Exploring the Integrated Role of miRNAs and lncRNAs in Regulating the Transcriptional Response to Amino Acids and Insulin-like Growth Factor 1 in Gilthead Sea Bream (Sparus aurata) Myoblasts
Abstract
1. Introduction
2. Results
2.1. Identification of miRNAs and lncRNAs in Gilthead Sea Bream Myoblasts
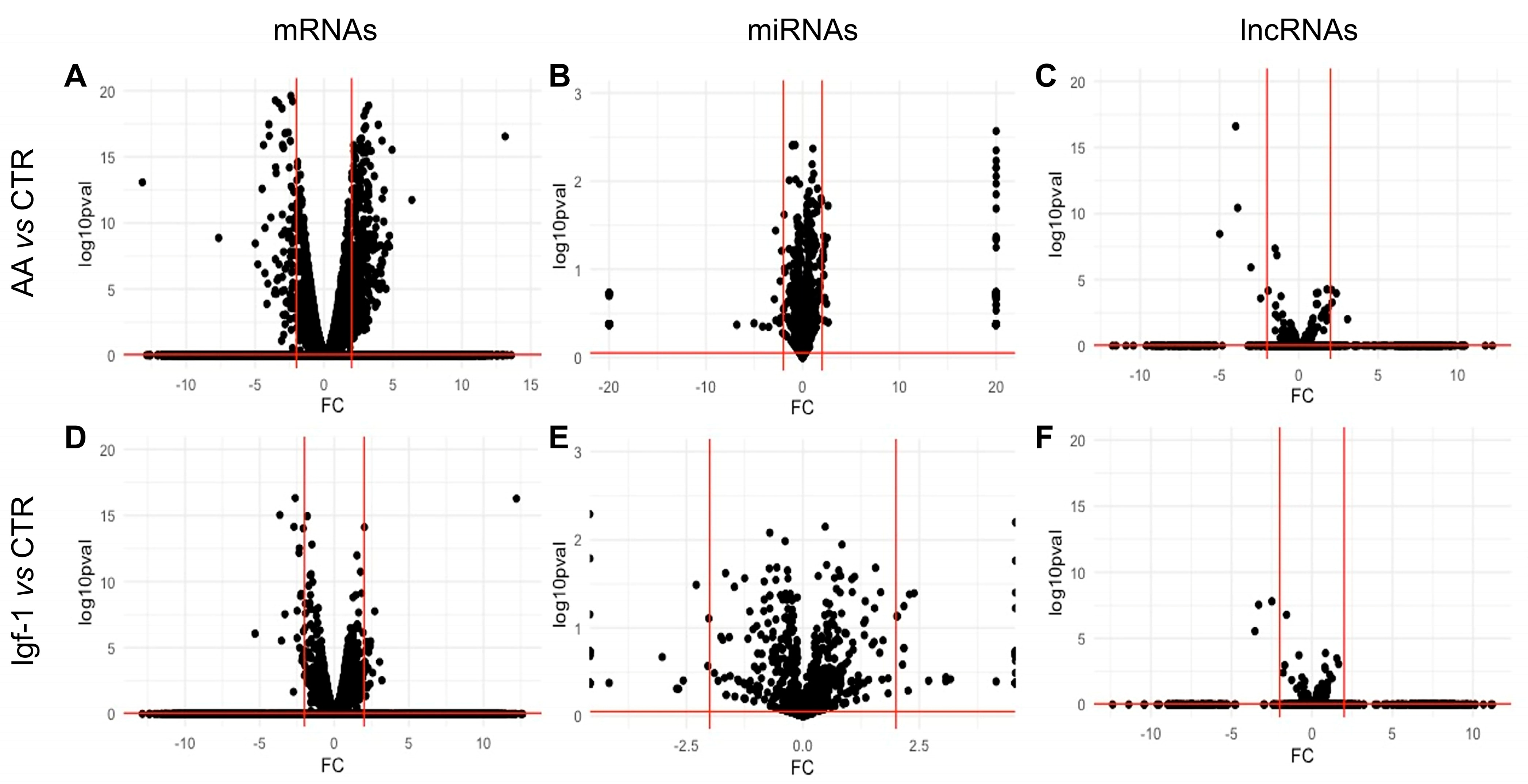
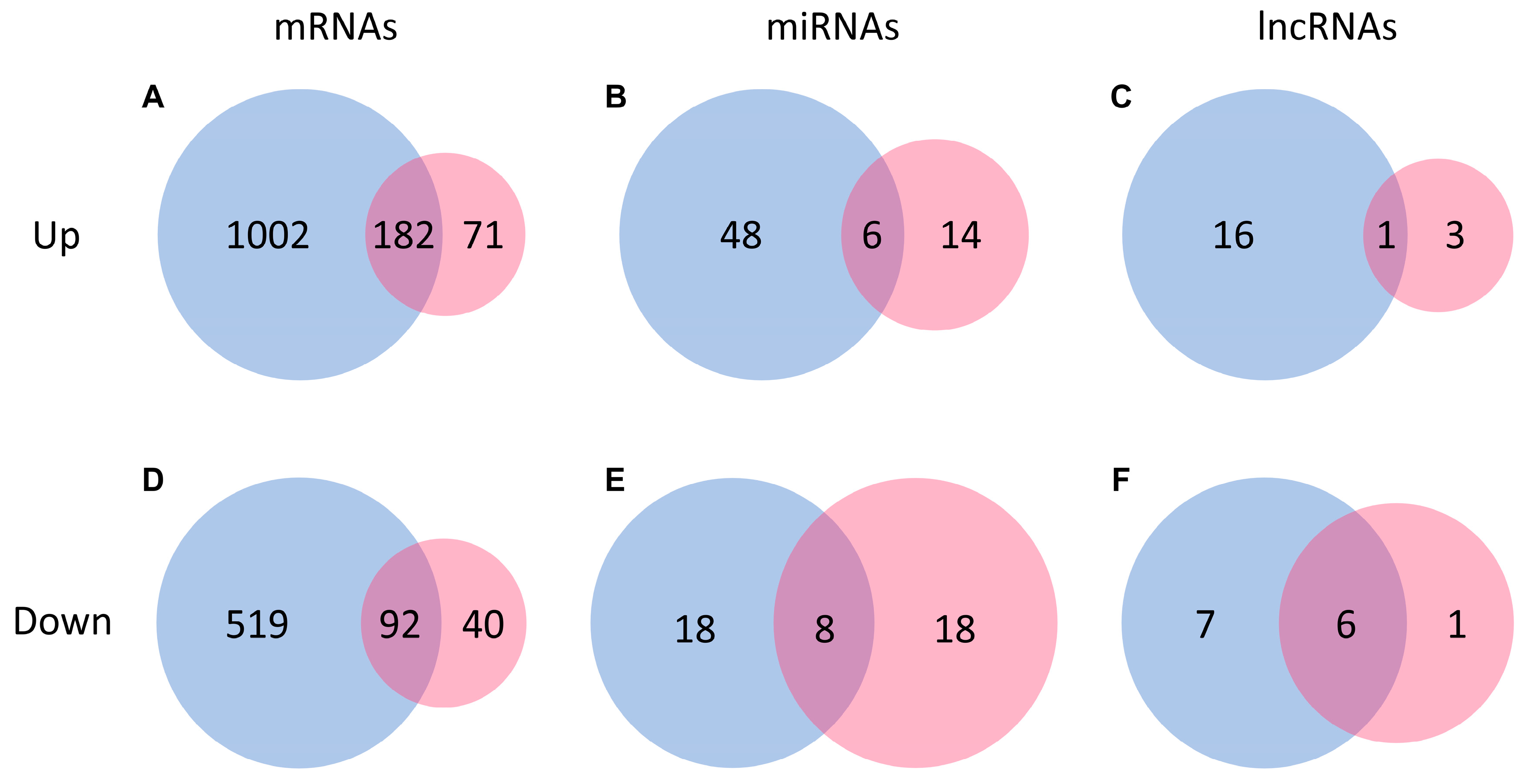
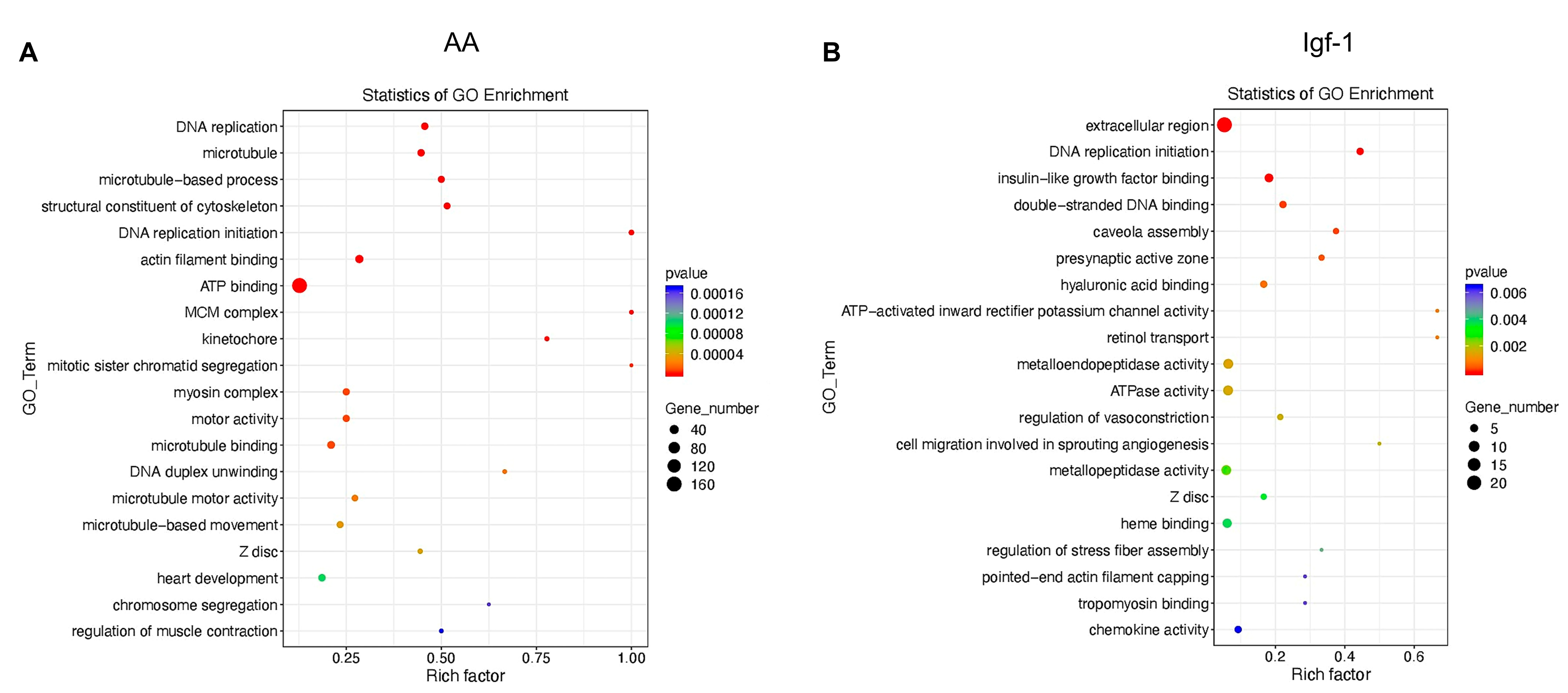
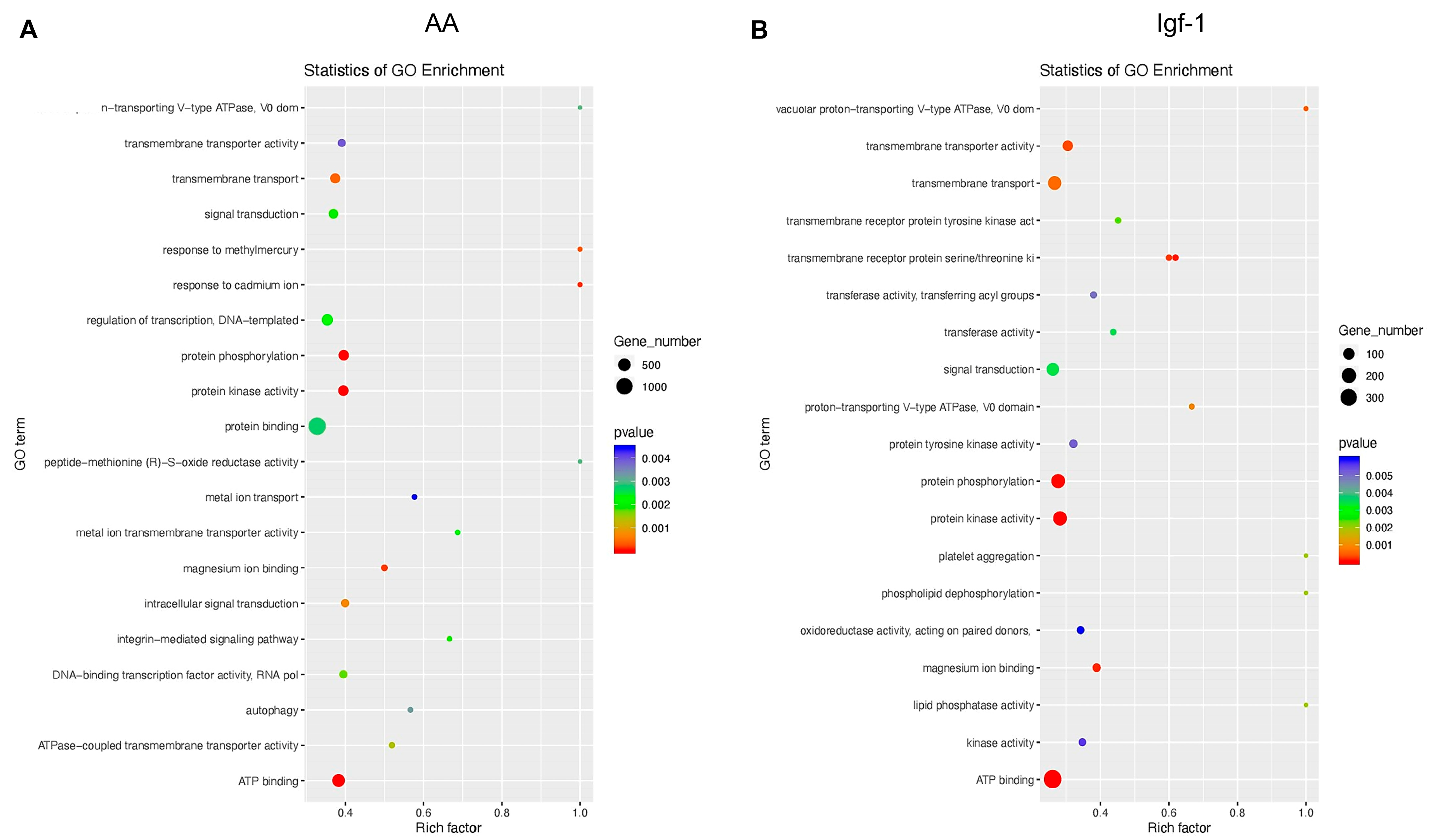
2.2. Transcriptomic Changes of mRNAs in Response to AA and Igf-1
2.3. Transcriptomic Analysis of ncRNAs
2.4. Predicted Interactions of miRNAs and lncRNAs with mRNAs Based on Transcriptomic Correlations and Bioinformatics Analysis
3. Discussion
4. Materials and Methods
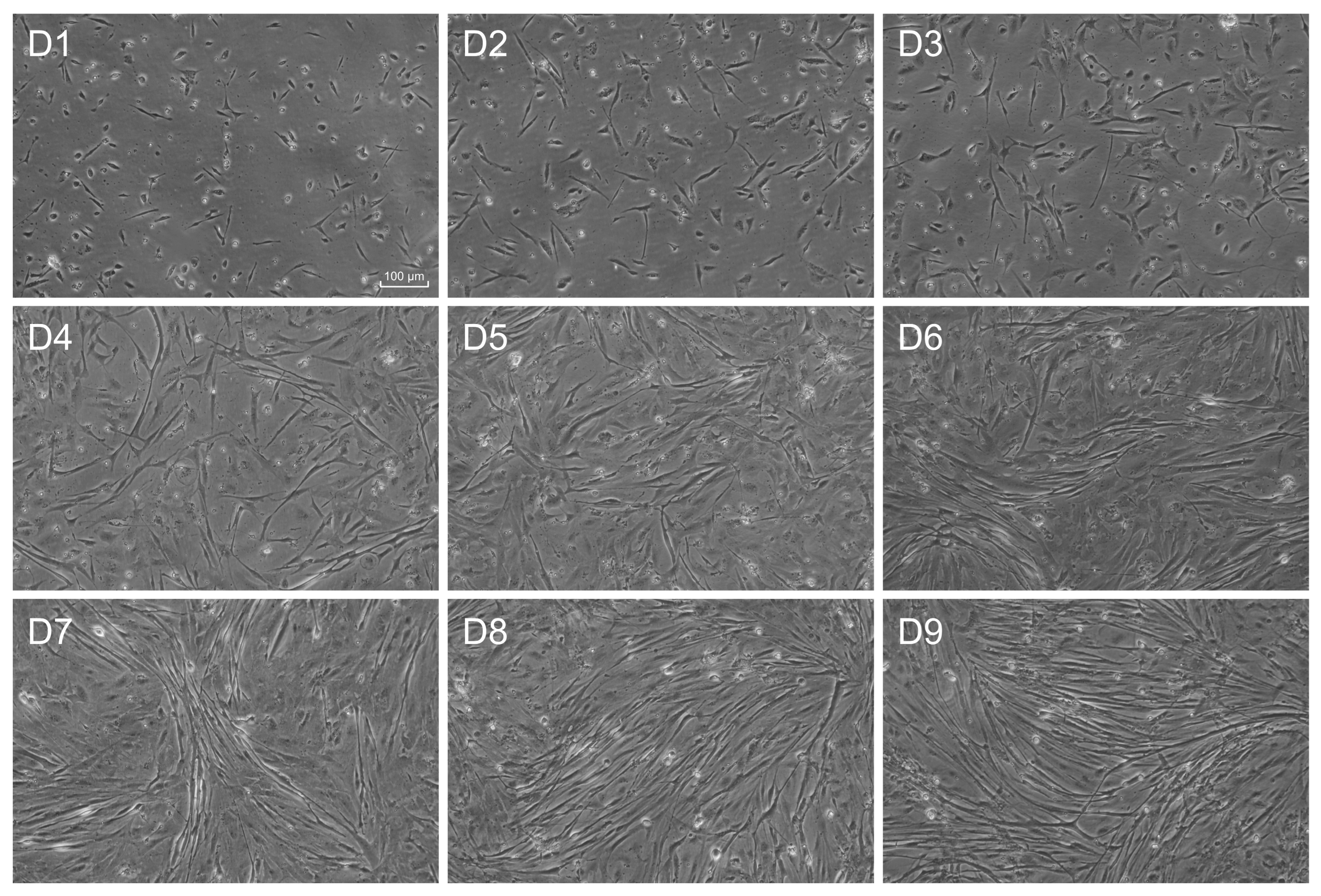
4.1. Gilthead Sea Bream Primary Myoblast Cell Culture and Treatments
4.2. RNA Extraction, Sequencing, and Bioinformatic Analyses
4.3. Validation of RNA-Seq Results by qPCR
4.4. Statistics of RNA-Seq Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mommsen, T.P. Paradigms of Growth in Fish. Comp. Biochem. Physiol.-B Biochem. Mol. Biol. 2001, 129, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.A. Environment and Plasticity of Myogenesis in Teleost Fish. J. Exp. Biol. 2006, 209, 2249–2264. [Google Scholar] [CrossRef] [PubMed]
- Weatherley, A.H.; Gill, H.S.; Lobo, A.F. Recruitment and Maximal Diameter of Axial Muscle Fibres in Teleosts and Their Relationship to Somatic Growth and Ultimate Size. J. Fish Biol. 1988, 33, 851–859. [Google Scholar] [CrossRef]
- Dal-Pai-Silva, M.; Zanella, B.T.T.; Duran, B.O.S.; Almeida, F.L.A.; Mareco, E.A.; de Paula, T.G. Cellular and Molecular Features of Skeletal Muscle Growth and Plasticity. In Biology and Physiology ofFreshwater Neotropical Fish; Baldisserotto, B., Urbinati, E.C., Cyrino, J.E.P., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 163–183. [Google Scholar] [CrossRef]
- Garcia de la serrana, D.; Codina, M.; Capilla, E.; Jiménez-Amilburu, V.; Navarro, I.; Du, S.J.; Johnston, I.A.; Gutiérrez, J. Characterisation and Expression of Myogenesis Regulatory Factors during in Vitro Myoblast Development and in Vivo Fasting in the Gilthead Sea Bream (Sparus aurata). Comp. Biochem. Physiol.-A Mol. Integr. Physiol. 2014, 167, 90–99. [Google Scholar] [CrossRef]
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The Myogenic Regulatory Factors, Determinants of Muscle Development, Cell Identity and Regeneration. Semin. Cell Dev. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Perelló-Amorós, M.; Otero-Tarrazón, A.; Jorge-Pedraza, V.; García-Pérez, I.; Sánchez-Moya, A.; Gabillard, J.C.; Moshayedi, F.; Navarro, I.; Capilla, E.; Fernández-Borràs, J.; et al. Myomaker and Myomixer Characterization in Gilthead Sea Bream under Different Myogenesis Conditions. Int. J. Mol. Sci. 2022, 23, 14639. [Google Scholar] [CrossRef]
- Lehka, L.; Rędowicz, M.J. Mechanisms Regulating Myoblast Fusion: A Multilevel Interplay. Semin. Cell Dev. Biol. 2020, 104, 81–92. [Google Scholar] [CrossRef]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-Rag Complex Targets MTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to MTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef]
- Manifava, M.; Smith, M.; Rotondo, S.; Walker, S.; Niewczas, I.; Zoncu, R.; Clark, J.; Ktistakis, N.T. Dynamics of MTORC1 Activation in Response to Amino Acids. Elife 2016, 5, e19960. [Google Scholar] [CrossRef]
- Yao, Y.; Jones, E.; Inoki, K. Lysosomal Regulation of MTORC1 by Amino Acids in Mammalian Cells. Biomolecules 2017, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Vélez, E.J.; Azizi, S.; Verheyden, D.; Salmerón, C.; Lutfi, E.; Sánchez-Moya, A.; Navarro, I.; Gutiérrez, J.; Capilla, E. Proteolytic Systems’ Expression during Myogenesis and Transcriptional Regulation by Amino Acids in Gilthead Sea Bream Cultured Muscle Cells. PLoS ONE 2017, 12, e0187339. [Google Scholar] [CrossRef] [PubMed]
- Seiliez, I.; Gabillard, J.C.; Riflade, M.; Sadoul, B.; Dias, K.; Avérous, J.; Tesseraud, S.; Skiba, S.; Panserat, S. Amino Acids Downregulate the Expression of Several Autophagy-Related Genes in Rainbow Trout Myoblasts. Autophagy 2012, 8, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, B.M.; Weber, G.M. Effects of Insulin-like Growth Factor-I, Insulin, and Leucine on Protein Turnover and Ubiquitin Ligase Expression in Rainbow Trout Primary Myocytes. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2010, 298, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, E.N.; Valdés, J.A.; Molina, A.; Björnsson, B.T. Regulation of Skeletal Muscle Growth in Fish by the Growth Hormone-Insulin-like Growth Factor System. Gen. Comp. Endocrinol. 2013, 192, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Ren, H.; Gao, S. Insulin-like Growth Factors (IGFs), IGF Receptors, and IGF-Binding Proteins: Roles in Skeletal Muscle Growth and Differentiation. Gen. Comp. Endocrinol. 2010, 167, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.W.; Duan, C.; Bern, H.A. Insulin-like Growth Factor Signaling in Fish. Int. Rev. Cytol. 2005, 243, 215–285. [Google Scholar] [CrossRef]
- Vélez, E.J.; Azizi, S.; Millán-Cubillo, A.; Fernández-Borràs, J.; Blasco, J.; Chan, S.J.; Calduch-Giner, J.A.; Pérez-Sánchez, J.; Navarro, I.; Capilla, E.; et al. Effects of Sustained Exercise on GH-IGFs Axis in Gilthead Sea Bream (Sparus Aurata). Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2016, 310, R313–R322. [Google Scholar] [CrossRef] [PubMed]
- Triantaphyllopoulos, K.A.; Cartas, D.; Miliou, H. Factors Influencing GH and IGF-I Gene Expression on Growth in Teleost Fish: How Can Aquaculture Industry Benefit? Rev. Aquac. 2020, 12, 1637–1662. [Google Scholar] [CrossRef]
- Güller, I.; Russell, A.P. MicroRNAs in Skeletal Muscle: Their Role and Regulation in Development, Disease and Function. J. Physiol. 2010, 588, 4075–4087. [Google Scholar] [CrossRef]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-Specific MicroRNAs in Skeletal Muscle Development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Martone, J.; Mariani, D.; Desideri, F.; Ballarino, M. Non-Coding RNAs Shaping Muscle. Front. Cell Dev. Biol. 2020, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- García-Pérez, I.; Molsosa-Solanas, A.; Perelló-Amorós, M.; Sarropoulou, E.; Blasco, J.; Gutiérrez, J.; Garcia de la serrana, D. The Emerging Role of Long Non-Coding RNAs in Development and Function of Gilthead Sea Bream (Sparus aurata) Fast Skeletal Muscle. Cells 2022, 11, 428. [Google Scholar] [CrossRef]
- Duran, B.O.S.; Zanella, B.T.T.; Perez, E.S.; Mareco, E.A.; Blasco, J.; Dal-Pai-Silva, M.; Garcia de la serrana, D. Amino Acids and IGF1 Regulation of Fish Muscle Growth Revealed by Transcriptome and MicroRNAome Integrative Analyses of Pacu (Piaractus mesopotamicus) Myotubes. Int. J. Mol. Sci. 2022, 23, 1180. [Google Scholar] [CrossRef] [PubMed]
- Paneru, B.; Ali, A.; Al-Tobasei, R.; Kenney, B.; Salem, M. Crosstalk among LncRNAs, MicroRNAs and MRNAs in the Muscle ‘Degradome’ of Rainbow Trout. Sci. Rep. 2018, 8, 8416. [Google Scholar] [CrossRef]
- Ali, A.; Al-Tobasei, R.; Kenney, B.; Leeds, T.D.; Salem, M. Integrated Analysis of LncRNA and MRNA Expression in Rainbow Trout Families Showing Variation in Muscle Growth and Fillet Quality Traits. Sci. Rep. 2018, 8, 12111. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Babiak, I. MicroRNA in Teleost Fish. Genome Biol. Evol. 2014, 6, 1911. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of Post-Transcriptional Regulation by MicroRNAs: Are the Answers in Sight? Nat. Rev. Genet. 2008 92 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNA Target Recognition and Regulatory Functions. Cell 2009, 136, 215. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.Z. Micromanagers of Gene Expression: The Potentially Widespread Influence of Metazoan MicroRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef]
- Koutsoulidou, A.; Mastroyiannopoulos, N.P.; Furling, D.; Uney, J.B.; Phylactou, L.A. Expression of MiR-1, MiR-133a, MiR-133b and MiR-206 Increases during Development of Human Skeletal Muscle. BMC Dev. Biol. 2011, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Mok, G.F.; Lozano-Velasco, E.; Münsterberg, A. MicroRNAs in Skeletal Muscle Development. Semin. Cell Dev. Biol. 2017, 72, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Gagan, J.; Dutta, A. MiR-206 and -486 Induce Myoblast Differentiation by Downregulating Pax7. Mol. Cell. Biol. 2011, 31, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.Z. MicroRNA-1 and MicroRNA-206 Regulate Skeletal Muscle Satellite Cell Proliferation and Differentiation by Repressing Pax7. J. Cell Biol. 2010, 190, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Winbanks, C.E.; Beyer, C.; Hagg, A.; Qian, H.; Sepulveda, P.V.; Gregorevic, P. MiR-206 Represses Hypertrophy of Myogenic Cells but Not Muscle Fibers via Inhibition of HDAC4. PLoS ONE 2013, 8, e73589. [Google Scholar] [CrossRef] [PubMed]
- Winbanks, C.E.; Wang, B.; Beyer, C.; Koh, P.; White, L.; Kantharidis, P.; Gregorevic, P. TGF-β Regulates MiR-206 and MiR-29 to Control Myogenic Differentiation through Regulation of HDAC4. J. Biol. Chem. 2011, 286, 13805–13814. [Google Scholar] [CrossRef]
- Yuasa, K.; Hagiwara, Y.; Ando, M.; Nakamura, A.; Takeda, S.; Hijikata, T. MicroRNA-206 Is Highly Expressed in Newly Formed Muscle Fibers: Implications Regarding Potential for Muscle Regeneration and Maturation in Muscular Dystrophy. Cell Struct. Funct. 2008, 33, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Williams, A.H.; Maxeiner, J.M.; Bezprozvannaya, S.; Shelton, J.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. MicroRNA-206 Promotes Skeletal Muscle Regeneration and Delays Progression of Duchenne Muscular Dystrophy in Mice. J. Clin. Investig. 2012, 122, 2054–2065. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The Role of MicroRNA-1 and MicroRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Luo, Y.; Wu, X.; Ling, Z.; Yuan, L.; Cheng, Y.; Chen, J.; Xiang, C. MicroRNA133a Targets Foxl2 and Promotes Differentiation of C2C12 into Myogenic Progenitor Cells. DNA Cell Biol. 2015, 34, 29. [Google Scholar] [CrossRef]
- Feng, Y.; Niu, L.L.; Wei, W.; Zhang, W.Y.; Li, X.Y.; Cao, J.H.; Zhao, S.H. A Feedback Circuit between MiR-133 and the ERK1/2 Pathway Involving an Exquisite Mechanism for Regulating Myoblast Proliferation and Differentiation. Cell Death Dis. 2013, 4, e934. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.J. The MyomiR Network in Skeletal Muscle Plasticity. Exerc. Sport Sci. Rev. 2011, 39, 150. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ono, Y.; Tan, S.C.; Chai, R.J.; Parkin, C.; Ingham, P.W. Prdm1a and MiR-499 Act Sequentially to Restrict Sox6 Activity to the Fast-Twitch Muscle Lineage in the Zebrafish Embryo. Development 2011, 138, 4399–4404. [Google Scholar] [CrossRef] [PubMed]
- van Rooij, E.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J.; Olson, E.N. A Family of MicroRNAs Encoded by Myosin Genes Governs Myosin Expression and Muscle Performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Guo, J.T.; Zhao, L.H.; Zhao, J.L. MicroRNA Expression Signature in Skeletal Muscle of Nile Tilapia. Aquaculture 2012, 364–365, 240–246. [Google Scholar] [CrossRef]
- Yan, X.; Ding, L.; Li, Y.; Zhang, X.; Liang, Y.; Sun, X.; Teng, C.B. Identification and Profiling of MicroRNAs from Skeletal Muscle of the Common Carp. PLoS ONE 2012, 7, e30925. [Google Scholar] [CrossRef] [PubMed]
- Duran, B.O.S.; Fernandez, G.J.; Mareco, E.A.; Moraes, L.N.; Salomão, R.A.S.; de Paula, T.G.; Santos, V.B.; Carvalho, R.F.; Dal-Pai-Silvca, M. Differential MicroRNA Expression in Fast- and Slow-Twitch Skeletal Muscle of Piaractus mesopotamicus during Growth. PLoS ONE 2015, 10, e0144481. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Abreu-Goodger, C.; Staton, A.A.; Stahlhut, C.; Shou, C.; Cheng, C.; Gerstein, M.; Enright, A.J.; Giraldez, A.J. Zebrafish MiR-1 and MiR-133 Shape Muscle Gene Expression and Regulate Sarcomeric Actin Organization. Genes Dev. 2009, 23, 619. [Google Scholar] [CrossRef]
- Huang, M.B.; Xu, H.; Xie, S.J.; Zhou, H.; Qu, L.H. Insulin-like Growth Factor-1 Receptor Is Regulated by MicroRNA-133 during Skeletal Myogenesis. PLoS ONE 2011, 6, e29173. [Google Scholar] [CrossRef]
- de Paula, T.G.; Zanella, B.T.T.; de Almeida Fantinatti, B.E.; de Moraes, L.N.; Duran, B.O.S.; de Oliveira, C.B.; Salomäo, R.A.S.; Da Silva, R.N.; Padovani, C.R.; Dos Santos, V.B.; et al. Food Restriction Increase the Expression of MTORC1 Complex Genes in the Skeletal Muscle of Juvenile Pacu (Piaractus mesopotamicus). PLoS ONE 2017, 12, e0177679. [Google Scholar] [CrossRef]
- Yan, B.; Zhu, C.D.; Guo, J.T.; Zhao, L.H.; Zhao, J.L. MiR-206 Regulates the Growth of the Teleost Tilapia (Oreochromis niloticus) through the Modulation of IGF-1 Gene Expression. J. Exp. Biol. 2013, 216, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, P.G.; Dias, M.C.; Carvalho, R.F.; Martins, C.; Pinhal, D. MicroRNA-499 Expression Distinctively Correlates to Target Genes Sox6 and Rod1 Profiles to Resolve the Skeletal Muscle Phenotype in Nile Tilapia. PLoS ONE 2015, 10, e0119804. [Google Scholar] [CrossRef] [PubMed]
- Duran, B.O.S.; Dal-Pai-Silva, M.; Garcia de la serrana, D. Rainbow Trout Slow Myoblast Cell Culture as a Model to Study Slow Skeletal Muscle, and the Characterization of Mir-133 and Mir-499 Families as a Case Study. J. Exp. Biol. 2020, 223, jeb216390. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Neguembor, M.V.; Jothi, M.; Gabellini, D. Long Noncoding RNAs, Emerging Players in Muscle\ndifferentiation and Disease. Skelet. Muscle 2014, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How Do LncRNAs Regulate Transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA–LncRNA Interactions. Methods Mol. Biol. 2016, 1402, 271–286. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional Gene Regulation by Long Noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef]
- Wang, S.; Zuo, H.; Jin, J.; Lv, W.; Xu, Z.; Fan, Y.; Zhang, J.; Zuo, B. Long Noncoding RNA Neat1 Modulates Myogenesis by Recruiting Ezh2. Cell Death Dis. 2019, 10, 505. [Google Scholar] [CrossRef]
- Mueller, A.C.; Cichewicz, M.A.; Dey, B.K.; Layer, R.; Reon, B.J.; Gagan, J.R.; Dutta, A. MUNC, a Long Noncoding RNA That Facilitates the Function of MyoD in Skeletal Myogenesis. Mol. Cell. Biol. 2015, 35, 498–513. [Google Scholar] [CrossRef]
- Cichewicz, M.A.; Kiran, M.; Przanowska, R.K.; Sobierajska, E.; Shibata, Y.; Dutta, A. MUNC, an Enhancer RNA Upstream from the MYOD Gene, Induces a Subgroup of Myogenic Transcripts in Trans Independently of MyoD. Mol. Cell. Biol. 2018, 38, e00655-17. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Y.; Li, T.; Ma, Z.; Jia, H.; Chen, Q.; Zhao, Y.; Zhai, L.; Zhong, R.; Li, C.; et al. Long Non-Coding RNA Linc-RAM Enhances Myogenic Differentiation by Interacting with MyoD. Nat. Commun. 2017, 8, 14016. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.; Han, Y.; Zhao, X.; Li, D.; Li, G. Long Non-Coding RNA Irm Enhances Myogenic Differentiation by Interacting with MEF2D. Cell Death Dis. 2019, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Hu, Q.; Egranov, S.D.; Xing, Z.; Zhang, Z.; Liang, K.; Ye, Y.; Pan, Y.; Chatterjee, S.S.; et al. Functional Significance of Gain-of-Function H19 LncRNA in Skeletal Muscle Differentiation and Anti-Obesity Effects. Genome Med. 2021, 13, 137. [Google Scholar] [CrossRef]
- Xu, H.; Cao, L.; Sun, B.; Wei, Y.; Liang, M. Transcriptomic Analysis of Potential “LncRNA-MRNA” Interactions in Liver of the Marine Teleost Cynoglossus Semilaevis Fed Diets with Different DHA/EPA Ratios. Front. Physiol. 2019, 10, 331. [Google Scholar] [CrossRef]
- Núñez-Acuña, G.; Détrée, C.; Gallardo-Escárate, C.; Gonçalves, A.T. Functional Diets Modulate LncRNA-Coding RNAs and Gene Interactions in the Intestine of Rainbow Trout Oncorhynchus mykiss. Mar. Biotechnol. 2017, 19, 287–300. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, S.; Wu, W.; Yu, F.; Chang, W.; Li, P.; Wang, K. Non-Coding RNAs Function as Immune Regulators in Teleost Fish. Front. Immunol. 2018, 9, 2801. [Google Scholar] [CrossRef]
- Cai, J.; Li, L.; Song, L.; Xie, L.; Luo, F.; Sun, S.; Chakraborty, T.; Zhou, L.; Wang, D. Effects of Long Term Antiprogestine Mifepristone (RU486) Exposure on Sexually Dimorphic LncRNA Expression and Gonadal Masculinization in Nile Tilapia (Oreochromis niloticus). Aquat. Toxicol. 2019, 215, 105289. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Muñoz, V.; Váldes, J.A.; Gallardo-Escárate, C. Transcriptome Profiling of Long Non-Coding RNAs During the Atlantic Salmon Smoltification Process. Mar. Biotechnol. 2021, 23, 308–320. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, J.; Liu, B.; Huang, Y.; Li, S.; Wen, H.; Zhang, M.; Li, J.; Li, Y.; He, F. Identification and Characterization of LncRNAs Related to the Muscle Growth and Development of Japanese Flounder (Paralichthys Olivaceus). Front. Genet. 2020, 11, 1034. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, M.; Lian, D.; Li, Y.; Li, Y.; Wang, J.; Deng, S.; Yu, K.; Lian, Z. Non-Coding RNA Regulates the Myogenesis of Skeletal Muscle Satellite Cells, Injury Repair and Diseases. Cells 2019, 8, 988. [Google Scholar] [CrossRef] [PubMed]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Montserrat, N.; Riera-Codina, M.; Capilla, E.; Navarro, I.; Gutiérrez, J. Contribution of in Vitro Myocytes Studies to Understanding Fish Muscle Physiology. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2016, 199, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Bower, N.I.; Johnston, I.A. Transcriptional Regulation of the IGF Signaling Pathway by Amino Acids and Insulin-like Growth Factors during Myogenesis in Atlantic Salmon. PLoS ONE 2010, 5, e11100. [Google Scholar] [CrossRef] [PubMed]
- Iresjö, B.M.; Diep, L.; Lundholm, K. Initiation of Muscle Protein Synthesis Was Unrelated to Simultaneously Upregulated Local Production of IGF-1 by Amino Acids in Non-Proliferating L6 Muscle Cells. PLoS ONE 2022, 17, e0270927. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Delafontaine, P. Mechanisms of IGF-1-Mediated Regulation of Skeletal Muscle Hypertrophy and Atrophy. Cells 2020, 9, 1970. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Mammucari, C. Regulation of Skeletal Muscle Growth by the IGF1-Akt/PKB Pathway: Insights from Genetic Models. Skelet. Muscle 2011, 1, 1–14. [Google Scholar] [CrossRef]
- Deldicque, L.; Theisen, D.; Francaux, M. Regulation of MTOR by Amino Acids and Resistance Exercise in Skeletal Muscle. Eur. J. Appl. Physiol. 2005, 94, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, E. Mechanisms of Amino Acid Sensing in MTOR Signaling Pathway. Nutr. Res. Pract. 2009, 3, 64. [Google Scholar] [CrossRef]
- Horsley, V.; Pavlath, G.K. Forming a Multinucleated Cell: Molecules That Regulate Myoblast Fusion. Cells Tissues Organs 2004, 176, 67–78. [Google Scholar] [CrossRef]
- Murgia, M.; Nogara, L.; Baraldo, M.; Reggiani, C.; Mann, M.; Schiaffino, S. Protein Profile of Fiber Types in Human Skeletal Muscle: A Single-Fiber Proteomics Study. Skelet. Muscle 2021, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Garcia de la serrana, D.; Macqueen, D.J. Insulin-like Growth Factor-Binding Proteins of Teleost Fishes. Front. Endocrinol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Salant, G.M.; Tat, K.L.; Goodrich, J.A.; Kugel, J.F. MiR-206 Knockout Shows It Is Critical for Myogenesis and Directly Regulates Newly Identified Target mRNAs. RNA Biol. 2020, 17, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Wang, X.; Sun, Q.; Papakonstantinou, E.; Sʼng, C.; Tamm, M.; Stolz, D.; Roth, M. IgE Downregulates PTEN through MicroRNA-21-5p and Stimulates Airway Smooth Muscle Cell Remodeling. Int. J. Mol. Sci. 2019, 20, 875. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Pan, Y.; Zhai, C.; Zhu, Y.; Ke, R.; Shi, W.; Wang, J.; Yan, X.; Su, X.; Song, Y.; et al. Activation of Peroxisome Proliferation–Activated Receptor-γ Inhibits Transforming Growth Factor-Β1-Induced Airway Smooth Muscle Cell Proliferation by Suppressing Smad–MiR-21 Signaling. J. Cell. Physiol. 2019, 234, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K.; Dandara, C. The Extracellular Matrix: Its Composition, Function, Remodeling, and Role in Tumorigenesis. Biomimetics 2023, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.; Ge, Y.; Chen, J. MicroRNA-146b Promotes Myogenic Differentiation and Modulates Multiple Gene Targets in Muscle Cells. PLoS ONE 2014, 9, e100657. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Q.; Wang, B.B.; Wu, W.J.; Wei, J.; Li, P.; Huang, R. MiR-22 Regulates C2C12 Myoblast Proliferation and Differentiation by Targeting TGFBR1. Eur. J. Cell Biol. 2018, 97, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Black, B.L.; Derynck, R. TGF-β Inhibits Muscle Differentiation through Functional Repression of Myogenic Transcription Factors by Smad3. Genes Dev. 2001, 15, 2950. [Google Scholar] [CrossRef]
- Girardi, F.; Taleb, A.; Ebrahimi, M.; Datye, A.; Gamage, D.G.; Peccate, C.; Giordani, L.; Millay, D.P.; Gilbert, P.M.; Cadot, B.; et al. TGFβ Signaling Curbs Cell Fusion and Muscle Regeneration. Nat. Commun. 2021, 12, 750. [Google Scholar] [CrossRef]
- Kim, H.K.; Lee, Y.S.; Sivaprasad, U.; Malhotra, A.; Dutta, A. Muscle-Specific MicroRNA MiR-206 Promotes Muscle Differentiation. J. Cell Biol. 2006, 174, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Shi, Y.; He, H.; Cai, M.; Xiao, W.; Yang, X.; Chen, S.; Jia, X.; Wang, J.; Lai, S. MiR-221 Modulates Skeletal Muscle Satellite Cells Proliferation and Differentiation. Vitr. Cell. Dev. Biol.-Anim. 2018, 54, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Kappeler, B.I.G.; Regitano, L.C.A.; Poleti, M.D.; Cesar, A.S.M.; Moreira, G.C.M.; Gasparin, G.; Coutinho, L.L. MiRNAs Differentially Expressed in Skeletal Muscle of Animals with Divergent Estimated Breeding Values for Beef Tenderness. BMC Mol. Biol. 2019, 20, 1. [Google Scholar] [CrossRef]
- Marceca, G.P.; Nigita, G.; Calore, F.; Croce, C.M. MicroRNAs in Skeletal Muscle and Hints on Their Potential Role in Muscle Wasting During Cancer Cachexia. Front. Oncol. 2020, 10, 607196. [Google Scholar] [CrossRef]
- Yang, J.H.; Chang, M.W.; Pandey, P.R.; Tsitsipatis, D.; Yang, X.; Martindale, J.L.; Munk, R.; De, S.; Abdelmohsen, K.; Gorospe, M. Interaction of OIP5-AS1 with MEF2C MRNA Promotes Myogenic Gene Expression. Nucleic Acids Res. 2020, 48, 12943–12956. [Google Scholar] [CrossRef]
- Dimartino, D.; Colantoni, A.; Ballarino, M.; Martone, J.; Mariani, D.; Danner, J.; Bruckmann, A.; Meister, G.; Morlando, M.; Bozzoni, I. The Long Non-Coding RNA Lnc-31 Interacts with Rock1 MRNA and Mediates Its YB-1-Dependent Translation. Cell Rep. 2018, 23, 733–740. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, J.; Jiang, R.; Wei, X.; Song, C.; Huang, Y.; Lan, X.; Lei, C.; Ma, Y.; Hu, L.; et al. Long Non-Coding RNA Profiling Reveals an Abundant MDNCR That Promotes Differentiation of Myoblasts by Sponging MiR-133a. Mol. Ther.-Nucleic Acids 2018, 12, 610–625. [Google Scholar] [CrossRef]
- Wang, Y.; Pang, W.J.; Wei, N.; Xiong, Y.; Wu, W.J.; Zhao, C.Z.; Shen, Q.W.; Yang, G.S. Identification, Stability and Expression of Sirt1 Antisense Long Non-Coding RNA. Gene 2014, 539, 117–124. [Google Scholar] [CrossRef]
- Song, C.; Wang, J.; Ma, Y.; Yang, Z.; Dong, D.; Li, H.; Yang, J.; Huang, Y.; Plath, M.; Ma, Y.; et al. Linc-Smad7 Promotes Myoblast Differentiation and Muscle Regeneration via Sponging MiR-125b. Epigenetics 2018, 13, 591. [Google Scholar] [CrossRef]
- Wang, S.; Jin, J.; Xu, Z.; Zuo, B. Functions and Regulatory Mechanisms of LncRNAs in Skeletal Myogenesis, Muscle Disease and Meat Production. Cells 2019, 8, 1107. [Google Scholar] [CrossRef]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long Non-Coding RNAs: Mechanism of Action and Functional Utility. Non-Coding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, J.; Xiao, J.; Yang, L.; Cai, M.; Shen, H.; Chen, X.; Ma, Y.; Hu, S.; Wang, Z.; et al. Lnc-Mg Is a Long Non-Coding RNA That Promotes Myogenesis. Nat. Commun. 2017, 8, 14718. [Google Scholar] [CrossRef] [PubMed]
- Militello, G.; Hosen, M.R.; Ponomareva, Y.; Gellert, P.; Weirick, T.; John, D.; Hindi, S.M.; Mamchaoui, K.; Mouly, V.; Döring, C.; et al. A Novel Long Non-Coding RNA Myolinc Regulates Myogenesis through TDP-43 and Filip1. J. Mol. Cell Biol. 2018, 10, 102–117. [Google Scholar] [CrossRef]
- Fauconneau, B.; Paboeuf, G. Effect of Fasting and Refeeding on in Vitro Muscle Cell Proliferation in Rainbow Trout (Oncorhynchus mykiss). Cell Tissue Res. 2000, 301, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Montserrat, N.; Sánchez-Gurmaches, J.; Garcia de la serrana, D.; Navarro, I.; Gutiérrez, J. IGF-I Binding and Receptor Signal Transduction in Primary Cell Culture of Muscle Cells of Gilthead Sea Bream: Changes throughout in Vitro Development. Cell Tissue Res. 2007, 330, 503–513. [Google Scholar] [CrossRef]
- Garcia de la serrana, D.; Johnston, I.A. Expression of Heat Shock Protein (Hsp90) Paralogues Is Regulated by Amino Acids in Skeletal Muscle of Atlantic Salmon. PLoS ONE 2013, 8, e74295. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 15 January 2023).
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA Target Prediction Easy, Fast and Flexible. Nucleic Acids Res. 2006, 34, W451-4. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegrant Graphics for Data Analysis, 2nd ed.; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]


| AA vs. CTR | ||||
|---|---|---|---|---|
| GO Term | Description | FDR | ||
| Upregulated | Biological Process | 0007049 | Cell cycle | 5.87 × 10−45 |
| 0006260 | DNA replication | 2.12 × 10−19 | ||
| 0007010 | Cytoskeleton organization | 4.82 × 10−08 | ||
| 0003012 | Muscle system process | 1.87 × 10−07 | ||
| 0042692 | Muscle cell differentiation | 3.52 × 10−05 | ||
| 0045214 | Sarcomere organization | 4.19 × 10−07 | ||
| Molecular Function | 0008092 | Cytoskeletal protein binding | 1.61 × 10−10 | |
| 0003688 | DNA replication origin binding | 3.47 × 10−07 | ||
| 0005515 | Protein binding | 8.75 × 10−06 | ||
| 0005524 | ATP binding | 0.0074 | ||
| 0016787 | Hydrolase activity | 0.026 | ||
| Cellular Component | 0043232 | Intracellular non-membrane-bounded organelle | 2.53 × 10−30 | |
| 0043292 | Contractile fiber | 2.15 × 10−18 | ||
| 0030017 | Sarcomere | 6.87 × 10−17 | ||
| 0005654 | Nucleoplasm | 0.0279 | ||
| Downregulated | Biological Process | 0032870 | Cellular response to hormone | 0.0043 |
| 0043473 | Pigmentation | 0.008 | ||
| 0034219 | Carbohydrate transmembrane transport | 0.0215 | ||
| Molecular Function | 0015293 | Symporter activity | 0.0088 | |
| 0008083 | Growth factor activity | 0.0430 | ||
| 0005125 | Cytokine activity | 0.0430 | ||
| 0005539 | Glycosaminoglycan binding | 0.0430 | ||
| Cellular Component | 0005576 | Extracellular region | 0.0110 | |
| 0110165 | Cellular anatomical entity | 0.0110 | ||
| 0031082 | BLOC complex | 0.0416 | ||
| Igf-1 vs. CTR | ||||
| Upregulated | Biological Process | 0042692 | Muscle cell differentiation | 0.0026 |
| 0055001 | Muscle cell development | 0.0120 | ||
| 0061061 | Muscle structure development | 0.0120 | ||
| 0009987 | Cellular process | 0.0120 | ||
| Cellular component | 0030016 | Myofibril | 0.00029 | |
| 0030017 | Sarcomere | 0.00029 | ||
| 0099512 | Supramolecular fiber | 0.00029 | ||
| 0015629 | Actin cytoskeleton | 0.0074 | ||
| Downregulated | Molecular Function | 0005539 | Glycosaminoglycan binding | 0.0250 |
| Cellular Component | 0005576 | Extracellular region | 0.0003 | |
| GO Term | Description | FDR | ||
|---|---|---|---|---|
| AA vs. CTR | ||||
| Upregulated | Biological Process | 0009888 | Tissue development | 0.034 |
| 0030163 | Protein catabolic process | 0.034 | ||
| 0097435 | Actin cytoskeleton organization | 0.034 | ||
| 0006260 | DNA replication | 0.034 | ||
| Molecular Function | 0004298 | Threonine-type endopeptidase activity | 0.001 | |
| Cellular Component | 0005622 | Intracellular | 0.0007 | |
| 0032991 | Protein-containing complex | 0.0086 | ||
| Downregulated | Biological Process | 0043473 | Pigmentation | 0.0002 |
| 0019262 | N-acetylneuraminate catabolic process | 0.026 | ||
| Molecular Function | 0016798 | Hydrolase activity, acting on glycosyl bonds | 0.031 | |
| 0005520 | Insulin-like growth factor binding | 0.041 | ||
| Cellular Component | 0110165 | Cellular anatomical entity | 0.010 | |
| 0012505 | Endomembrane system | 0.039 | ||
| 0005773 | Vacuole | 0.000 | ||
| Igf-1 vs. CTR | ||||
| Upregulated | Biological Process | 0048731 | System development | 0.045 |
| 0006259 | DNA metabolic process | 0.045 | ||
| 0055001 | Muscle cell development | 0.008 | ||
| Cellular Component | 0005856 | Cytoskeleton | 0.019 | |
| 0030017 | Sarcomere | 0.000 | ||
| Downregulated | Cellular Component | 0005576 | Extracellular regions | 0.000 |
| lncRNAs ID | miRNAs | Correlation Index | Energy (ndG) |
|---|---|---|---|
| ENSSAUG00010001802 | miR-27a; miR-29d; miR-29b | −0.88; −0.86; −0.87 | −29.1; −26.2; −27.1 |
| ENSSAUG00010017848 | miR-122; miR-92a; miR-29a; miR-29d; miR-29b; miR-203a; miR-25; miR-31 | −0.88; −0.80; −0.86; −0.92; −0.84; −0.91; −0.84; −0.81 | −26.7; −32.5; −32.2; −32.2; −26.6; −28.1; −27.7; −28.6 |
| ENSSAUG00010024948 | miR-122; miR-92a; miR-10c; miR-10d; miR-27a; miR-29b; miR-31 | −0.92; −0.89; −0.87; −0.87; −0.82; −0.90; −0.93 | −26.0; −27.6; −25.5; −25.5; −30.6; −32.3; −26.0 |
| ENSSAUG00010012228 | miR-338; miR-133a; miR-133b; miR-206; miR-17a; miR-125a; miR-106; mir-217 | −0.80; −0.92; −0.91; −0.87; −0.80; −0.82; −0.90; −0.91 | −27.1; −26.1; −26.1; −28.6; −27.8; −28.6; −28.1; −29.0 |
| ENSSAUG00010000237 | miR-125b | −0.83 | −28.5 |
| ENSSAUG00010012182 | miR-7a; miR-338; miR-133a; miR-133b; miR-206; miR-106; miR-17a; miR-125a | −0.93; −0.80; −0.92; −0.91; −0.87; −0.90; −0.80; −0.82 | −29.0; −27.1; −26.1; −26.1; −28.6; −28.6; −27.8; −28.6 |
| ENSSAUG00010012549 | miR-17a | −0.80 | −26.5 |
| ENSSAUG00010015941 | miR-206; miR-17a; miR-125b; mir-145; miR-454 | −0.86; −0.83; −0.84; −0.83; −0.86 | −26.4; −30.2; −30.2; −25.6; −29.7 |
| ENSSAUG00010016074 | miR-15a; miR-19b; miR-217; miR-34 | −0.89; −0.81; −0.85; −0.81 | −27; −25.3; −27.6; −27.5 |
| ENSSAUG00010016143 | miR-133a | −0.82 | −25.1 |
| ENSSAUG00010017089 | miR-206; miR-106; miR-128; miR-17a | −0.95; −0.82; −0.88; −0.91 | −28.6; −26.8; −30.7; −27.8 |
| ENSSAUG00010016280 | miR-122; miR-92a; miR-25 | −0.86; −0.81; −0.80 | −29.6; −27.8; −25.1 |
| ENSSAUG00010002983 | miR-15a | −0.90 | −31.2 |
| ENSSAUG00010008657 | miR-338; miR-15a; miR-34; miR-7147 | −0.88; −0.85; −0.88; −0.90 | −33.3; −29.4; −25.6; −25.8 |
| ENSSAUG00010022074 | miR-128; miR-365; miR-454; miR-19a; miR-15a; miR-34; miR-7147 | −0.83; −0.83; −0.82; −0.82; −0.80; −0.83; −0.86 | −31.3; −29.7; −25.9; −27.5; −32.9; −28.6; −27.2 |
| ENSSAUG00010013187 | miR-30e; miR-29a; miR-29d; miR-22b; miR-30a | −0.91; −0.90; −0.83; −0.80; −0.90 | −26.8; −28.8; −28.8; −28.3; −29.5 |
| ENSSAUG00010013622 | miR-30e; miR-29d; miR-8160ba; miR-30a | −0.85; −0.84; −0.90; −0.82 | −27.0; −27.3; −25.2; −26.5; |
| ENSSAUG00010015504 | miR-27d; miR-30a | −0.85; −0.81 | −29.1; −30.1 |
| ENSSAUG00010016109 | miR-30e; miR-25; miR-27d; miR-27a | −0.81; −0.82; −0.84; −0.85 | −31.7; −25.8; −25.5; −26.0 |
| ENSSAUG00010001416 | miR-29b | −0.83 | −25.2 |
| ENSSAUG00010017066 | let-7g | −0.81 | −28.0 |
| ENSSAUG00010002786 | miR-10926; miR-29d; miR-8160ba | −0.80; −0.83; −0.97 | −28.1; −28.7; −26.3 |
| ENSSAUG00010004711 | miR-8160ba | −0.89 | −27.1 |
| ENSSAUG00010026349 | miR-10926; miR-22b; miR-29a; miR-29d; miR-551; miR-8160ba | −0.81; −0.82; −0.82; −0.82; −0.81; −0.82 | −29.4; −27; −26.2; −26.2; −25.6; −25.6 |
| ENSSAUG00010009596 | miR-128; mir-365; miR-7550 | −0.95; −0.90; −0.83 | −27.7; −27.3; −25.5 |
| ENSSAUG00010015789 | miR-128; miR-365; miR-125b | −0.89; −0.82; −0.81 | −28.9; −32.7; −25.2 |
| ENSSAUG00010020704 | miR-128; miR-365; miR-26b; miR-454; miR-19a; miR-15a; miR-34; miR-7147 | −0.83; −0.83; −0.86; −0.82; −0.82; −0.80; −0.83; −0.85 | −31.3; −29.2; −26.3; −25.9; −27.5; −32.9; −28.6; −27.2 |
| ENSSAUG00010010920 | miR-139; miR-27d; miR-8160ba | −0.81; −0.85; −0.83 | −28.5; −26.1; −25.9 |
| ENSSAUG00010003663 | miR-15a; miR-301b; miR-33b; miR-34; miR-7147 | −0.91; −0.81; −0.81; −0.84; −0.85 | −29.1; −25.3; −26.1; −33.2; −28.1 |
| ENSSAUG00010016209 | miR-27a; miR-122; miR-92a | −0.85; −0.86; −0.81 | −26.0; −29.6; −27.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Pérez, I.; Duran, B.O.S.; Dal-Pai-Silva, M.; Garcia de la serrana, D. Exploring the Integrated Role of miRNAs and lncRNAs in Regulating the Transcriptional Response to Amino Acids and Insulin-like Growth Factor 1 in Gilthead Sea Bream (Sparus aurata) Myoblasts. Int. J. Mol. Sci. 2024, 25, 3894. https://doi.org/10.3390/ijms25073894
García-Pérez I, Duran BOS, Dal-Pai-Silva M, Garcia de la serrana D. Exploring the Integrated Role of miRNAs and lncRNAs in Regulating the Transcriptional Response to Amino Acids and Insulin-like Growth Factor 1 in Gilthead Sea Bream (Sparus aurata) Myoblasts. International Journal of Molecular Sciences. 2024; 25(7):3894. https://doi.org/10.3390/ijms25073894
Chicago/Turabian StyleGarcía-Pérez, Isabel, Bruno Oliveira Silva Duran, Maeli Dal-Pai-Silva, and Daniel Garcia de la serrana. 2024. "Exploring the Integrated Role of miRNAs and lncRNAs in Regulating the Transcriptional Response to Amino Acids and Insulin-like Growth Factor 1 in Gilthead Sea Bream (Sparus aurata) Myoblasts" International Journal of Molecular Sciences 25, no. 7: 3894. https://doi.org/10.3390/ijms25073894
APA StyleGarcía-Pérez, I., Duran, B. O. S., Dal-Pai-Silva, M., & Garcia de la serrana, D. (2024). Exploring the Integrated Role of miRNAs and lncRNAs in Regulating the Transcriptional Response to Amino Acids and Insulin-like Growth Factor 1 in Gilthead Sea Bream (Sparus aurata) Myoblasts. International Journal of Molecular Sciences, 25(7), 3894. https://doi.org/10.3390/ijms25073894










