Reprogramming Chromosome Ends by Functional Histone Acetylation
Abstract
1. Introduction
2. Results
2.1. Human ZSCAN4 Co-Localizes with the Telomere Region
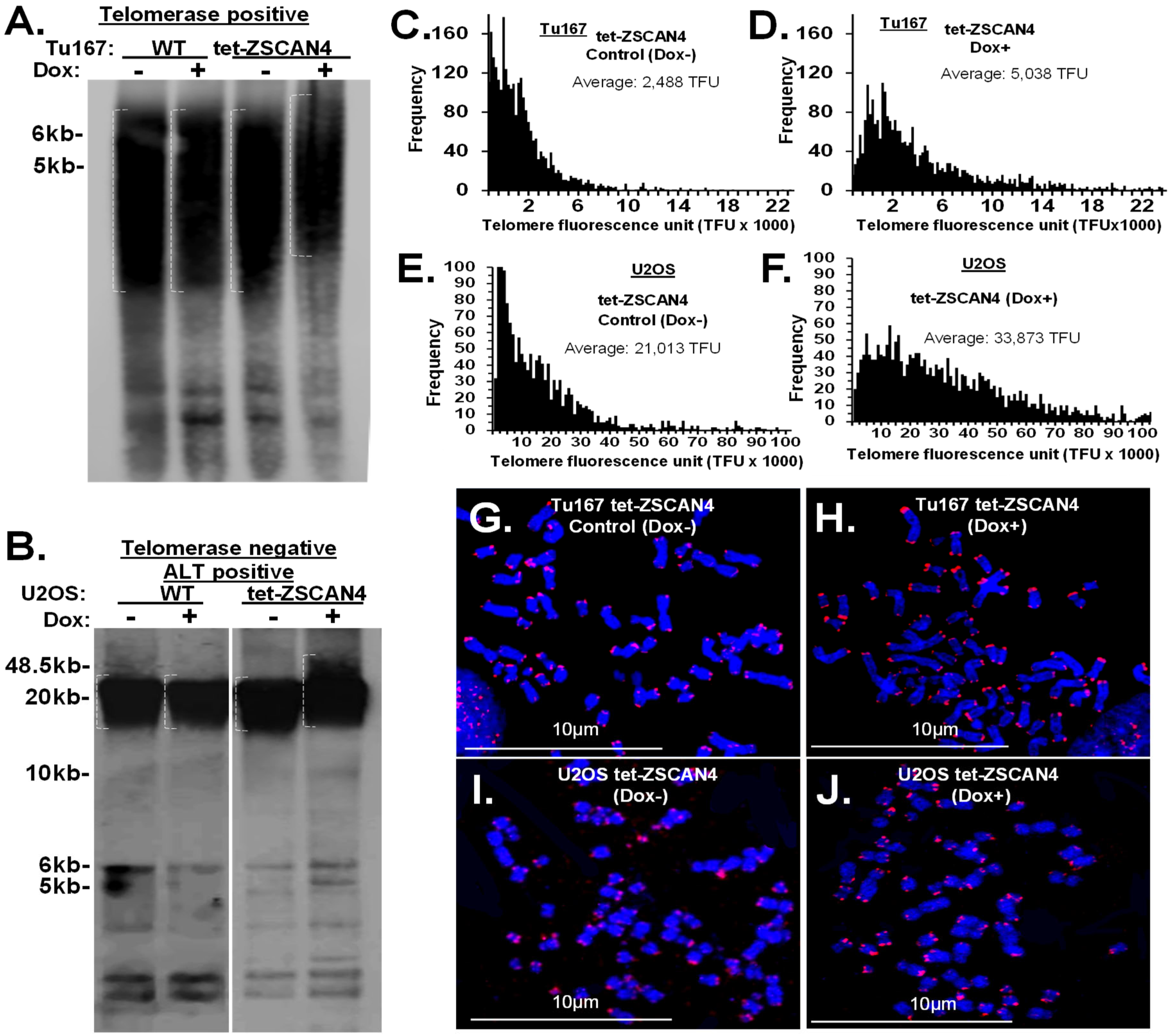
2.2. Human ZSCAN4 Facilitates Telomere Extension
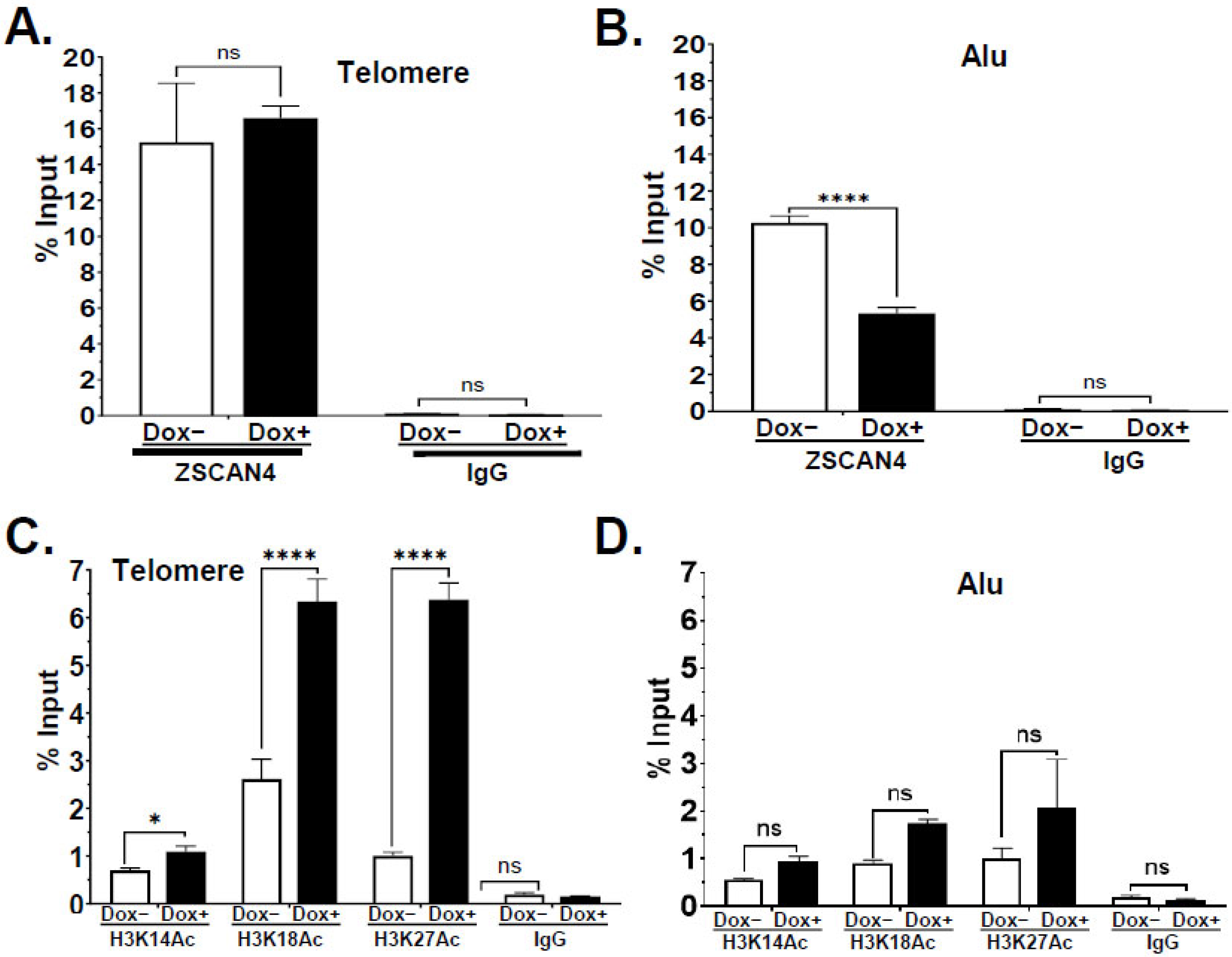
2.3. Telomere-Specific Next-Gen ChIP-Seq Reveals Significant Enrichment of ZSCAN4
2.4. ZSCAN4 Facilitates H3 Histone Acetylation at the Telomere Region
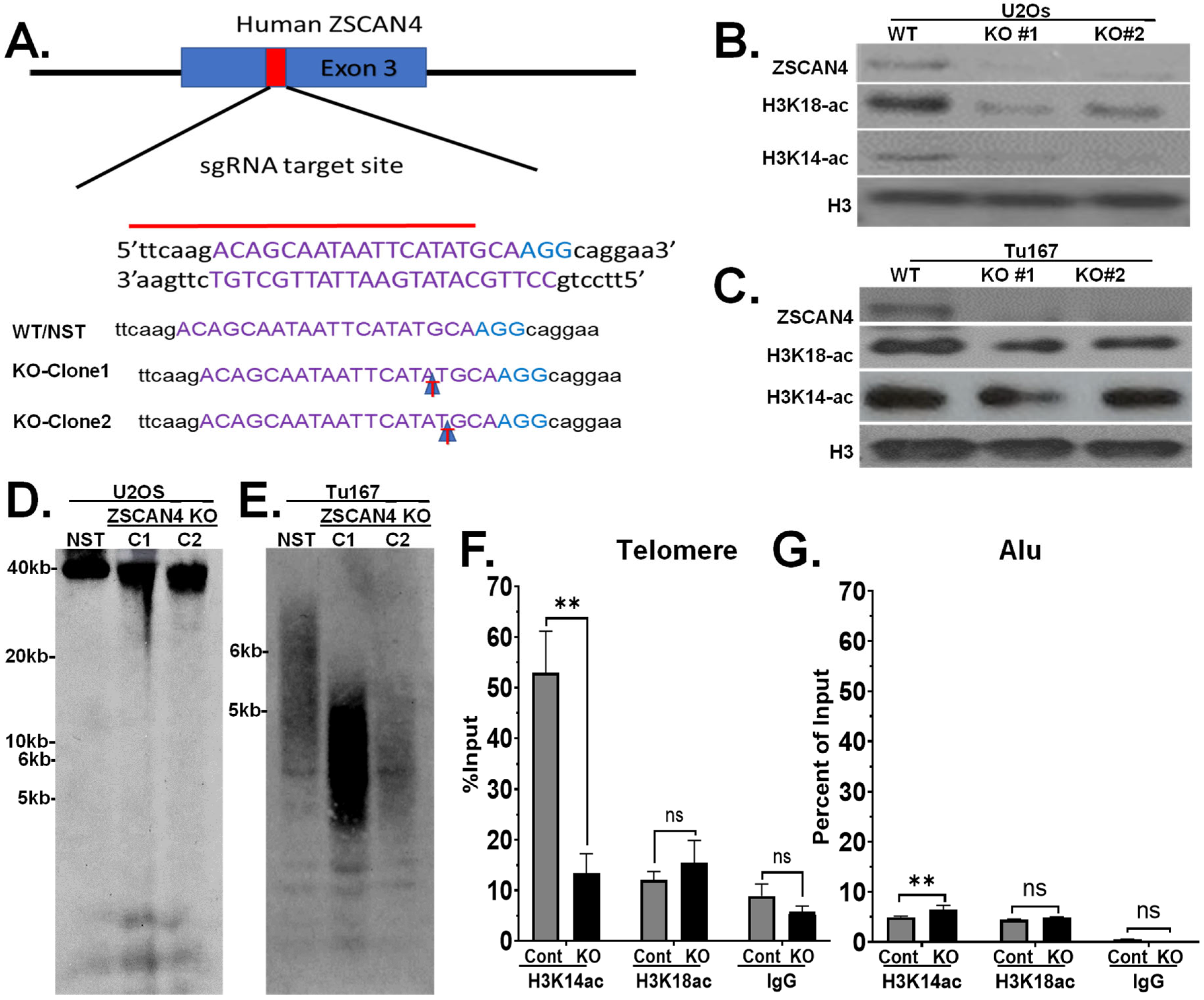
2.5. ZSCAN4 Knockout Leads to Telomere Shortening and a Decrease in H3K14ac
2.6. Telomere Factors Are Maintained during ZSCAN4 Induction
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Generation of Doxycycline Inducible ZSCAN4 Cells
4.3. Generation of ZSCAN4 CRISPR/Cas9 Knockout Clones
4.4. Telomere Quantitative Fluorescence In Situ Hybridization (Q-FISH)
4.5. Co-Immunohistochemistry with Telomere FISH
4.6. Terminal Restriction Fragment (TRF) Length Southern Blot Analysis
4.7. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.8. Quantitative Polymerase Chain Reaction (qPCR) Analyses
4.9. Chromatin Immunoprecipitation (ChIP)-qPCR
4.10. ZSCAN4-ChIP Followed by Next Generation Sequencing Analyses
4.11. Dot Blot Analyses
4.12. Telomerase Activity Measurement
4.13. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Stanulis-Praeger, B.M. Cellular senescence revisited: A review. Mech. Ageing Dev. 1987, 38, 1–48. [Google Scholar] [CrossRef]
- Campisi, J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001, 11, S27–S31. [Google Scholar] [CrossRef]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Allsopp, R.C.; Vaziri, H.; Patterson, C.; Goldstein, S.; Younglai, E.V.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA 1992, 89, 10114–10118. [Google Scholar] [CrossRef]
- Olovnikov, A. A theory of marginotomy: The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef]
- Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007, 8, 299–309. [Google Scholar] [CrossRef]
- Zalzman, M.; Falco, G.; Sharova, L.V.; Nishiyama, A.; Thomas, M.; Lee, S.-L.; Stagg, C.A.; Hoang, H.G.; Yang, H.-T.; Indig, F.E.; et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 2010, 464, 858–863. [Google Scholar] [CrossRef]
- Park, H.-S.; Hwang, I.; Choi, K.-A.; Jeong, H.; Lee, J.-Y.; Hong, S. Generation of induced pluripotent stem cells without genetic defects by small molecules. Biomaterials 2015, 39, 47–58. [Google Scholar] [CrossRef]
- Falco, G.; Lee, S.-L.; Stanghellini, I.; Bassey, U.C.; Hamatani, T.; Ko, M.S. Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007, 307, 539–550. [Google Scholar] [CrossRef]
- Dan, J.; Zhou, Z.; Wang, F.; Wang, H.; Guo, R.; Keefe, D.L.; Liu, L. Zscan4 Contributes to Telomere Maintenance in Telomerase-Deficient Late Generation Mouse ESCs and Human ALT Cancer Cells. Cells 2022, 11, 456. [Google Scholar] [CrossRef]
- Dan, J.; Rousseau, P.; Hardikar, S.; Veland, N.; Wong, J.; Autexier, C.; Chen, T. Zscan4 Inhibits Maintenance DNA Methylation to Facilitate Telomere Elongation in Mouse Embryonic Stem Cells. Cell Rep. 2017, 20, 1936–1949. [Google Scholar] [CrossRef]
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic Genome Activation During the Maternal-to-Zygotic Transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef]
- Horton, J.K.; Wilson, S.H. Strategic Combination of DNA-Damaging Agent and PARP Inhibitor Results in Enhanced Cytotoxicity. Front. Oncol. 2013, 3, 257. [Google Scholar] [CrossRef]
- Jiang, J.; Lv, W.; Ye, X.; Wang, L.; Zhang, M.; Yang, H.; Okuka, M.; Zhou, C.; Zhang, X.; Liu, L.; et al. Zscan4 promotes genomic stability during reprogramming and dramat-ically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013, 23, 92–106. [Google Scholar] [CrossRef]
- Nakai-Futatsugi, Y.; Niwa, H. Zscan4 Is Activated after Telomere Shortening in Mouse Embryonic Stem Cells. Stem Cell Rep. 2016, 6, 483–495. [Google Scholar] [CrossRef]
- Hirata, T.; Amano, T.; Nakatake, Y.; Amano, M.; Piao, Y.; Hoang, H.G.; Ko, M.S.H. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci. Rep. 2012, 2, 208. [Google Scholar] [CrossRef]
- Amano, T.; Hirata, T.; Falco, G.; Monti, M.; Sharova, L.V.; Amano, M.; Sheer, S.; Hoang, H.G.; Piao, Y.; Stagg, C.A.; et al. Zscan4 restores the developmental potency of embryonic stem cells. Nat. Commun. 2013, 4, 1966. [Google Scholar] [CrossRef]
- Storm, M.P.; Kumpfmueller, B.; Bone, H.K.; Buchholz, M.; Ripoll, Y.S.; Chaudhuri, J.B.; Niwa, H.; Tosh, D.; Welham, M.J. Zscan4 Is Regulated by PI3-Kinase and DNA-Damaging Agents and Directly Interacts with the Transcriptional Repressors LSD1 and CtBP2 in Mouse Embryonic Stem Cells. PLoS ONE 2014, 9, e89821. [Google Scholar] [CrossRef]
- Akiyama, T.; Xin, L.; Oda, M.; Sharov, A.A.; Amano, M.; Piao, Y.; Cadet, J.S.; Dudekula, D.B.; Qian, Y.; Wang, W.; et al. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015, 22, 307–318. [Google Scholar] [CrossRef]
- Lee, K.; Gollahon, L.S. Zscan4 interacts directly with human Rap1 in cancer cells regardless of telomerase status. Cancer Biol. Ther. 2014, 15, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Gollahon, L.S. ZSCAN4 and TRF1: A functionally indirect interaction in cancer cells independent of telomerase activity. Biochem. Biophys. Res. Commun. 2015, 466, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Fu, D.; Xu, Q.; Cong, X.; Wu, C.; Zhong, X.; Ma, Y.; Lv, Z.; Chen, F.; Han, L.; et al. The senescence-associated secretory phenotype is potentiated by feed-forward regulatory mechanisms involving Zscan4 and TAK1. Nat. Commun. 2018, 9, 1723. [Google Scholar] [CrossRef] [PubMed]
- Portney, B.A.; Arad, M.; Gupta, A.; Brown, R.A.; Khatri, R.; Lin, P.N.; Hebert, A.M.; Angster, K.H.; Silipino, L.E.; Meltzer, W.A.; et al. ZSCAN4 facilitates chromatin remodeling and promotes the cancer stem cell phenotype. Oncogene 2020, 39, 4970–4982. [Google Scholar] [CrossRef] [PubMed]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Guan, J.Z.; Koyanagi, M.; Higuchi, Y.; Makino, N. Aging-associated alteration of telomere length and subtelomeric status in female patients with Parkinson’s disease. J. Neurogenet. 2012, 26, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Dan, J.; Yang, J.; Liu, Y.; Xiao, A.; Liu, L. Roles for Histone Acetylation in Regulation of Telomere Elongation and Two-cell State in Mouse ES Cells. J. Cell Physiol. 2015, 230, 2337–2344. [Google Scholar] [CrossRef] [PubMed]
- Poon, S.S.; Martens, U.M.; Ward, R.K.; Lansdorp, P.M. Telomere length measurements using digital fluorescence microscopy. Cytometry 1999, 36, 267–278. [Google Scholar] [CrossRef]
- Portney, B.A.; Khatri, R.; Meltzer, W.A.; Mariano, J.M.; Zalzman, M. ZSCAN4 is negatively regulated by the ubiquitin-proteasome system and the E3 ubiquitin ligase RNF20. Biochem. Biophys. Res. Commun. 2018, 498, 72–78. [Google Scholar] [CrossRef]
- Srinivasan, R.; Nady, N.; Arora, N.; Hsieh, L.J.; Swigut, T.; Narlikar, G.J.; Wossidlo, M.; Wysocka, J. Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage. Sci. Adv. 2020, 6, eaaz9115. [Google Scholar] [CrossRef]
- Feuerbach, L.; Sieverling, L.; Deeg, K.I.; Ginsbach, P.; Hutter, B.; Buchhalter, I.; Northcott, P.A.; Mughal, S.S.; Chudasama, P.; Glimm, H.; et al. TelomereHunter–In silico estimation of telomere content and composition from cancer genomes. BMC Bioinform. 2019, 20, 272. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Ishiguro, K.-I.; Chikazawa, N.; Ko, S.B.H.; Yukawa, M.; Ko, M.S.H. ZSCAN4-binding motif-TGCACAC is conserved and enriched in CA/TG microsatellites in both mouse and human genomes. DNA Res. 2024, 31, dsad029. [Google Scholar] [CrossRef] [PubMed]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Wang, L.; Zhu, X.; Zhang, Y.; Ou, Q.; Ma, A.; Sheng, F.; Wei, X. Dai, Y. Li, G.; et al. Global mapping of binding sites for phic31 integrase in transgenic maden-darby bovine kidney cells using ChIP-seq. Hereditas 2019, 156, 3. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.D.; Zamudio-Hurtado, A.; Clawson, H.; Kent, W.J.; Haussler, D.; Salama, S.R.; Haeussler, M. The UCSC repeat browser allows discovery and visualization of evolutionary conflict across repeat families. Mob. DNA 2020, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Azuara, V.; Perry, P.; Sauer, S.; Spivakov, M.; Jørgensen, H.F.; John, R.M.; Gouti, M.; Casanova, M.; Warnes, G.; Merkenschlager, M.; et al. Chromatin signatures of pluripotent cell lines. Nature 2006, 8, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Meshorer, E.; Misteli, T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006, 7, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Bibikova, M.; Laurent, L.C.; Ren, B.; Loring, J.F.; Fan, J.-B. Unraveling Epigenetic Regulation in Embryonic Stem Cells. Cell Stem Cell 2008, 2, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Gaspar-Maia, A.; Alajem, A.; Meshorer, E.; Ramalho-Santos, M. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011, 12, 36–47. [Google Scholar] [CrossRef]
- Orkin, S.H.; Hochedlinger, K. Chromatin Connections to Pluripotency and Cellular Reprogramming. Cell 2011, 145, 835–850. [Google Scholar] [CrossRef]
- de Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef]
- Palm, W.; de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H. Epigenetic regulation of telomere chromatin integrity in pluripotent embryonic stem cells. Epigenomics 2010, 2, 639–655. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Greider, C.W.; Henderson, E.; Lee, M.S.; Shampay, J.; Shippen-Lentz, D. Recognition and elongation of telomeres by telomerase. Genome 1989, 31, 553–560. [Google Scholar] [CrossRef]
- Benetti, R.; García-Cao, M.; A Blasco, M. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 2007, 39, 243–250. [Google Scholar] [CrossRef]
- Eckersley-Maslin, M.A.; Svensson, V.; Krueger, C.; Stubbs, T.M.; Giehr, P.; Krueger, F.; Miragaia, R.J.; Kyriakopoulos, C.; Berrens, R.V.; Milagre, I.; et al. MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Rep. 2016, 17, 179–192. [Google Scholar] [CrossRef]
- Liu, T.-J.; Wang, M.; Breau, R.L.; Henderson, Y.; El-Naggar, A.K.; Steck, K.D.; Sicard, M.W.; Clayman, G.L. Apoptosis induction by E2F-1 via adenoviral-mediated gene transfer results in growth suppression of head and neck squamous cell carcinoma cell lines. Cancer Gene Ther. 1999, 6, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Sano, D.; Pickering, C.R.; Jasser, S.A.; Henderson, Y.C.; Clayman, G.L.; Sturgis, E.M.; Ow, T.J.; Lotan, R.; Carey, T.E.; et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin. Cancer Res. 2011, 17, 7248–7264. [Google Scholar] [CrossRef]
- Kunická, Z.; Mucha, I.; Fajkus, J. Telomerase activity in head and neck cancer. Anticancer Res. 2008, 28, 3125–3129. [Google Scholar]
- Gupta, A.; Hwang, B.-J.; Benyamien-Roufaeil, D.; Jain, S.; Liu, S.; Gonzales, R.; Brown, R.A.; Zalzman, M.; Lu, A.-L. Mammalian MutY Homolog (MYH or MUTYH) is Critical for Telomere Integrity under Oxidative Stress. OBM Geriatr. 2022, 6, 1. [Google Scholar] [CrossRef]
- Hwang, B.-J.; Jin, J.; Gunther, R.; Madabushi, A.; Shi, G.; Wilson, G.M.; Lu, A.-L. Association of the Rad9–Rad1–Hus1 checkpoint clamp with MYH DNA glycosylase and DNA. DNA Repair 2015, 31, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Rasband, W.S. ImageJ; U.S. National Institutes of Health: Bethesda, MD, USA, 1997. Available online: https://imagej.nih.gov/ij/ (accessed on 27 March 2024).
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meltzer, W.A.; Gupta, A.; Lin, P.N.; Brown, R.A.; Benyamien-Roufaeil, D.S.; Khatri, R.; Mahurkar, A.A.; Song, Y.; Taylor, R.J.; Zalzman, M. Reprogramming Chromosome Ends by Functional Histone Acetylation. Int. J. Mol. Sci. 2024, 25, 3898. https://doi.org/10.3390/ijms25073898
Meltzer WA, Gupta A, Lin PN, Brown RA, Benyamien-Roufaeil DS, Khatri R, Mahurkar AA, Song Y, Taylor RJ, Zalzman M. Reprogramming Chromosome Ends by Functional Histone Acetylation. International Journal of Molecular Sciences. 2024; 25(7):3898. https://doi.org/10.3390/ijms25073898
Chicago/Turabian StyleMeltzer, W. Alex, Aditi Gupta, Phyo Nay Lin, Robert A. Brown, Daniel S. Benyamien-Roufaeil, Raju Khatri, Anup A. Mahurkar, Yang Song, Rodney J. Taylor, and Michal Zalzman. 2024. "Reprogramming Chromosome Ends by Functional Histone Acetylation" International Journal of Molecular Sciences 25, no. 7: 3898. https://doi.org/10.3390/ijms25073898
APA StyleMeltzer, W. A., Gupta, A., Lin, P. N., Brown, R. A., Benyamien-Roufaeil, D. S., Khatri, R., Mahurkar, A. A., Song, Y., Taylor, R. J., & Zalzman, M. (2024). Reprogramming Chromosome Ends by Functional Histone Acetylation. International Journal of Molecular Sciences, 25(7), 3898. https://doi.org/10.3390/ijms25073898





