PFDN4 as a Prognostic Marker Was Associated with Chemotherapy Resistance through CREBP1/AURKA Pathway in Triple-Negative Breast Cancer
Abstract
:1. Introduction
2. Results
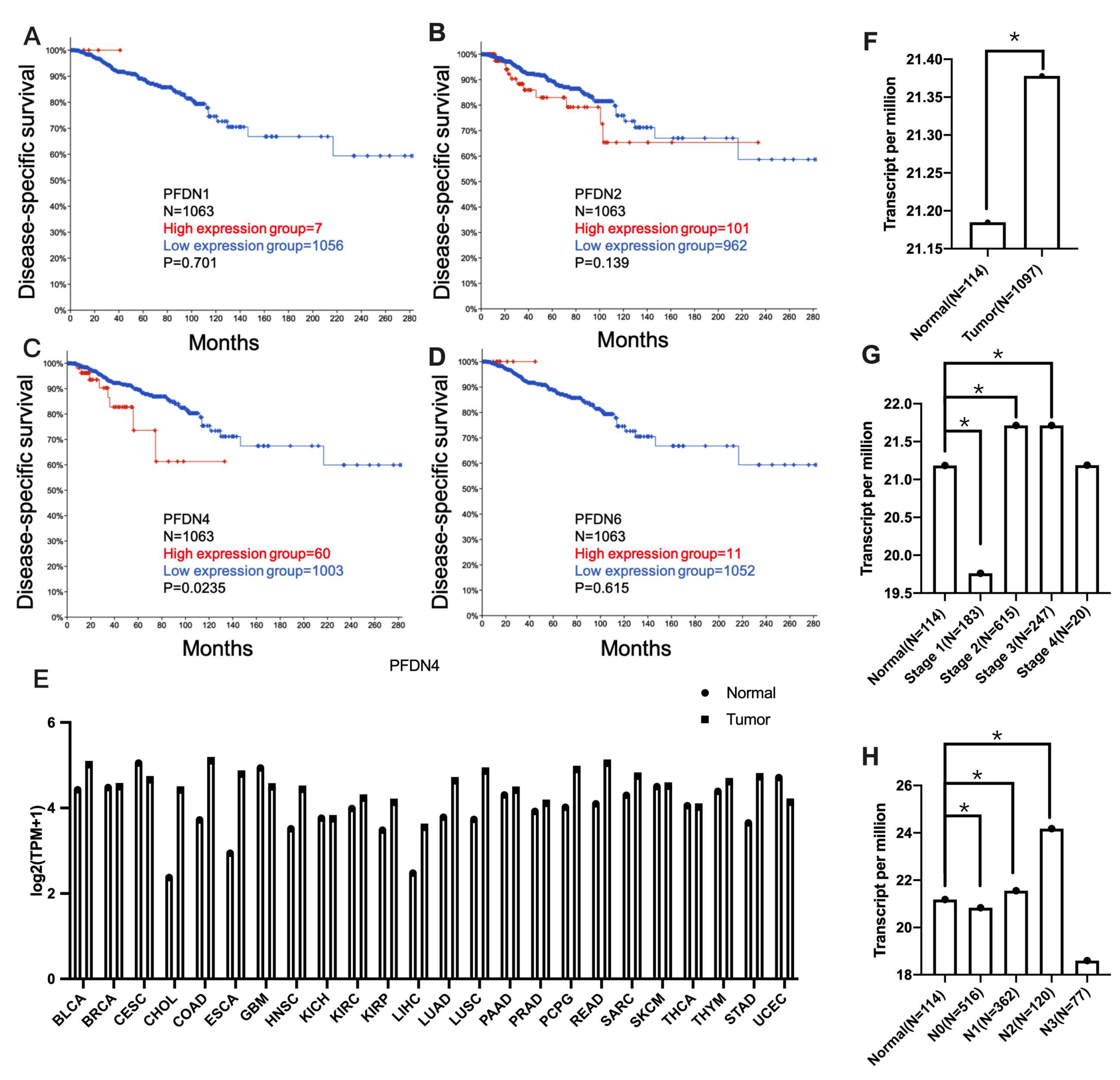
2.1. Association of PFDN with Disease-Specific Survival and Clinicopathological Features in Breast Cancer Subjects
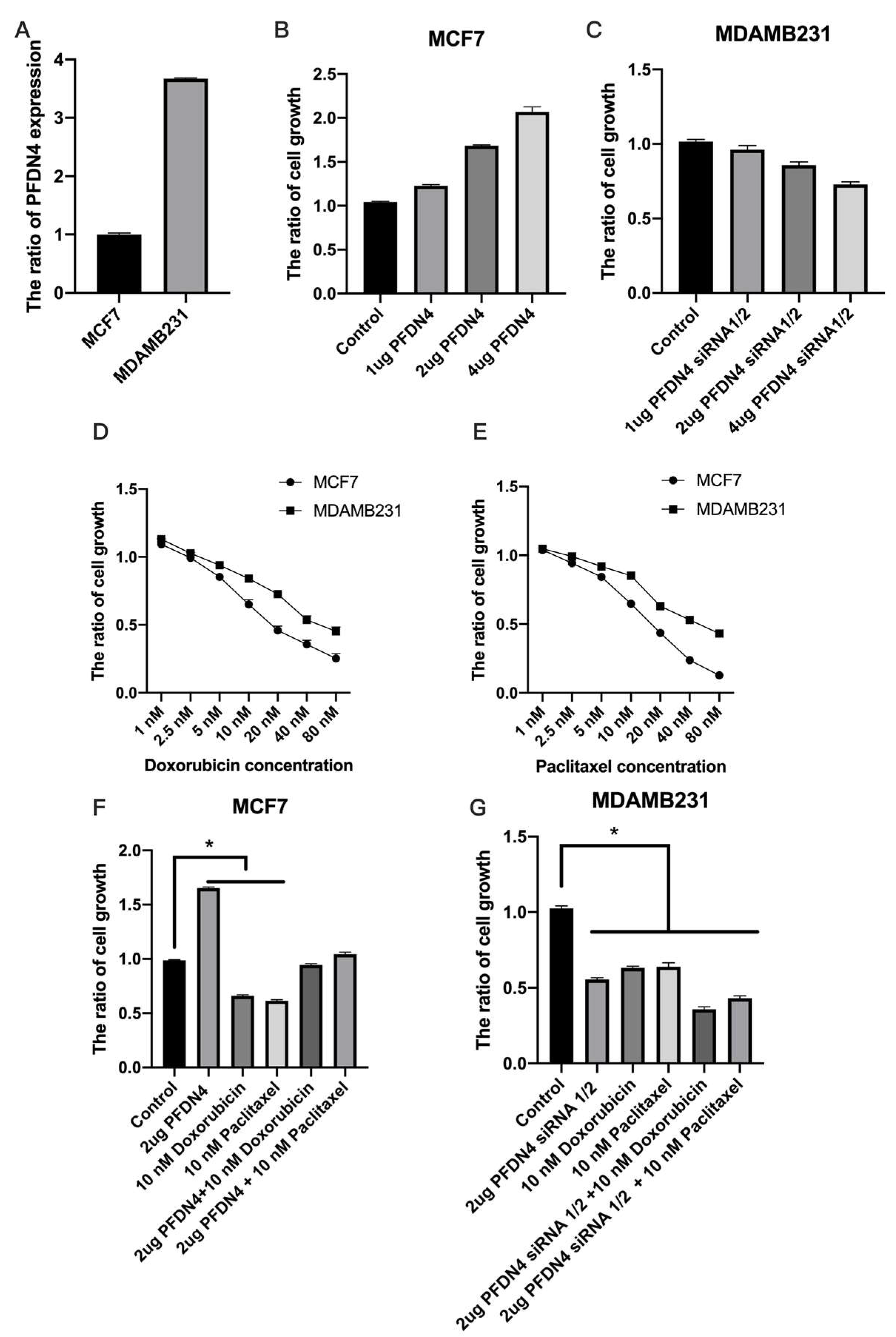
2.2. PFDN4 Mediates Cell Growth and Cell Cycle in Triple-Negative Breast Cancer Cell Lines
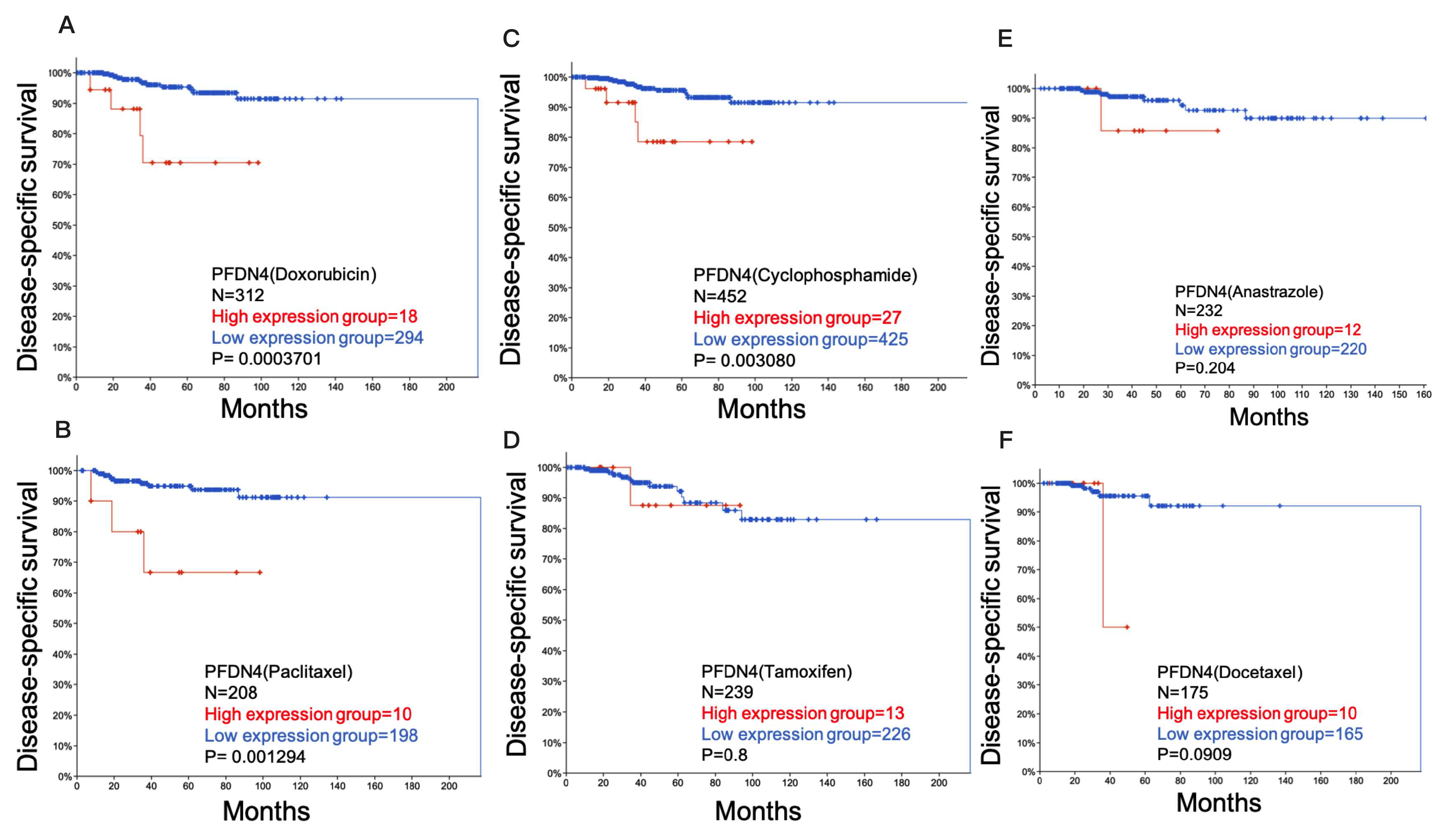
2.3. Role of PFDN4 in Chemotherapy Resistance
2.4. PFDN4 Antagonizes the Effects of Doxorubicin and Paclitaxel in Breast Cancer Cell Lines
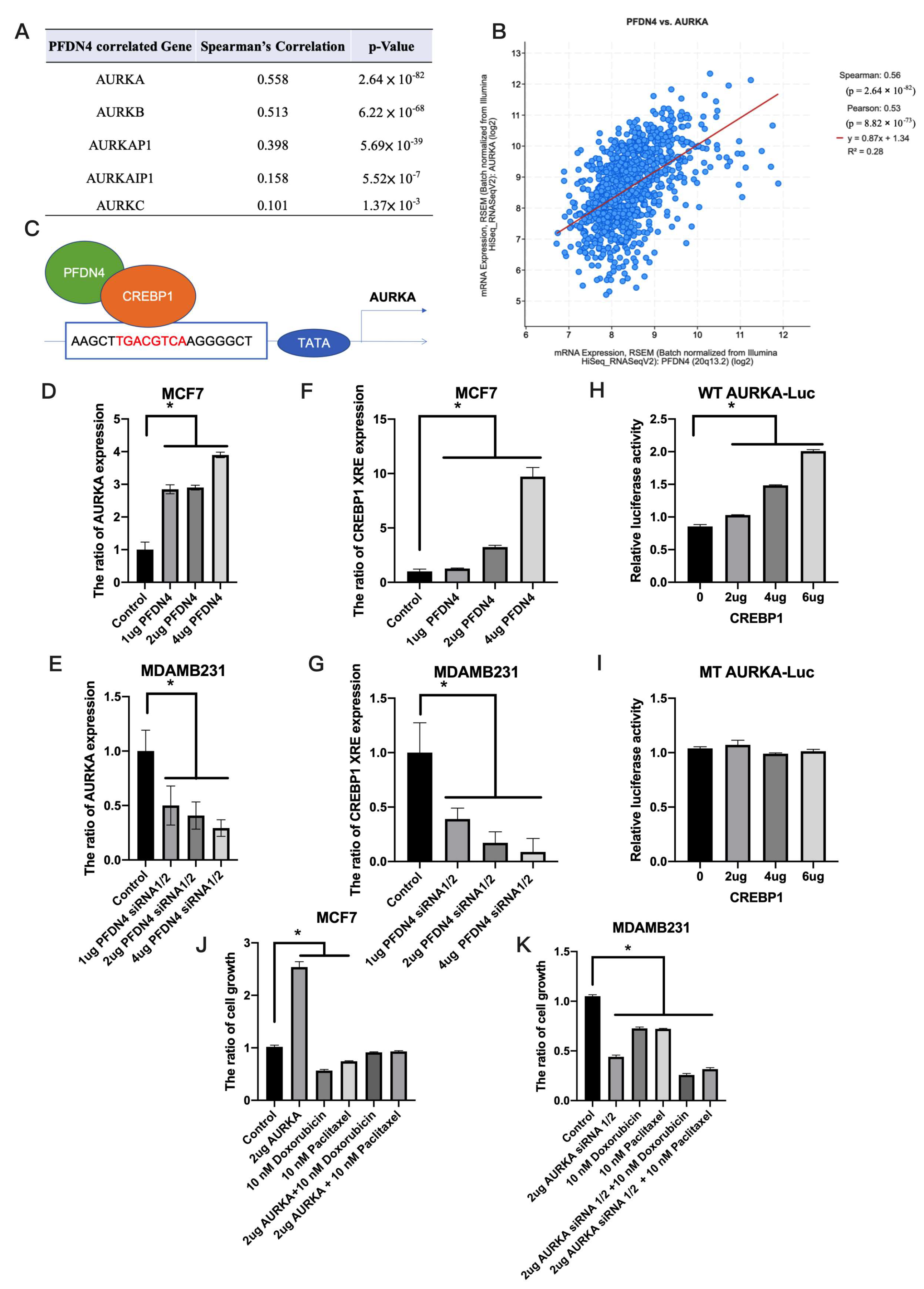
2.5. PFDN4 Regulates Breast Cancer Chemotherapy Resistance through AURKA
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Transfection
4.3. Cell Growth
4.4. QPCR and Chromatin Immunoprecipitation Quantitative Real-Time PCR
4.5. UALCAN
4.6. cBioPortal
4.7. Gene Effect Scores
4.8. Luciferase Report Assay
4.9. Flow Cytometry
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sancar, A.; Lindsey-Boltz, L.A.; Unsal-Kacmaz, K.; Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Cortes, J.; Dent, R.; Pusztai, L.; McArthur, H.; Kummel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; et al. Event-free Survival with Pembrolizumab in Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2022, 386, 556–567. [Google Scholar] [CrossRef]
- Longley, D.B.; Johnston, P.G. Molecular mechanisms of drug resistance. J. Pathol. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Sahlan, M.; Zako, T.; Yohda, M. Prefoldin, a jellyfish-like molecular chaperone: Functional cooperation with a group II chaperonin and beyond. Biophys. Rev. 2018, 10, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Shi, W.; Tang, Y.; Liu, Y.; He, K.; Hu, Y.; Li, J.; Yang, Y.; Song, J. Prefoldin 1 promotes EMT and lung cancer progression by suppressing cyclin A expression. Oncogene 2017, 36, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Alldinger, I.; Dittert, D.; Peiper, M.; Fusco, A.; Chiappetta, G.; Staub, E.; Lohr, M.; Jesnowski, R.; Baretton, G.; Ockert, D.; et al. Gene expression analysis of pancreatic cell lines reveals genes overexpressed in pancreatic cancer. Pancreatology 2005, 5, 370–379. [Google Scholar] [CrossRef]
- Asperger, A.; Renner, C.; Menzel, M.; Gebhardt, R.; Meixensberger, J.; Gaunitz, F. Identification of factors involved in the anti-tumor activity of carnosine on glioblastomas using a proteomics approach. Cancer Investig. 2011, 29, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Lopez, V.; Gonzalez-Peramato, P.; Suela, J.; Serrano, A.; Algaba, F.; Cigudosa, J.C.; Vidal, A.; Bellmunt, J.; Heredero, O.; Sanchez-Carbayo, M. Identification of prefoldin amplification (1q23.3–q24.1) in bladder cancer using comparative genomic hybridization (CGH) arrays of urinary DNA. J. Transl. Med. 2013, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Ostrov, D.A.; Barnes, C.L.; Smith, L.E.; Binns, S.; Brusko, T.M.; Brown, A.C.; Quint, P.S.; Litherland, S.A.; Roopenian, D.C.; Iczkowski, K.A. Characterization of HKE2: An ancient antigen encoded in the major histocompatibility complex. Tissue Antigens 2007, 69, 181–188. [Google Scholar] [CrossRef]
- Narita, R.; Kitaura, H.; Torii, A.; Tashiro, E.; Miyazawa, M.; Ariga, H.; Iguchi-Ariga, S.M. Rabring7 degrades c-Myc through complex formation with MM-1. PLoS ONE 2012, 7, e41891. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Iseda, T.; Hino, O. Identification of a novel protein (VBP-1) binding to the von Hippel-Lindau (VHL) tumor suppressor gene product. Cancer Res. 1996, 56, 2881–2885. [Google Scholar] [PubMed]
- Collins, C.; Volik, S.; Kowbel, D.; Ginzinger, D.; Ylstra, B.; Cloutier, T.; Hawkins, T.; Predki, P.; Martin, C.; Wernick, M.; et al. Comprehensive genome sequence analysis of a breast cancer amplicon. Genome Res. 2001, 11, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, G.; Hager, J.H.; Volik, S.; Hariono, S.; Wernick, M.; Moore, D.; Nowak, N.; Albertson, D.G.; Pinkel, D.; Collins, C.; et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat. Genet. 2001, 29, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Mimori, K.; Nishida, N.; Tokuoka, M.; Akita, H.; Sekimoto, M.; Doki, Y.; Mori, M. Abnormal expression of PFDN4 in colorectal cancer: A novel marker for prognosis. Ann. Surg. Oncol. 2010, 17, 3030–3036. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Sunamura, M.; Horii, A. Molecular mechanisms of pancreatic carcinogenesis. Cancer Sci. 2006, 97, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nikonova, A.S.; Astsaturov, I.; Serebriiskii, I.G.; Dunbrack, R.L., Jr.; Golemis, E.A. Aurora A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 2013, 70, 661–687. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Stati, G.; Passaretta, F.; Gindraux, F.; Centurione, L.; Di Pietro, R. The Role of the CREB Protein Family Members and the Related Transcription Factors in Radioresistance Mechanisms. Life 2021, 11, 1437. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Wang, L.; Agyin, J.; Tang, Y.; Lin, S.; Yeh, I.T.; De, K.; Sun, L.Z. Doxorubicin in combination with a small TGFbeta inhibitor: A potential novel therapy for metastatic breast cancer in mouse models. PLoS ONE 2010, 5, e10365. [Google Scholar] [CrossRef]
- Gupta, N.; Gupta, P.; Srivastava, S.K. Penfluridol overcomes paclitaxel resistance in metastatic breast cancer. Sci. Rep. 2019, 9, 5066. [Google Scholar] [CrossRef]
- Turton, N.J.; Judah, D.J.; Riley, J.; Davies, R.; Lipson, D.; Styles, J.A.; Smith, A.G.; Gant, T.W. Gene expression and amplification in breast carcinoma cells with intrinsic and acquired doxorubicin resistance. Oncogene 2001, 20, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Ajabnoor, G.M.; Crook, T.; Coley, H.M. Paclitaxel resistance is associated with switch from apoptotic to autophagic cell death in MCF-7 breast cancer cells. Cell Death Dis. 2012, 3, e260. [Google Scholar] [CrossRef] [PubMed]
- Sikov, W.M.; Berry, D.A.; Perou, C.M.; Singh, B.; Cirrincione, C.T.; Tolaney, S.M.; Kuzma, C.S.; Pluard, T.J.; Somlo, G.; Port, E.R.; et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J. Clin. Oncol. 2015, 33, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Nami, B.; Wang, Z. Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers 2018, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Harrouk, W.; Codrington, A.; Vinson, R.; Robaire, B.; Hales, B.F. Paternal exposure to cyclophosphamide induces DNA damage and alters the expression of DNA repair genes in the rat preimplantation embryo. Mutat. Res. 2000, 461, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Swift, L.P.; Rephaeli, A.; Nudelman, A.; Phillips, D.R.; Cutts, S.M. Doxorubicin-DNA adducts induce a non-topoisomerase II—Mediated form of cell death. Cancer Res. 2006, 66, 4863–4871. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, J.; Yang, X.; Guan, S.; Feng, H.; Han, D.; Lu, J.; Ou, B.; Jin, R.; Sun, J.; et al. PFDN1, an indicator for colorectal cancer prognosis, enhances tumor cell proliferation and motility through cytoskeletal reorganization. Med. Oncol. 2015, 32, 264. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Ding, Z.; Chen, T.; Wu, H.; Song, J.; Xiang, Z.; Yang, C.; Wang, S.; Xiong, B. PFDN2 promotes cell cycle progression via the hnRNPD-MYBL2 axis in gastric cancer. Front. Oncol. 2023, 13, 1164070. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Maeda, Y.; Kitaura, H.; Taira, T.; Iguchi-Ariga, S.M.; Ariga, H. MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc. J. Biol. Chem. 1998, 273, 29794–29800. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, J. Prefoldin 6 promotes glioma progression via the AKT signalling pathway. Cell Biol. Int. 2023, 47, 52–62. [Google Scholar] [CrossRef]
- Roth, A.D.; Fazio, N.; Stupp, R.; Falk, S.; Bernhard, J.; Saletti, P.; Koberle, D.; Borner, M.M.; Rufibach, K.; Maibach, R.; et al. Docetaxel, cisplatin, and fluorouracil; docetaxel and cisplatin; and epirubicin, cisplatin, and fluorouracil as systemic treatment for advanced gastric carcinoma: A randomized phase II trial of the Swiss Group for Clinical Cancer Research. J. Clin. Oncol. 2007, 25, 3217–3223. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Belkhiri, A.; El-Rifai, W. The aurora kinase A regulates GSK-3beta in gastric cancer cells. Oncogene 2009, 28, 866–875. [Google Scholar] [CrossRef]
- Jin, S.; Wang, X.; Tong, T.; Zhang, D.; Shi, J.; Chen, J.; Zhan, Q. Aurora-A enhances malignant development of esophageal squamous cell carcinoma (ESCC) by phosphorylating beta-catenin. Mol. Oncol. 2015, 9, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Nikhil, K.; Viccaro, K.; Chang, L.; Jacobsen, M.; Sandusky, G.; Shah, K. The Aurora-A-Twist1 axis promotes highly aggressive phenotypes in pancreatic carcinoma. J. Cell Sci. 2017, 130, 1078–1093. [Google Scholar] [CrossRef]
- Zheng, X.Q.; Guo, J.P.; Yang, H.; Kanai, M.; He, L.L.; Li, Y.Y.; Koomen, J.M.; Minton, S.; Gao, M.; Ren, X.B.; et al. Aurora-A is a determinant of tamoxifen sensitivity through phosphorylation of ERalpha in breast cancer. Oncogene 2014, 33, 4985–4996. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, J.; Depatie, C.; Ohmichi, M.; Mercola, D. The activation of c-Jun NH2-terminal kinase (JNK) by DNA-damaging agents serves to promote drug resistance via activating transcription factor 2 (ATF2)-dependent enhanced DNA repair. J. Biol. Chem. 2003, 278, 20582–20592. [Google Scholar] [CrossRef]
- Rudraraju, B.; Droog, M.; Abdel-Fatah, T.M.; Zwart, W.; Giannoudis, A.; Malki, M.I.; Moore, D.; Patel, H.; Shaw, J.; Ellis, I.O.; et al. Phosphorylation of activating transcription factor-2 (ATF-2) within the activation domain is a key determinant of sensitivity to tamoxifen in breast cancer. Breast Cancer Res. Treat. 2014, 147, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Meyer, A.S.; Weiler, S.M.E.; Rupp, C.; Toth, M.; Sticht, C.; Singer, S.; Thomann, S.; Roessler, S.; Schorpp-Kistner, M.; et al. Cytoplasmic localization of the cell polarity factor scribble supports liver tumor formation and tumor cell invasiveness. Hepatology 2018, 67, 1842–1856. [Google Scholar] [CrossRef]
- Wu, D.S.; Chen, C.; Wu, Z.J.; Liu, B.; Gao, L.; Yang, Q.; Chen, W.; Chen, J.M.; Bao, Y.; Qu, L.; et al. ATF2 predicts poor prognosis and promotes malignant phenotypes in renal cell carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 108. [Google Scholar] [CrossRef]
- Gomaa, A.; Peng, D.; Chen, Z.; Soutto, M.; Abouelezz, K.; Corvalan, A.; El-Rifai, W. Epigenetic regulation of AURKA by miR-4715-3p in upper gastrointestinal cancers. Sci. Rep. 2019, 9, 16970. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Dempster, J.M.; Boyle, I.; Vazquez, F.; Root, D.E.; Boehm, J.S.; Hahn, W.C.; Tsherniak, A.; McFarland, J.M. Chronos: A cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021, 22, 343. [Google Scholar] [CrossRef] [PubMed]
- Pacini, C.; Dempster, J.M.; Boyle, I.; Goncalves, E.; Najgebauer, H.; Karakoc, E.; van der Meer, D.; Barthorpe, A.; Lightfoot, H.; Jaaks, P.; et al. Integrated cross-study datasets of genetic dependencies in cancer. Nat. Commun. 2021, 12, 1661. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.-H.; Yeh, C.-H.; Wu, C.-W.; Hsu, C.-Y.; Tsai, E.-M.; Hung, C.-M.; Wang, Y.-W.; Hsieh, T.-H. PFDN4 as a Prognostic Marker Was Associated with Chemotherapy Resistance through CREBP1/AURKA Pathway in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2024, 25, 3906. https://doi.org/10.3390/ijms25073906
Wang S-H, Yeh C-H, Wu C-W, Hsu C-Y, Tsai E-M, Hung C-M, Wang Y-W, Hsieh T-H. PFDN4 as a Prognostic Marker Was Associated with Chemotherapy Resistance through CREBP1/AURKA Pathway in Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2024; 25(7):3906. https://doi.org/10.3390/ijms25073906
Chicago/Turabian StyleWang, Shih-Ho, Cheng-Hsi Yeh, Chia-Wei Wu, Chia-Yi Hsu, Eing-Mei Tsai, Chao-Ming Hung, Yi-Wen Wang, and Tsung-Hua Hsieh. 2024. "PFDN4 as a Prognostic Marker Was Associated with Chemotherapy Resistance through CREBP1/AURKA Pathway in Triple-Negative Breast Cancer" International Journal of Molecular Sciences 25, no. 7: 3906. https://doi.org/10.3390/ijms25073906
APA StyleWang, S.-H., Yeh, C.-H., Wu, C.-W., Hsu, C.-Y., Tsai, E.-M., Hung, C.-M., Wang, Y.-W., & Hsieh, T.-H. (2024). PFDN4 as a Prognostic Marker Was Associated with Chemotherapy Resistance through CREBP1/AURKA Pathway in Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 25(7), 3906. https://doi.org/10.3390/ijms25073906




