Applications of Flow Cytometry in Drug Discovery and Translational Research
Abstract
:1. Introduction
2. Hit Identification and Lead Optimization
3. Translational Research Informing the Path to the Clinic
4. Quantitative Pharmacokinetic/Pharmacodynamic Evaluation
5. Limitations and Opportunities for Flow Cytometric Techniques to Inform Early Clinical Decision-Making
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McKinnon, K.M. Flow Cytometry: An Overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, M.H.; Nolan, G.P. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, D.L.; Reinin, G.; Chua, E. Full Spectrum Flow Cytometry as a Powerful Technology for Cancer Immunotherapy Research. Front. Mol. Biosci. 2021, 7, 612801. [Google Scholar] [CrossRef] [PubMed]
- Rees, P.; Summers, H.D.; Filby, A.; Carpenter, A.E.; Doan, M. Imaging Flow Cytometry. Nat. Rev. Methods Prim. 2022, 2, 86. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.; Hamers, A.A.J.; Pillai, A.B. CyTOF® for the Masses. Front. Immunol. 2022, 13, 815828. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, P.; Xu, J.; Shao, W.; Yang, C.; Cui, Y. Hydrodynamic Flow Cytometer Performance Enhancement by Two-Dimensional Acoustic Focusing. Biomed. Microdevices 2020, 22, 27. [Google Scholar] [CrossRef] [PubMed]
- Tuijnenburg, P.; de Kerk, D.J.A.; Jansen, M.H.; Morris, B.; Lieftink, C.; Beijersbergen, R.L.; van Leeuwen, E.M.; Kuijpers, T.W. High-throughput Compound Screen Reveals MTOR Inhibitors as Potential Therapeutics to Reduce (Auto)Antibody Production by Human Plasma Cells. Eur. J. Immunol. 2020, 50, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Su, Y.; Kang, B.H.; Fan, Z.; Dong, T.; Brown, D.R.; Cheah, J.; Wittrup, K.D.; Chen, J. High-Throughput Phenotypic Screen and Transcriptional Analysis Identify New Compounds and Targets for Macrophage Reprogramming. Nat. Commun. 2021, 12, 773. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Brengdahl, J.; Lindqvist, M.; Gehrmann, U.; Ericson, E.; von Berg, S.; Ripa, L.; Malhotra, R. A Phenotypic Screening Approach Using Human Treg Cells Identified Regulators of Forkhead Box P3 Expression. ACS Chem. Biol. 2019, 14, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Buranda, T.; Gineste, C.; Wu, Y.; Bondu, V.; Perez, D.; Lake, K.R.; Edwards, B.S.; Sklar, L.A. A High-Throughput Flow Cytometry Screen Identifies Molecules That Inhibit Hantavirus Cell Entry. SLAS Discov. 2018, 23, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Schardt, J.S.; Pornnoppadol, G.; Desai, A.A.; Park, K.S.; Zupancic, J.M.; Makowski, E.K.; Smith, M.D.; Chen, H.; Barbosa, M.G.d.M.; Cascalho, M.; et al. Discovery and Characterization of High-Affinity, Potent SARS-CoV-2 Neutralizing Antibodies via Single B Cell Screening. Sci. Rep. 2021, 11, 20738. [Google Scholar] [CrossRef] [PubMed]
- Joslin, J.; Gilligan, J.; Anderson, P.; Garcia, C.; Sharif, O.; Hampton, J.; Cohen, S.; King, M.; Zhou, B.; Jiang, S.; et al. A Fully Automated High-Throughput Flow Cytometry Screening System Enabling Phenotypic Drug Discovery. SLAS Discov. 2018, 23, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Goenaga, A.-L.; Harms, B.D.; Zou, H.; Lou, J.; Conrad, F.; Adams, G.P.; Schoeberl, B.; Nielsen, U.B.; Marks, J.D. Impact of Intrinsic Affinity on Functional Binding and Biological Activity of EGFR Antibodies. Mol. Cancer Ther. 2012, 11, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Floc’h, A.L.; Allinne, J.; Nagashima, K.; Scott, G.; Birchard, D.; Asrat, S.; Bai, Y.; Lim, W.K.; Martin, J.; Huang, T.; et al. Dual Blockade of IL-4 and IL-13 with Dupilumab, an IL-4Rα Antibody, Is Required to Broadly Inhibit Type 2 Inflammation. Allergy 2020, 75, 1188–1204. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, K.; Georgiev, P.; Wells, S.; Xu, H.; Lacey, B.M.; Xu, Z.; Laskey, J.; Mcleod, R.; Methot, J.L.; et al. Pharmacological Inhibition of Hematopoietic Progenitor Kinase 1 Positively Regulates T-Cell Function. PLoS ONE 2020, 15, e0243145. [Google Scholar] [CrossRef] [PubMed]
- Linnane, E.; Davey, P.; Zhang, P.; Puri, S.; Edbrooke, M.; Chiarparin, E.; Revenko, A.S.; Macleod, A.R.; Norman, J.C.; Ross, S.J. Differential Uptake, Kinetics and Mechanisms of Intracellular Trafficking of next-Generation Antisense Oligonucleotides across Human Cancer Cell Lines. Nucleic Acids Res. 2019, 47, 4375–4392. [Google Scholar] [CrossRef] [PubMed]
- Revenko, A.; Carnevalli, L.S.; Sinclair, C.; Johnson, B.; Peter, A.; Taylor, M.; Hettrick, L.; Chapman, M.; Klein, S.; Solanki, A.; et al. Direct Targeting of FOXP3 in Tregs with AZD8701, a Novel Antisense Oligonucleotide to Relieve Immunosuppression in Cancer. J. Immunother. Cancer 2022, 10, e003892. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.Y.; Milton, M.N. The Determination and Interpretation of the Therapeutic Index in Drug Development. Nat. Rev. Drug Discov. 2012, 11, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Guo, C.; Li, Y.; Liu, K.; Zhao, Q.; Ouyang, L. Identification of Claudin-6 as a Molecular Biomarker in Pan-Cancer Through Multiple Omics Integrative Analysis. Front. Cell Dev. Biol. 2021, 9, 726656. [Google Scholar] [CrossRef]
- Screnci, B.; Stafford, L.J.; Barnes, T.; Shema, K.; Gilman, S.; Wright, R.; Absi, S.A.; Phillips, T.; Azuelos, C.; Slovik, K.; et al. Antibody Specificity against Highly Conserved Membrane Protein Claudin 6 Driven by Single Atomic Contact Point. iScience 2022, 25, 105665. [Google Scholar] [CrossRef]
- McDermott, M.S.J.; O’Brien, N.A.; Hoffstrom, B.; Gong, K.; Lu, M.; Zhang, J.; Luo, T.; Liang, M.; Jia, W.; Hong, J.; et al. Preclinical Efficacy of the Antibody-Drug-Conjugate CLDN6-23-ADC for the Treatment of CLDN6 Positive Solid Tumors. Clin. Cancer Res. 2023, 29, 2131–2143. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Properties of FDA-Approved Small Molecule Protein Kinase Inhibitors: A 2023 Update. Pharmacol. Res. 2023, 187, 106552. [Google Scholar] [CrossRef] [PubMed]
- Bendels, S.; Bissantz, C.; Fasching, B.; Gerebtzoff, G.; Guba, W.; Kansy, M.; Migeon, J.; Mohr, S.; Peters, J.-U.; Tillier, F.; et al. Safety Screening in Early Drug Discovery: An Optimized Assay Panel. J. Pharmacol. Toxicol. Methods 2019, 99, 106609. [Google Scholar] [CrossRef] [PubMed]
- Bowes, J.; Brown, A.J.; Hamon, J.; Jarolimek, W.; Sridhar, A.; Waldron, G.; Whitebread, S. Reducing Safety-Related Drug Attrition: The Use of in Vitro Pharmacological Profiling. Nat. Rev. Drug Discov. 2012, 11, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.; Rhedin, M.; Hendrickx, R.; Berglund, S.; Piras, A.; Blomgran, P.; Cavallin, A.; Collins, M.; Dahl, G.; Dekkak, B.; et al. Characterization of Selective and Potent JAK1 Inhibitors Intended for the Inhaled Treatment of Asthma. Drug Des. Dev. Ther. 2022, 16, 2901–2917. [Google Scholar] [CrossRef] [PubMed]
- Jain, T.; Sun, T.; Durand, S.; Hall, A.; Houston, N.R.; Nett, J.H.; Sharkey, B.; Bobrowicz, B.; Caffry, I.; Yu, Y.; et al. Biophysical Properties of the Clinical-Stage Antibody Landscape. Proc. Natl. Acad. Sci. USA 2017, 114, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Makowski, E.K.; Wu, L.; Desai, A.A.; Tessier, P.M. Highly Sensitive Detection of Antibody Nonspecific Interactions Using Flow Cytometry. mAbs 2021, 13, 1951426. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, M.E.; Patfield, S.; Hughes, A.C.; Hernlem, B.; He, X. A Novel Shiga Toxin 2a Neutralizing Antibody Therapeutic with Low Immunogenicity and High Efficacy. Antimicrob. Agents Chemother. 2024, 68, e0059823. [Google Scholar] [CrossRef] [PubMed]
- Roopenian, D.C.; Akilesh, S. FcRn: The Neonatal Fc Receptor Comes of Age. Nat. Rev. Immunol. 2007, 7, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Reynolds, H.M.; Lumicisi, B.; Bryson, C.J. Immunogenicity of Protein Therapeutics: The Key Causes, Consequences and Challenges. SelfNonself 2010, 1, 314–322. [Google Scholar] [CrossRef]
- Ko, S.; Park, S.; Sohn, M.H.; Jo, M.; Ko, B.J.; Na, J.-H.; Yoo, H.; Jeong, A.L.; Ha, K.; Woo, J.R.; et al. An Fc Variant with Two Mutations Confers Prolonged Serum Half-Life and Enhanced Effector Functions on IgG Antibodies. Exp. Mol. Med. 2022, 54, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Mandrup, O.A.; Ong, S.C.; Lykkemark, S.; Dinesen, A.; Rudnik-Jansen, I.; Dagnæs-Hansen, N.F.; Andersen, J.T.; Alvarez-Vallina, L.; Howard, K.A. Programmable Half-Life and Anti-Tumour Effects of Bispecific T-Cell Engager-Albumin Fusions with Tuned FcRn Affinity. Commun. Biol. 2021, 4, 310. [Google Scholar] [CrossRef]
- Nath, N.; Godat, B.; Zimprich, C.; Dwight, S.J.; Corona, C.; McDougall, M.; Urh, M. Homogeneous Plate Based Antibody Internalization Assay Using PH Sensor Fluorescent Dye. J. Immunol. Methods 2016, 431, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Parameswaran, N.; Luo, L.; Zhang, L.; Chen, J.; DiFilippo, F.P.; Androjna, C.; Fox, D.A.; Ondrejka, S.L.; Hsi, E.D.; Jagadeesh, D.; et al. CD6-Targeted Antibody-Drug Conjugate as a New Therapeutic Agent for T Cell Lymphoma. Leukemia 2023, 37, 2050–2057. [Google Scholar] [CrossRef]
- Yang, M.-C.; Shia, C.-S.; Li, W.-F.; Wang, C.-C.; Chen, I.-J.; Huang, T.-Y.; Chen, Y.-J.; Chang, H.-W.; Lu, C.-H.; Wu, Y.-C.; et al. Preclinical Studies of OBI-999: A Novel Globo H–Targeting Antibody–Drug Conjugate. Mol. Cancer Ther. 2021, 20, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Phanse, Y.; Ramer-Tait, A.E.; Friend, S.L.; Carrillo-Conde, B.; Lueth, P.; Oster, C.J.; Phillips, G.J.; Narasimhan, B.; Wannemuehler, M.J.; Bellaire, B.H. Analyzing Cellular Internalization of Nanoparticles and Bacteria by Multi-Spectral Imaging Flow Cytometry. J. Vis. Exp. 2012, 64, e3884. [Google Scholar] [CrossRef]
- Sharma, S.; Li, Z.; Bussing, D.; Shah, D.K. Evaluation of Quantitative Relationship Between Target Expression and ADC Exposure Inside Cancer Cells. Drug Metab. Dispos. 2020, 48, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Li, S.; Han, S.; Shi, C.; Zhang, Y. Antibody Drug Conjugate: The “Biological Missile” for Targeted Cancer Therapy. Signal Transduct. Target. Ther. 2022, 7, 93. [Google Scholar] [CrossRef]
- Kopp, A.; Hofsess, S.; Cardillo, T.M.; Govindan, S.V.; Donnell, J.; Thurber, G.M. Antibody–Drug Conjugate Sacituzumab Govitecan Drives Efficient Tissue Penetration and Rapid Intracellular Drug Release. Mol. Cancer Ther. 2022, 22, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Sharma, S.; Shah, D.K. Quantitative Characterization of in Vitro Bystander Effect of Antibody-Drug Conjugates. J. Pharmacokinet. Pharmacodyn. 2016, 43, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Perry, S.R.; Muniz-Medina, V.; Wang, X.; Wetzel, L.K.; Rebelatto, M.C.; Hinrichs, M.J.M.; Bezabeh, B.Z.; Fleming, R.L.; Dimasi, N.; et al. A Biparatopic HER2-Targeting Antibody-Drug Conjugate Induces Tumor Regression in Primary Models Refractory to or Ineligible for HER2-Targeted Therapy. Cancer Cell 2016, 29, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Maio, M.; Grob, J.-J.; Aamdal, S.; Bondarenko, I.; Robert, C.; Thomas, L.; Garbe, C.; Chiarion-Sileni, V.; Testori, A.; Chen, T.-T.; et al. Five-Year Survival Rates for Treatment-Naive Patients with Advanced Melanoma Who Received Ipilimumab Plus Dacarbazine in a Phase III Trial. J. Clin. Oncol. 2015, 33, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Mosely, S.I.S.; Prime, J.E.; Sainson, R.C.A.; Koopmann, J.-O.; Wang, D.Y.Q.; Greenawalt, D.M.; Ahdesmaki, M.J.; Leyland, R.; Mullins, S.; Pacelli, L.; et al. Rational Selection of Syngeneic Preclinical Tumor Models for Immunotherapeutic Drug Discovery. Cancer Immunol. Res. 2017, 5, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Dovedi, S.J.; Elder, M.J.; Yang, C.; Sitnikova, S.I.; Irving, L.; Hansen, A.; Hair, J.; Jones, D.C.; Hasani, S.; Wang, B.; et al. Design and Efficacy of a Monovalent Bispecific PD-1/CTLA4 Antibody That Enhances CTLA4 Blockade on PD-1+ Activated T Cells. Cancer Discov. 2021, 11, 1100–1117. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.A.; Hughes, A.M.; Walton, J.; Coenen-Stass, A.M.L.; Magiera, L.; Mooney, L.; Bell, S.; Staniszewska, A.D.; Sandin, L.C.; Barry, S.T.; et al. Longitudinal Immune Characterization of Syngeneic Tumor Models to Enable Model Selection for Immune Oncology Drug Discovery. J. Immunother. Cancer 2019, 7, 328. [Google Scholar] [CrossRef]
- Leyland, R.; Watkins, A.; Mulgrew, K.A.; Holoweckyj, N.; Bamber, L.; Tigue, N.J.; Offer, E.; Andrews, J.; Yan, L.; Mullins, S.; et al. A Novel Murine GITR Ligand Fusion Protein Induces Antitumor Activity as a Monotherapy That Is Further Enhanced in Combination with an OX40 Agonist. Clin. Cancer Res. 2017, 23, 3416–3427. [Google Scholar] [CrossRef] [PubMed]
- Borodovsky, A.; Barbon, C.M.; Wang, Y.; Ye, M.; Prickett, L.; Chandra, D.; Shaw, J.; Deng, N.; Sachsenmeier, K.; Clarke, J.D.; et al. Small Molecule AZD4635 Inhibitor of A2AR Signaling Rescues Immune Cell Function Including CD103+ Dendritic Cells Enhancing Anti-Tumor Immunity. J. Immunother. Cancer 2020, 8, e000417. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.D.; Pollizzi, K.N.; Heikamp, E.B.; Horton, M.R. Regulation of Immune Responses by MTOR. Annu. Rev. Immunol. 2012, 30, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Langdon, S.; Hughes, A.; Taylor, M.A.; Kuczynski, E.A.; Mele, D.A.; Delpuech, O.; Jarvis, L.; Staniszewska, A.; Cosulich, S.; Carnevalli, L.S.; et al. Combination of Dual MTORC1/2 Inhibition and Immune-Checkpoint Blockade Potentiates Anti-Tumour Immunity. OncoImmunology 2018, 7, e1458810. [Google Scholar] [CrossRef] [PubMed]
- Carnevalli, L.S.; Sinclair, C.; Taylor, M.A.; Gutierrez, P.M.; Langdon, S.; Coenen-Stass, A.M.L.; Mooney, L.; Hughes, A.; Jarvis, L.; Staniszewska, A.; et al. PI3Kα/δ Inhibition Promotes Anti-Tumor Immunity through Direct Enhancement of Effector CD8+ T-Cell Activity. J. Immunother. Cancer 2018, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Sceneay, J.; Sinclair, C. The Future of Immune Checkpoint Combinations with Tumor-Targeted Small Molecule Drugs. Emerg. Top. Life Sci. 2021, 5, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Wichroski, M.; Benci, J.; Liu, S.-Q.; Chupak, L.; Fang, J.; Cao, C.; Wang, C.; Onorato, J.; Qiu, H.; Shan, Y.; et al. DGKα/ζ Inhibitors Combine with PD-1 Checkpoint Therapy to Promote T Cell–Mediated Antitumor Immunity. Sci. Transl. Med. 2023, 15, eadh1892. [Google Scholar] [CrossRef] [PubMed]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Campesato, L.F.; Budhu, S.; Tchaicha, J.; Weng, C.-H.; Gigoux, M.; Cohen, I.J.; Redmond, D.; Mangarin, L.; Pourpe, S.; Liu, C.; et al. Blockade of the AHR Restricts a Treg-Macrophage Suppressive Axis Induced by L-Kynurenine. Nat. Commun. 2020, 11, 4011. [Google Scholar] [CrossRef] [PubMed]
- McGovern, K.; Castro, A.C.; Cavanaugh, J.; Coma, S.; Walsh, M.; Tchaicha, J.; Syed, S.; Natarajan, P.; Manfredi, M.; Zhang, X.M.; et al. Discovery and Characterization of a Novel Aryl Hydrocarbon Receptor Inhibitor, IK-175, and Its Inhibitory Activity on Tumor Immune Suppression. Mol. Cancer Ther. 2022, 21, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, E.A.; Carnevalli, L.; Sinclair, C. Longitudinal Tracking of T Cell Lymphomas in Mice Using Flow Cytometry. STAR Protoc. 2023, 4, 102144. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, E.A.; Morlino, G.; Peter, A.; Coenen-Stass, A.M.L.; Moss, J.I.; Wali, N.; Delpuech, O.; Reddy, A.; Solanki, A.; Sinclair, C.; et al. A Preclinical Model of Peripheral T-cell Lymphoma GATA3 Reveals DNA Damage Response Pathway Vulnerability. EMBO Mol. Med. 2022, 14, e15816. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, X.; Lavezzi, S.M.; Arshad, U.; Sharma, R. Concept and Application of the Probability of Pharmacological Success (PoPS) as a Decision Tool in Drug Development: A Position Paper. J. Transl. Med. 2023, 21, 17. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, C.K.; Ebrahimi-Nik, H.; Iracheta-Vellve, A.; Hamel, K.M.; Olander, K.E.; Davis, T.G.R.; McGuire, K.A.; Halvorsen, G.T.; Avila, O.I.; Patel, C.H.; et al. The PTPN2/PTPN1 Inhibitor ABBV-CLS-484 Unleashes Potent Anti-Tumour Immunity. Nature 2023, 622, 850–862. [Google Scholar] [CrossRef] [PubMed]
- Casey, K.A.; Guo, X.; Smith, M.A.; Wang, S.; Sinibaldi, D.; Sanjuan, M.A.; Wang, L.; Illei, G.G.; White, W.I. Type I Interferon Receptor Blockade with Anifrolumab Corrects Innate and Adaptive Immune Perturbations of SLE. Lupus Sci. Med. 2018, 5, e000286. [Google Scholar] [CrossRef] [PubMed]
- Kasturi, S.P.; Kozlowski, P.A.; Nakaya, H.I.; Burger, M.C.; Russo, P.; Pham, M.; Kovalenkov, Y.; Silveira, E.L.V.; Havenar-Daughton, C.; Burton, S.L.; et al. Adjuvanting a Simian Immunodeficiency Virus Vaccine with Toll-Like Receptor Ligands Encapsulated in Nanoparticles Induces Persistent Antibody Responses and Enhanced Protection in TRIM5α Restrictive Macaques. J. Virol. 2017, 91, e01844-16. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.S.; Scott, M.K.D.; Hagan, T.; Li, C.; Feng, Y.; Wimmers, F.; Grigoryan, L.; Trisal, M.; Edara, V.V.; Lai, L.; et al. Systems Vaccinology of the BNT162b2 MRNA Vaccine in Humans. Nature 2021, 596, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Rosa, S.C.D. Vaccine Applications of Flow Cytometry. Methods 2012, 57, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Kumari, D.; Singh, S.; Kumari, M.; Gupta, H.; Chauhan, D.; Singh, K.; Eslavath, M.R.; Bhushan, B.; Dogra, V.; Bargotya, M.; et al. Flow Cytometry Profiling of Cellular Immune Response in COVID-19 Infected, Recovered and Vaccinated Individuals. Immunobiology 2023, 228, 152392. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B. Learning Immunology from the Yellow Fever Vaccine: Innate Immunity to Systems Vaccinology. Nat. Rev. Immunol. 2009, 9, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Van de Sande, B.; Lee, J.S.; Mutasa-Gottgens, E.; Naughton, B.; Bacon, W.; Manning, J.; Wang, Y.; Pollard, J.; Mendez, M.; Hill, J.; et al. Applications of Single-Cell RNA Sequencing in Drug Discovery and Development. Nat. Rev. Drug Discov. 2023, 22, 496–520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Z.; Skrzypczynska, K.M.; Fang, Q.; Zhang, W.; O’Brien, S.A.; He, Y.; Wang, L.; Zhang, Q.; Kim, A.; et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 2020, 181, 442–459.e29. [Google Scholar] [CrossRef] [PubMed]
- Martini, E.; Kunderfranco, P.; Peano, C.; Carullo, P.; Cremonesi, M.; Schorn, T.; Carriero, R.; Termanini, A.; Colombo, F.S.; Jachetti, E.; et al. Single-Cell Sequencing of Mouse Heart Immune Infiltrate in Pressure Overload–Driven Heart Failure Reveals Extent of Immune Activation. Circulation 2019, 140, 2089–2107. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Netto, K.G.; Zhou, L.; Liu, X.; Wang, M.; Zhang, G.; Foster, P.S.; Li, F.; Yang, M. Single-Cell Transcriptomic Analysis Reveals the Immune Landscape of Lung in Steroid-Resistant Asthma Exacerbation. Proc. Natl. Acad. Sci. USA 2021, 118, e2005590118. [Google Scholar] [CrossRef] [PubMed]
- Carraro, G.; Langerman, J.; Sabri, S.; Lorenzana, Z.; Purkayastha, A.; Zhang, G.; Konda, B.; Aros, C.J.; Calvert, B.A.; Szymaniak, A.; et al. Transcriptional Analysis of Cystic Fibrosis Airways at Single-Cell Resolution Reveals Altered Epithelial Cell States and Composition. Nat. Med. 2021, 27, 806–814. [Google Scholar] [CrossRef]
- Skånland, S.S. Phospho Flow Cytometry with Fluorescent Cell Barcoding for Single Cell Signaling Analysis and Biomarker Discovery. J. Vis. Exp. 2018, 140, e58386. [Google Scholar] [CrossRef]
- Sinclair, C.; Bains, I.; Yates, A.J.; Seddon, B. Asymmetric Thymocyte Death Underlies the CD4:CD8 T-Cell Ratio in the Adaptive Immune System. Proc. Natl. Acad. Sci. USA 2013, 110, E2905–E2914. [Google Scholar] [CrossRef] [PubMed]
- Hogan, T.; Shuvaev, A.; Commenges, D.; Yates, A.; Callard, R.; Thiebaut, R.; Seddon, B. Clonally Diverse T Cell Homeostasis Is Maintained by a Common Program of Cell-Cycle Control. J. Immunol. 2013, 190, 3985–3993. [Google Scholar] [CrossRef] [PubMed]
- Hogan, T.; Nowicka, M.; Cownden, D.; Pearson, C.F.; Yates, A.J.; Seddon, B. Differential Impact of Self and Environmental Antigens on the Ontogeny and Maintenance of CD4+ T Cell Memory. eLife 2019, 8, e48901. [Google Scholar] [CrossRef] [PubMed]
- Verheijen, M.; Rane, S.; Pearson, C.; Yates, A.J.; Seddon, B. Fate Mapping Quantifies the Dynamics of B Cell Development and Activation throughout Life. Cell Rep. 2020, 33, 108376. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 Programs the Development and Function of CD4+ CD25+ Regulatory T Cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. FOXP3+ Treg as a Therapeutic Target for Promoting Anti-Tumor Immunity. Expert Opin. Ther. Targets 2018, 22, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Wildin, R.S.; Ramsdell, F.; Peake, J.; Faravelli, F.; Casanova, J.-L.; Buist, N.; Levy-Lahad, E.; Mazzella, M.; Goulet, O.; Perroni, L.; et al. X-Linked Neonatal Diabetes Mellitus, Enteropathy and Endocrinopathy Syndrome Is the Human Equivalent of Mouse Scurfy. Nat. Genet. 2001, 27, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.L.; Ochs, H.D. IPEX Is a Unique X-Linked Syndrome Characterized by Immune Dysfunction, Polyendocrinopathy, Enteropathy, and a Variety of Autoimmune Phenomena. Curr. Opin. Pediatr. 2001, 13, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Sarnaik, A.A.; Hamid, O.; Khushalani, N.I.; Lewis, K.D.; Medina, T.; Kluger, H.M.; Thomas, S.S.; Domingo-Musibay, E.; Pavlick, A.C.; Whitman, E.D.; et al. Lifileucel, a Tumor-Infiltrating Lymphocyte Therapy, in Metastatic Melanoma. J. Clin. Oncol. 2021, 39, 2656–2666. [Google Scholar] [CrossRef] [PubMed]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric Antigen Receptor T Cells for Sustained Remissions in Leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.T.; Maude, S.L.; Porter, D.L.; Frey, N.; Wood, P.; Han, X.; Waldron, E.; Chakraborty, A.; Awasthi, R.; Levine, B.L.; et al. Cellular Kinetics of CTL019 in Relapsed/Refractory B-Cell Acute Lymphoblastic Leukemia and Chronic Lymphocytic Leukemia. Blood 2017, 130, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.T.; Waldron, E.; Grupp, S.A.; Levine, J.E.; Laetsch, T.W.; Pulsipher, M.A.; Boyer, M.W.; August, K.J.; Hamilton, J.; Awasthi, R.; et al. Clinical Pharmacology of Tisagenlecleucel in B-Cell Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2018, 24, 6175–6184. [Google Scholar] [CrossRef] [PubMed]
- Neelapu, S.S. CAR-T Efficacy: Is Conditioning the Key? Blood 2019, 133, 1799–1800. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Munoz, J.; Goy, A.; Locke, F.L.; Jacobson, C.A.; Hill, B.T.; Timmerman, J.M.; Holmes, H.; Jaglowski, S.; Flinn, I.W.; et al. KTE-X19 CAR T-Cell Therapy in Relapsed or Refractory Mantle-Cell Lymphoma. N. Engl. J. Med. 2020, 382, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, J.; Hirayama, A.V.; Purushe, J.; Hay, K.A.; Lymp, J.; Li, D.H.; Yeung, C.C.S.; Sheih, A.; Pender, B.S.; Hawkins, R.M.; et al. Feasibility and Efficacy of CD19-Targeted CAR T Cells with Concurrent Ibrutinib for CLL after Ibrutinib Failure. Blood 2020, 135, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Fraietta, J.A.; Lacey, S.F.; Orlando, E.J.; Pruteanu-Malinici, I.; Gohil, M.; Lundh, S.; Boesteanu, A.C.; Wang, Y.; O’Connor, R.S.; Hwang, W.-T.; et al. Determinants of Response and Resistance to CD19 Chimeric Antigen Receptor (CAR) T Cell Therapy of Chronic Lymphocytic Leukemia. Nat. Med. 2018, 24, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Kvistborg, P.; Shu, C.J.; Heemskerk, B.; Fankhauser, M.; Thrue, C.A.; Toebes, M.; van Rooij, N.; Linnemann, C.; van Buuren, M.M.; Urbanus, J.H.; et al. TIL Therapy Broadens the Tumor-Reactive CD8+ T Cell Compartment in Melanoma Patients. Oncoimmunology 2012, 1, 409–418. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, J.H.; Heemskerk, B.; van Rooij, N.; Gomez-Eerland, R.; Michels, S.; van Zon, M.; de Boer, R.; Bakker, N.A.M.; Jorritsma-Smit, A.; van Buuren, M.M.; et al. Tumor Infiltrating Lymphocytes (TIL) Therapy in Metastatic Melanoma: Boosting of Neoantigen-Specific T Cell Reactivity and Long-Term Follow-Up. J. Immunother. Cancer 2020, 8, e000848. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, L.; Masoumi, E.; Fallah-Mehrjardi, K.; Mirzaei, H.R.; Hadjati, J. Prolonged Persistence of Chimeric Antigen Receptor (CAR) T Cell in Adoptive Cancer Immunotherapy: Challenges and Ways Forward. Front. Immunol. 2020, 11, 702. [Google Scholar] [CrossRef] [PubMed]
- Guzman, G.; Reed, M.R.; Bielamowicz, K.; Koss, B.; Rodriguez, A. CAR-T Therapies in Solid Tumors: Opportunities and Challenges. Curr. Oncol. Rep. 2023, 25, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Bakker, D.S.; van der Wal, M.M.; Heeb, L.E.; Giovannone, B.; Asamoah, M.; Delemarre, E.M.; Drylewicz, J.; Nierkens, S.; Boyman, O.; de Bruin-Weller, M.S.; et al. Early and Long-Term Effects of Dupilumab Treatment on Circulating T-Cell Functions in Patients with Moderate-to-Severe Atopic Dermatitis. J. Investig. Dermatol. 2021, 141, 1943–1953.e13. [Google Scholar] [CrossRef] [PubMed]
- Diks, A.M.; Khatri, I.; Oosten, L.E.; de Mooij, B.; Groenland, R.J.; Teodosio, C.; Perez-Andres, M.; Orfao, A.; Berbers, G.A.M.; Zwaginga, J.J.; et al. Highly Sensitive Flow Cytometry Allows Monitoring of Changes in Circulating Immune Cells in Blood After Tdap Booster Vaccination. Front. Immunol. 2021, 12, 666953. [Google Scholar] [CrossRef]
- Stern, L.; McGuire, H.; Avdic, S.; Rizzetto, S.; Groth, B.F.d.S.; Luciani, F.; Slobedman, B.; Blyth, E. Mass Cytometry for the Assessment of Immune Reconstitution After Hematopoietic Stem Cell Transplantation. Front. Immunol. 2018, 9, 1672. [Google Scholar] [CrossRef] [PubMed]
- Morgan, P.; Brown, D.G.; Lennard, S.; Anderton, M.J.; Barrett, J.C.; Eriksson, U.; Fidock, M.; Hamrén, B.; Johnson, A.; March, R.E.; et al. Impact of a Five-Dimensional Framework on R&D Productivity at AstraZeneca. Nat. Rev. Drug Discov. 2018, 17, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.H.; Deng, Q.; Ting, N. Proof of Concept: Drug Selection? Or Dose Selection? Thoughts on Multiplicity Issues. Ther. Innov. Regul. Sci. 2021, 55, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Morgan, P.; Graaf, P.H.V.D.; Arrowsmith, J.; Feltner, D.E.; Drummond, K.S.; Wegner, C.D.; Street, S.D.A. Can the Flow of Medicines Be Improved? Fundamental Pharmacokinetic and Pharmacological Principles toward Improving Phase II Survival. Drug Discov. Today 2012, 17, 419–424. [Google Scholar] [CrossRef]
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of Clinical Trial Success Rates and Related Parameters. Biostatistics 2019, 20, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Schilling, H.-L.; Glehr, G.; Kapinsky, M.; Ahrens, N.; Riquelme, P.; Cordero, L.; Bitterer, F.; Schlitt, H.J.; Geissler, E.K.; Haferkamp, S.; et al. Development of a Flow Cytometry Assay to Predict Immune Checkpoint Blockade-Related Complications. Front. Immunol. 2021, 12, 765644. [Google Scholar] [CrossRef] [PubMed]
- Gadalla, R.; Noamani, B.; MacLeod, B.L.; Dickson, R.J.; Guo, M.; Xu, W.; Lukhele, S.; Elsaesser, H.J.; Razak, A.R.A.; Hirano, N.; et al. Validation of CyTOF Against Flow Cytometry for Immunological Studies and Monitoring of Human Cancer Clinical Trials. Front. Oncol. 2019, 9, 415. [Google Scholar] [CrossRef] [PubMed]
- Fricker, M.; Qin, L.; Niessen, N.; Baines, K.J.; McDonald, V.M.; Scott, H.A.; Simpson, J.L.; Gibson, P.G. Relationship of Sputum Mast Cells with Clinical and Inflammatory Characteristics of Asthma. Clin. Exp. Allergy 2020, 50, 696–707. [Google Scholar] [CrossRef]
- Gonzalez-Vivo, M.; Tiirikainen, M.K.L.; Andreu, M.; Fernandez-Clotet, A.; López-García, A.; Gonzalo, F.M.; Rodriguez, L.A.; de Jesús-Gil, C.; Ruiz-Romeu, E.; Nicolàs, L.S.-D.S.; et al. Memory T Cell Subpopulations as Early Predictors of Remission to Vedolizumab in Ulcerative Colitis. Front. Med. 2022, 9, 837294. [Google Scholar] [CrossRef]
- Liang, M.; Schwickart, M.; Schneider, A.K.; Vainshtein, I.; Nagro, C.D.; Standifer, N.; Roskos, L.K. Receptor Occupancy Assessment by Flow Cytometry as a Pharmacodynamic Biomarker in Biopharmaceutical Development. Cytom. Part B Clin. Cytom. 2016, 90, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Lagoo, A.S. How to Design and Validate a Clinical Flow Cytometry Assay. Clin. Lab. Med. 2023, 43, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Devitt, K.A.; Oldaker, T.; Shah, K.; Illingworth, A. Summary of Validation Considerations with Real-life Examples Using Both Qualitative and Semiquantitative Flow Cytometry Assays. Cytom. Part B Clin. Cytom. 2023, 104, 374–391. [Google Scholar] [CrossRef]
- Audia, A.; Bannish, G.; Bunting, R.; Riveley, C. Flow Cytometry and Receptor Occupancy in Immune-Oncology. Expert Opin. Biol. Ther. 2022, 22, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Cabanski, M.; Oldaker, T.; Stewart, J.J.; Selliah, N.; Eck, S.; Green, C.; Litwin, V.; Vitaliti, A. Flow Cytometric Method Transfer: Recommendations for Best Practice. Cytom. Part B Clin. Cytom. 2021, 100, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Brestoff, J.R.; Frater, J.L. Contemporary Challenges in Clinical Flow Cytometry: Small Samples, Big Data, Little Time. J. Appl. Lab. Med. 2022, 7, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Kalina, T.; Flores-Montero, J.; van der Velden, V.H.J.; Martin-Ayuso, M.; Böttcher, S.; Ritgen, M.; Almeida, J.; Lhermitte, L.; Asnafi, V.; Mendonça, A.; et al. EuroFlow Standardization of Flow Cytometer Instrument Settings and Immunophenotyping Protocols. Leukemia 2012, 26, 1986–2010. [Google Scholar] [CrossRef] [PubMed]
- Krutzik, P.O.; Nolan, G.P. Intracellular Phospho-protein Staining Techniques for Flow Cytometry: Monitoring Single Cell Signaling Events. Cytom. Part A 2003, 55A, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Olson, W.C.; Smolkin, M.E.; Farris, E.M.; Fink, R.J.; Czarkowski, A.R.; Fink, J.H.; Chianese-Bullock, K.A.; Slingluff, C.L. Shipping Blood to a Central Laboratory in Multicenter Clinical Trials: Effect of Ambient Temperature on Specimen Temperature, and Effects of Temperature on Mononuclear Cell Yield, Viability and Immunologic Function. J. Transl. Med. 2011, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Magallon, R.E.; Harmacek, L.D.; Arger, N.K.; Grewal, P.; Powers, L.; Werner, B.R.; Barkes, B.Q.; Li, L.; MacPhail, K.; Gillespie, M.; et al. Standardization of Flow Cytometry and Cell Sorting to Enable a Transcriptomic Analysis in a Multi-Site Sarcoidosis Study. PLoS ONE 2023, 18, e0281210. [Google Scholar] [CrossRef] [PubMed]
- Brestoff, J.R. Full Spectrum Flow Cytometry in the Clinical Laboratory. Int. J. Lab. Hematol. 2023, 45, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Baumgaertner, P.; Sankar, M.; Herrera, F.; Benedetti, F.; Barras, D.; Thierry, A.-C.; Dangaj, D.; Kandalaft, L.E.; Coukos, G.; Xenarios, I.; et al. Unsupervised Analysis of Flow Cytometry Data in a Clinical Setting Captures Cell Diversity and Allows Population Discovery. Front. Immunol. 2021, 12, 633910. [Google Scholar] [CrossRef] [PubMed]
- Kamysheva, A.L.; Fastovets, D.V.; Kruglikov, R.N.; Sokolov, A.A.; Fefler, A.S.; Bolshakova, A.A.; Radko, A.; Krauz, I.E.; Yong, S.T.; Goldberg, M.; et al. Machine Learning (ML)-Enabled Automation for High-Throughput Data Processing in Flow Cytometry. Blood 2023, 142, 905. [Google Scholar] [CrossRef]
- Xu, C.; Yang, J.; Kosters, A.; Babcock, B.R.; Qiu, P.; Ghosn, E.E.B. Comprehensive Multi-Omics Single-Cell Data Integration Reveals Greater Heterogeneity in the Human Immune System. iScience 2022, 25, 105123. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, J.M.; Luke, J.J.; Guglietta, S.; Krieg, C. High Throughput Multi-Omics Approaches for Clinical Trial Evaluation and Drug Discovery. Front. Immunol. 2021, 12, 590742. [Google Scholar] [CrossRef] [PubMed]

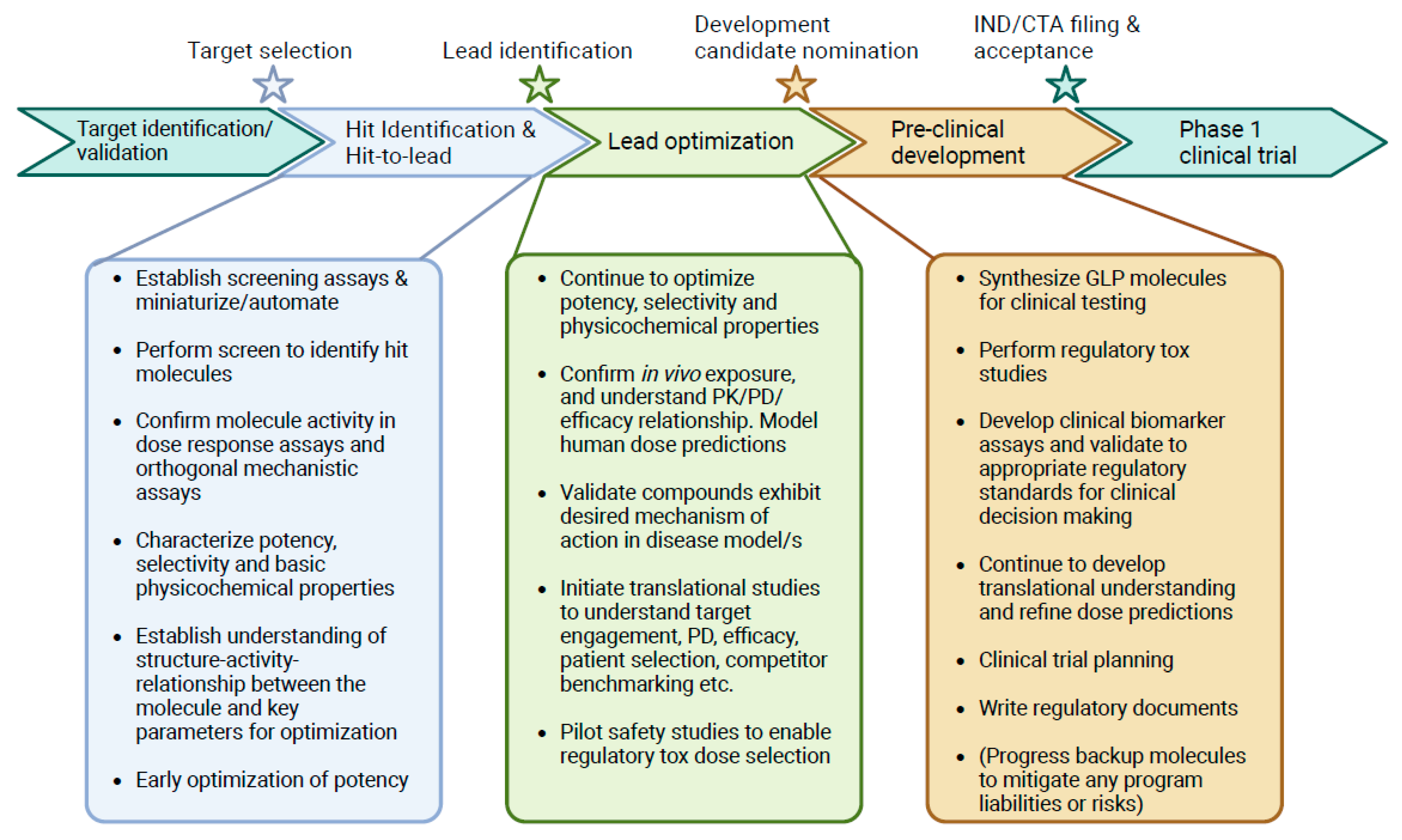
| Therapeutic Property | Examples of Parameters Considered | Objective |
|---|---|---|
| Potency |
| Early screening hits typically lack sufficient potency for clinical activity at a feasible dose and require iterative optimization to improve potency. |
| Safety and selectivity |
| Optimize primary target specificity while minimizing secondary interactions conferring safety risks. |
| Pharmacokinetics and drug exposure |
| Optimization of properties that affect drug exposure in target tissues. These typically include characteristics related to absorption, distribution, metabolism, and excretion (ADME) of a molecule. |
| Therapeutic functionality |
| Properties that affect the molecular mechanism of action. |
| Lead Author | Disease Indication | Target | Modality | Use of Flow Cytometry |
|---|---|---|---|---|
| Tuijnenburg [7] | Auto-immunity | MTOR pathway (via phenotypic screen) | Small molecule | Phenotypic screen for regulators of auto-antibody production |
| Schardt [11] | Virology | SARS-CoV-2 neutralizing antibodies | Antibody | Sorting of memory-specific B cell clones for expansion/antibody production |
| Zhou [13] | Oncology | EGFR | Antibody | Characterization and rank ordering of antibodies based on binding affinity |
| Revenko [17] | Oncology | FOXP3 | ASOs | Potency ranking of ASOs in primary cells |
| McDermott [21] | Oncology | CLDN6 | Antibody/ADC | Characterize selectivity versus related family members |
| Nilsson [25] | Oncology | JAK1 | Small molecule | Characterize selectivity (JAK1 vs. JAK2) |
| Kirkland [28] | Virology | Shiga toxin | Antibody | Evaluate immune cell activation |
| Mandrup [32] | Immuno-Oncology | N/A | Bi-specific antibody | Evaluate the effect of half-life modifications on efficacy |
| Parameswaran [34] | Oncology | CD6 | ADC | Measure internalization efficiency |
| Li [41] | Oncology | HER2 | ADC | ADC payload release versus target and bystander cells |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullas, S.; Sinclair, C. Applications of Flow Cytometry in Drug Discovery and Translational Research. Int. J. Mol. Sci. 2024, 25, 3851. https://doi.org/10.3390/ijms25073851
Ullas S, Sinclair C. Applications of Flow Cytometry in Drug Discovery and Translational Research. International Journal of Molecular Sciences. 2024; 25(7):3851. https://doi.org/10.3390/ijms25073851
Chicago/Turabian StyleUllas, Sumana, and Charles Sinclair. 2024. "Applications of Flow Cytometry in Drug Discovery and Translational Research" International Journal of Molecular Sciences 25, no. 7: 3851. https://doi.org/10.3390/ijms25073851
APA StyleUllas, S., & Sinclair, C. (2024). Applications of Flow Cytometry in Drug Discovery and Translational Research. International Journal of Molecular Sciences, 25(7), 3851. https://doi.org/10.3390/ijms25073851





