Methylmercury Induces Apoptosis in Mouse C17.2 Neural Stem Cells through the Induction of OSGIN1 Expression by NRF2
Abstract
:1. Introduction
2. Results
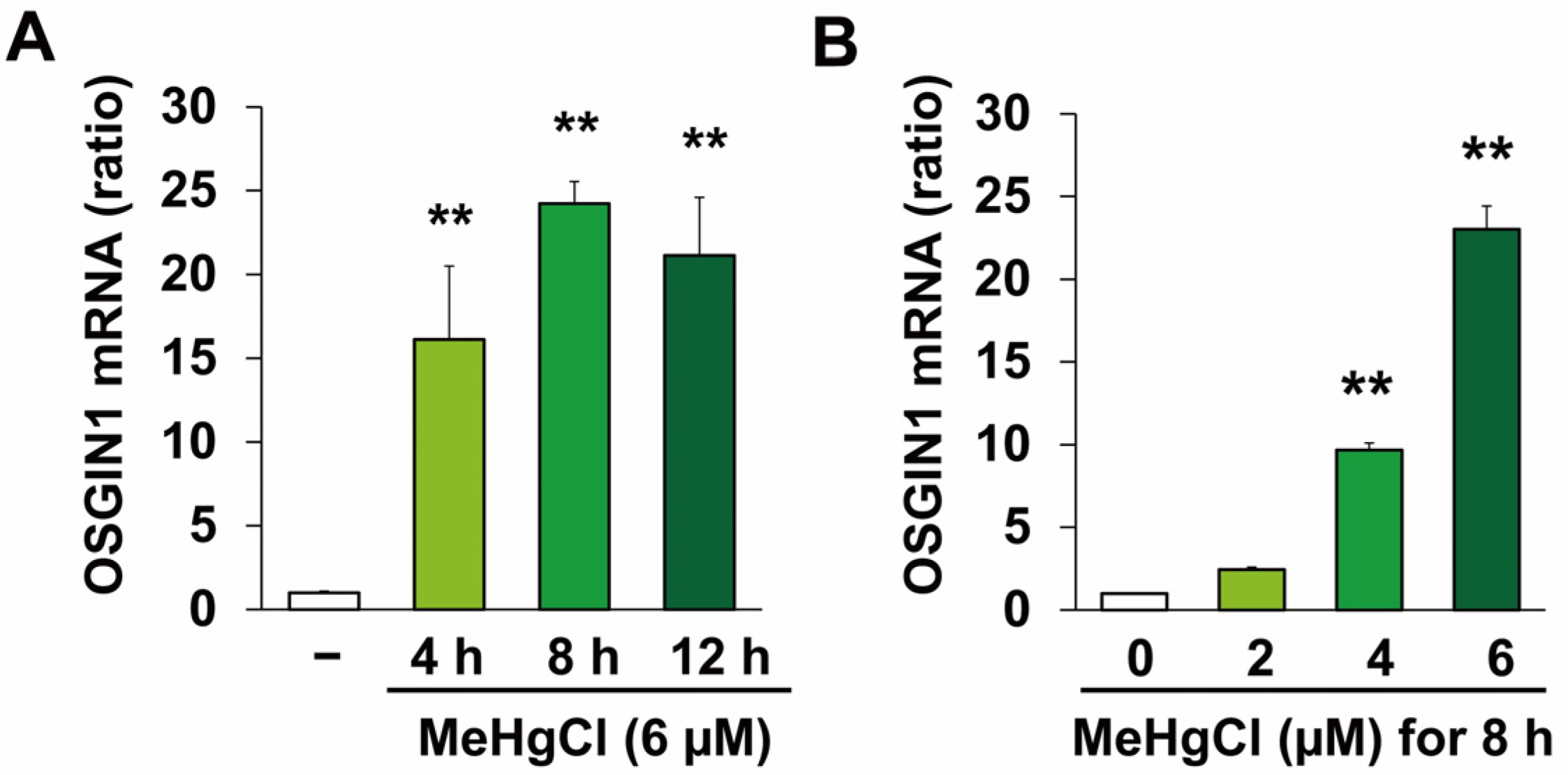
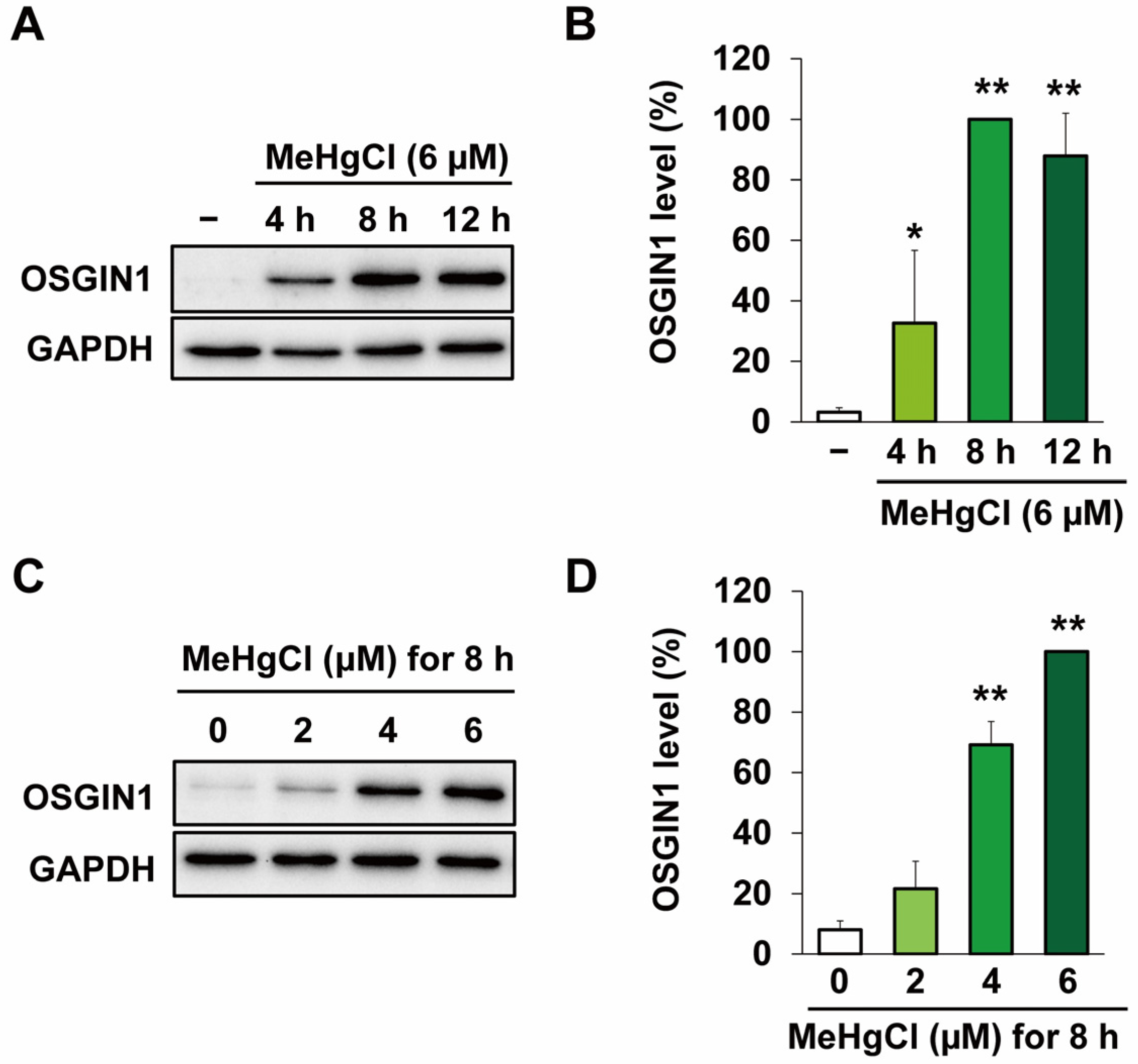
2.1. Methylmercury Induces OSGIN1 Expression in C17.2 Cells
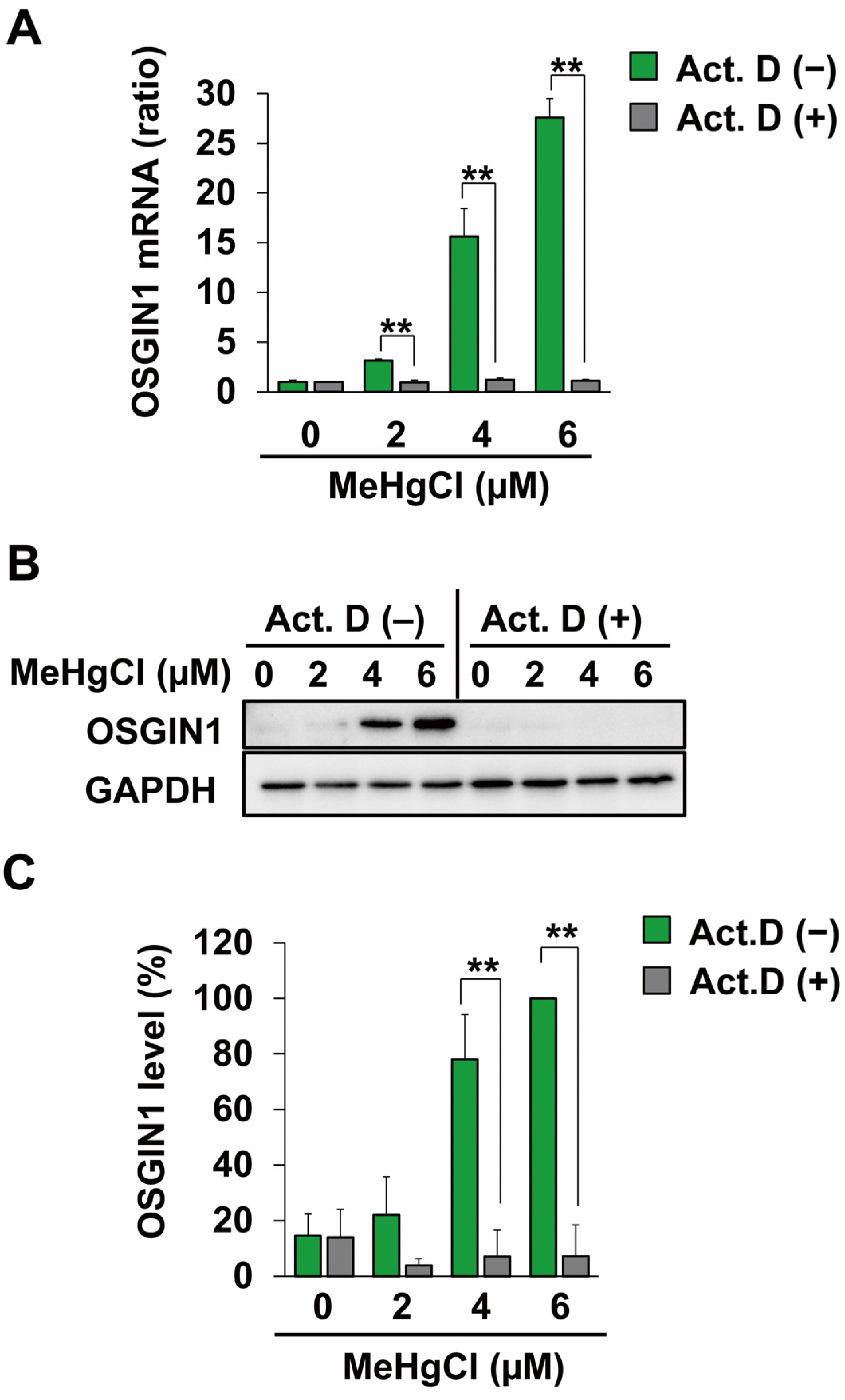
2.2. Methylmercury Induces OSGIN1 Expression by Promoting Its Transcription
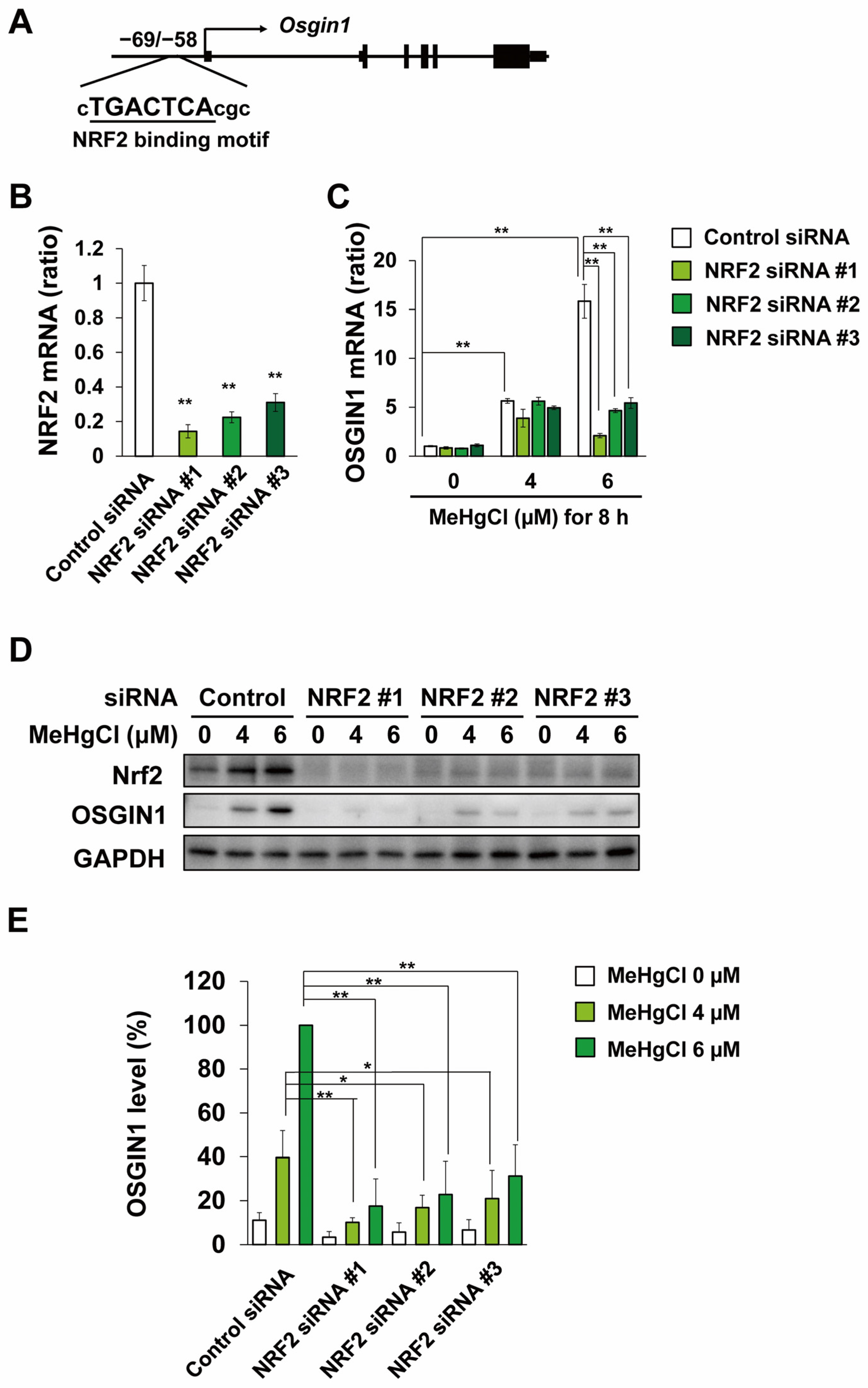
2.3. Methylmercury Induces OSGIN1 Expression in C17.2 Cells in an NRF2-Dependent Manner
2.4. OSGIN1 Is Involved in Apoptosis Caused by Methylmercury
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Quantitative PCR (qPCR)
4.3. Western Blotting
4.4. siRNA Transfection
4.5. Plasmid Construction and Transfection
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eto, K. Review Article: Pathology of Minamata Disease. Toxicol. Pathol. 1997, 25, 614–623. [Google Scholar] [CrossRef]
- Farina, M.; Rocha, J.B.T.; Aschner, M. Mechanisms of Methylmercury-Induced Neurotoxicity: Evidence from Experimental Studies. Life Sci. 2011, 89, 555–563. [Google Scholar] [CrossRef]
- Nagashima, K.; Fujii, Y.; Tsukamoto, T.; Nukuzuma, S.; Satoh, M.; Fujita, M.; Fujioka, Y.; Akagi, H. Apoptotic Process of Cerebellar Degeneration in Experimental Methylmercury Intoxication of Rats. Acta Neuropathol. 1996, 91, 72–77. [Google Scholar] [CrossRef]
- Hoang, V.A.T.; Do, H.T.T.; Agusa, T.; Koriyama, C.; Akiba, S.; Ishibashi, Y.; Sakamoto, M.; Yamamoto, M. Hair Mercury Levels in Relation to Fish Consumption among Vietnamese in Hanoi. J. Toxicol. Sci. 2017, 42, 651–662. [Google Scholar] [CrossRef]
- Driscoll, C.T.; Mason, R.P.; Chan, H.M.; Jacob, D.J.; Pirrone, N. Mercury as a Global Pollutant: Sources, Pathways, and Effects. Environ. Sci. Technol. 2013, 47, 4967–4983. [Google Scholar] [CrossRef] [PubMed]
- Santos-Sacramento, L.; Arrifano, G.P.; Lopes-Araújo, A.; Augusto-Oliveira, M.; Albuquerque-Santos, R.; Takeda, P.Y.; Souza-Monteiro, J.R.; Macchi, B.M.; do Nascimento, J.L.M.; Lima, R.R.; et al. Human Neurotoxicity of Mercury in the Amazon: A Scoping Review with Insights and Critical Considerations. Ecotoxicol. Environ. Saf. 2021, 208, 111686. [Google Scholar] [CrossRef] [PubMed]
- Meneses, H.d.N.d.M.; Oliveira-da-Costa, M.; Basta, P.C.; Morais, C.G.; Pereira, R.J.B.; de Souza, S.M.S.; Hacon, S.d.S. Mercury Contamination: A Growing Threat to Riverine and Urban Communities in the Brazilian Amazon. Int. J. Environ. Res. Public Health 2022, 19, 2816. [Google Scholar] [CrossRef]
- Santos-Lima, C.d.; Mourão, D.d.S.; Carvalho, C.F.d.; Souza-Marques, B.; Vega, C.M.; Gonçalves, R.A.; Argollo, N.; Menezes-Filho, J.A.; Abreu, N.; de Souza Hacon, S.. Neuropsychological Effects of Mercury Exposure in Children and Adolescents of the Amazon Region, Brazil. Neurotoxicology 2020, 79, 48–57. [Google Scholar] [CrossRef]
- Jacobson, J.L.; Muckle, G.; Ayotte, P.; Dewailly, É.; Jacobson, S.W. Relation of Prenatal Methylmercury Exposure from Environmental Sources to Childhood IQ. Environ. Health Perspect. 2015, 123, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, N.; Murata, K.; Iwai-Shimada, M.; Yaginuma-Sakurai, K.; Satoh, H.; Nakai, K. Psychomotor Ability in Children Prenatally Exposed to Methylmercury: The 18-Month Follow-up of Tohoku Study of Child Development. Tohoku J. Exp. Med. 2017, 242, 1–8. [Google Scholar] [CrossRef]
- Tatsuta, N.; Nakai, K.; Sakamoto, M.; Murata, K.; Satoh, H. Methylmercury Exposure and Developmental Outcomes in Tohoku Study of Child Development at 18 Months of Age. Toxics 2018, 6, 49. [Google Scholar] [CrossRef] [PubMed]
- Iwai-Shimada, M.; Takahashi, T.; Kim, M.; Fujimura, M.; Ito, H.; Toyama, T.; Naganuma, A.; Hwang, G.-W. Methylmercury Induces the Expression of TNF-α Selectively in the Brain of Mice. Sci. Rep. 2016, 6, 38294. [Google Scholar] [CrossRef] [PubMed]
- Toyama, T.; Xu, S.; Nakano, R.; Hasegawa, T.; Endo, N.; Takahashi, T.; Lee, J.Y.; Naganuma, A.; Hwang, G.W. The Nuclear Protein HOXB13 Enhances Methylmercury Toxicity by Inducing Oncostatin M and Promoting Its Binding to TNFR3 in Cultured Cells. Cells 2020, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Kim, M.S.; Iwai-Shimada, M.; Hoshi, T.; Fujimura, M.; Toyama, T.; Fujiwara, Y.; Naganuma, A.; Hwang, G.W. Induction of Chemokine CCL3 by NF-ΚB Reduces Methylmercury Toxicity in C17.2 Mouse Neural Stem Cells. Environ. Toxicol. Pharmacol. 2019, 71, 103216. [Google Scholar] [CrossRef]
- Sato, M.; Toyama, T.; Kim, M.S.; Lee, J.Y.; Hoshi, T.; Miura, N.; Naganuma, A.; Hwang, G.W. Increased Putrescine Levels Due to ODC1 Overexpression Prevents Mitochondrial Dysfunction-Related Apoptosis Induced by Methylmercury. Life Sci. 2020, 256, 118031. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Toyama, T.; Lee, J.Y.; Miura, N.; Naganuma, A.; Hwang, G.W. Activation of Ornithine Decarboxylase Protects against Methylmercury Toxicity by Increasing Putrescine. Toxicol. Appl. Pharmacol. 2018, 356, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Toyama, T.; Wang, Y.; Kim, M.S.; Takahashi, T.; Naganuma, A.; Hwang, G.W. Increased Expression of TCF3, Transcription Factor 3, Is a Defense Response against Methylmercury Toxicity in Mouse Neuronal C17.2 Cells. Toxicol. Res. 2021, 37, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Toyama, T.; Yoshida, E.; Shinkai, Y.; Kumagai, Y. DNA Microarray Analysis of Human Neuroblastoma SH-SY5Y Cells Exposed to Methylmercury. J. Toxicol. Sci. 2011, 36, 843–845. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Peng, S.; He, L.; Wang, Y.; Li, Y.; Ma, D.; Wang, Y.; Sun, L.; Zheng, H.; Yang, W.; et al. Methylmercury Cytotoxicity and Possible Mechanisms in Human Trophoblastic HTR-8/SVneo Cells. Ecotoxicol. Environ. Saf. 2021, 207, 111520. [Google Scholar] [CrossRef]
- Yu, X.; Robinson, J.F.; Sidhu, J.S.; Hong, S.; Faustman, E.M. A System-Based Comparison of Gene Expression Reveals Alterations in Oxidative Stress, Disruption of Ubiquitin-Proteasome System and Altered Cell Cycle Regulation after Exposure to Cadmium and Methylmercury in Mouse Embryonic Fibroblast. Toxicol. Sci. 2010, 114, 356–377. [Google Scholar] [CrossRef]
- Ishihara, Y.; Tsuji, M.; Kawamoto, T.; Yamazaki, T. Involvement of Reactive Oxygen Species Derived from Mitochondria in Neuronal Injury Elicited by Methylmercury. J. Clin. Biochem. Nutr. 2016, 59, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Edoff, K.; Raciti, M.; Moors, M.; Sundström, E.; Ceccatelli, S. Gestational Age and Sex Influence the Susceptibility of Human Neural Progenitor Cells to Low Levels of MeHg. Neurotox. Res. 2017, 32, 683–693. [Google Scholar] [CrossRef]
- Tamm, C.; Duckworth, J.; Hermanson, O.; Ceccatelli, S. High Susceptibility of Neural Stem Cells to Methylmercury Toxicity: Effects on Cell Survival and Neuronal Differentiation. J. Neurochem. 2006, 97, 69–78. [Google Scholar] [CrossRef]
- Watanabe, J.; Nakamachi, T.; Ohtaki, H.; Naganuma, A.; Shioda, S.; Nakajo, S. Low Dose of Methylmercury (MeHg) Exposure Induces Caspase Mediated-Apoptosis in Cultured Neural Progenitor Cells. J. Toxicol. Sci. 2013, 38, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Yen, C.; Tang, F.; Lee, K.; Liu, S.; Wu, C.; Hsieh, S.; Su, C.; Kuo, C.; Chen, Y. Methylmercury Exposure Induces ROS / Akt Inactivation-Triggered Endoplasmic Reticulum Stress-Regulated Neuronal Cell Apoptosis. Toxicology 2019, 425, 152245. [Google Scholar] [CrossRef]
- Brennan, M.S.; Matos, M.F.; Richter, K.E.; Li, B.; Scannevin, R.H. The NRF2 Transcriptional Target, OSGIN1, Contributes to Monomethyl Fumarate-Mediated Cytoprotection in Human Astrocytes. Sci. Rep. 2017, 7, 42054. [Google Scholar] [CrossRef]
- Li, R.; Chen, W.; Yanes, R.; Lee, S.; Berliner, J.A. OKL38 Is an Oxidative Stress Response Gene Stimulated by Oxidized Phospholipids. J. Lipid Res. 2007, 48, 709–715. [Google Scholar] [CrossRef]
- Toyama, T.; Sumi, D.; Shinkai, Y.; Yasutake, A.; Taguchi, K.; Tong, K.I.; Yamamoto, M.; Kumagai, Y. Cytoprotective Role of Nrf2/Keap1 System in Methylmercury Toxicity. Biochem. Biophys. Res. Commun. 2007, 363, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Kanda, H.; Shinkai, Y.; Toyama, T. The Role of the Keap1/Nrf2 Pathway in the Cellular Response to Methylmercury. Oxidative Med. Cell. Longev. 2013, 2013, 848279. [Google Scholar] [CrossRef]
- Algahtani, M.M.; Ahmad, S.F.; Alkharashi, L.A.; Al-Harbi, N.O.; Alanazi, W.A.; Alhamed, A.S.; Attia, S.M.; Bakheet, S.A.; Ibrahim, K.E.; Nadeem, A. Exposure to Methylmercury at Juvenile Stage Worsens Autism-like Symptoms in Adult BTBR T+tf/J Mice Due to Lack of Nuclear Factor Erythroid 2-Related Factor 2 Signaling Upregulation in Periphery and Brain. Toxics 2023, 11, 546. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, H.; Yin, Z.; Aschner, M.; Cai, J. Methylmercury Toxicity and Nrf2-Dependent Detoxification in Astrocytes. Toxicol. Sci. 2009, 107, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Yao, H.; Gan, F.; Tokarski, A.; Wang, Y. Interaction of OKL38 and P53 in Regulating Mitochondrial Structure and Function. PLoS ONE 2012, 7, e43362. [Google Scholar] [CrossRef] [PubMed]
- Satta, S.; Beal, R.; Smith, R.; Luo, X.; Ferris, G.R.; Langford-Smith, A.; Teasdale, J.; Ajime, T.T.; Serré, J.; Hazell, G.; et al. A Nrf2-OSGIN1&2-HSP70 Axis Mediates Cigarette Smoke-Induced Endothelial Detachment: Implications for Plaque Erosion. Cardiovasc. Res. 2023, 119, 1869–1882. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Zhu, H.; Liu, H.; Wang, M.; Chu, H.; Zhang, Z. METTL3 Regulates PM2.5-Induced Cell Injury by Targeting OSGIN1 in Human Airway Epithelial Cells. J. Hazard. Mater. 2021, 415, 125573. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.; Shen, Y.; Chen, H.-W.; Liu, K.; Chang, J.; Chen, P.; Lin, C.; Yao, H.; Li, C. Docosahexaenoic Acid Increases the Expression of Oxidative Stress-Induced Growth Inhibitor 1 through the PI3K/Akt/Nrf2 Signaling Pathway in Breast Cancer Cells. Food Chem. Toxicol. 2017, 108, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Oguro, A.; Fujita, K.; Ishihara, Y.; Yamamoto, M.; Yamazaki, T. DHA and Its Metabolites Have a Protective Role against Methylmercury-Induced Neurotoxicity in Mouse Primary Neuron and SH-SY5Y Cells. Int. J. Mol. Sci. 2021, 22, 3213. [Google Scholar] [CrossRef]
- Fujimura, M.; Usuki, F. Methylmercury-Mediated Oxidative Stress and Activation of the Cellular Protective System. Antioxidants 2020, 9, 1004. [Google Scholar] [CrossRef]
- Pant, A.; Dasgupta, D.; Tripathi, A.; Pyaram, K. Beyond Antioxidation: Keap1-Nrf2 in the Development and Effector Functions of Adaptive Immune Cells. ImmunoHorizons 2023, 7, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.H.; Lii, C.K.; Wang, T.S.; Liu, K.L.; Chen, H.W.; Huang, C.S.; Li, C.C. Docosahexaenoic Acid Promotes the Formation of Autophagosomes in MCF-7 Breast Cancer Cells through Oxidative Stress-Induced Growth Inhibitor 1 Mediated Activation of AMPK/MTOR Pathway. Food Chem. Toxicol. 2021, 154, 112318. [Google Scholar] [CrossRef]
- Toyama, T.; Shinkai, Y.; Yasutake, A.; Uchida, K.; Yamamoto, M.; Kumagai, Y. Isothiocyanates Reduce Mercury Accumulation via an Nrf2-Dependent Mechanism during Exposure of Mice to Methylmercury. Environ. Health Perspect. 2011, 119, 1117–1122. [Google Scholar] [CrossRef]
- Snyder, E.Y.; Deitcher, D.L.; Walsh, C.; Arnold-Aldea, S.; Hartwieg, E.A.; Cepko, C.L. Multipotent Neural Cell Lines Can Engraft and Participate in Development of Mouse Cerebellum. Cell 1992, 68, 33–51. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamashita, N.; Uchiyama, M.; Yamagata, R.; Hwang, G.-W. Methylmercury Induces Apoptosis in Mouse C17.2 Neural Stem Cells through the Induction of OSGIN1 Expression by NRF2. Int. J. Mol. Sci. 2024, 25, 3886. https://doi.org/10.3390/ijms25073886
Yamashita N, Uchiyama M, Yamagata R, Hwang G-W. Methylmercury Induces Apoptosis in Mouse C17.2 Neural Stem Cells through the Induction of OSGIN1 Expression by NRF2. International Journal of Molecular Sciences. 2024; 25(7):3886. https://doi.org/10.3390/ijms25073886
Chicago/Turabian StyleYamashita, Naoya, Marino Uchiyama, Ryota Yamagata, and Gi-Wook Hwang. 2024. "Methylmercury Induces Apoptosis in Mouse C17.2 Neural Stem Cells through the Induction of OSGIN1 Expression by NRF2" International Journal of Molecular Sciences 25, no. 7: 3886. https://doi.org/10.3390/ijms25073886




