel Tool for Constructing Multiple Protein-Coding DNA Alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks Ratio: Diagnosing the Form of Sequence Evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Sun, X.; Wu, G.; He, X.; Liu, W.; Wang, Y.; Sun, Q.; Zhao, Y.; Xu, D.; Dai, X.; et al. Genome-Wide Identification and Expression Profiling of the ABF Transcription Factor Family in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2024, 25, 3783. https://doi.org/10.3390/ijms25073783
Yang F, Sun X, Wu G, He X, Liu W, Wang Y, Sun Q, Zhao Y, Xu D, Dai X, et al. Genome-Wide Identification and Expression Profiling of the ABF Transcription Factor Family in Wheat (Triticum aestivum L.). International Journal of Molecular Sciences. 2024; 25(7):3783. https://doi.org/10.3390/ijms25073783
Chicago/Turabian StyleYang, Fuhui, Xuelian Sun, Gang Wu, Xiaoyan He, Wenxing Liu, Yongmei Wang, Qingyi Sun, Yan Zhao, Dengan Xu, Xuehuan Dai, and et al. 2024. "Genome-Wide Identification and Expression Profiling of the ABF Transcription Factor Family in Wheat (Triticum aestivum L.)" International Journal of Molecular Sciences 25, no. 7: 3783. https://doi.org/10.3390/ijms25073783
APA StyleYang, F., Sun, X., Wu, G., He, X., Liu, W., Wang, Y., Sun, Q., Zhao, Y., Xu, D., Dai, X., Ma, W., & Zeng, J. (2024). Genome-Wide Identification and Expression Profiling of the ABF Transcription Factor Family in Wheat (Triticum aestivum L.). International Journal of Molecular Sciences, 25(7), 3783. https://doi.org/10.3390/ijms25073783





