Targeting Group 3 Medulloblastoma by the Anti-PRUNE-1 and Anti-LSD1/KDM1A Epigenetic Molecules
Abstract
:1. Introduction
2. Results
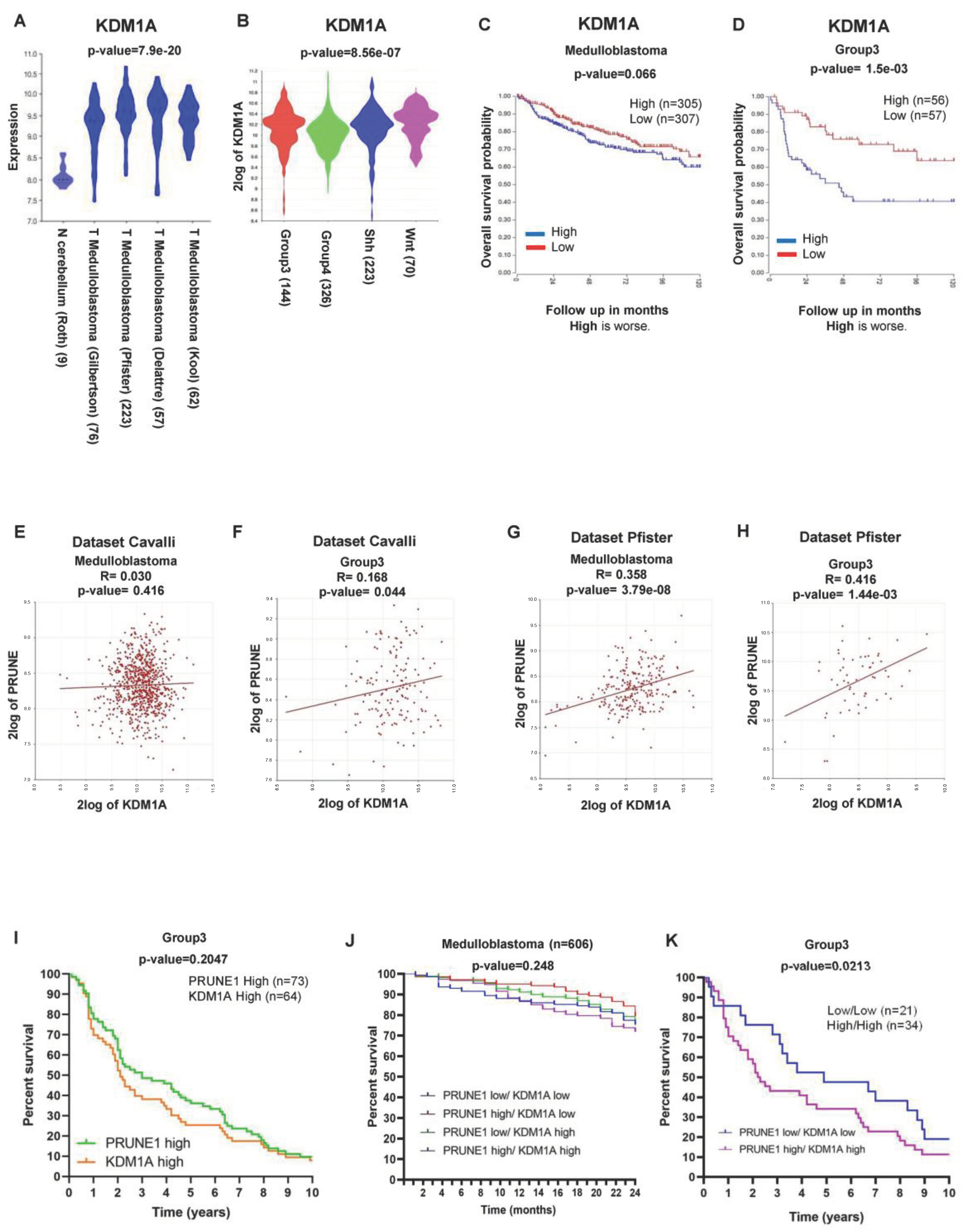
2.1. LSD1/KDM1A Expression and Epigenetically Regulation of PRUNE-1 in Medulloblastoma
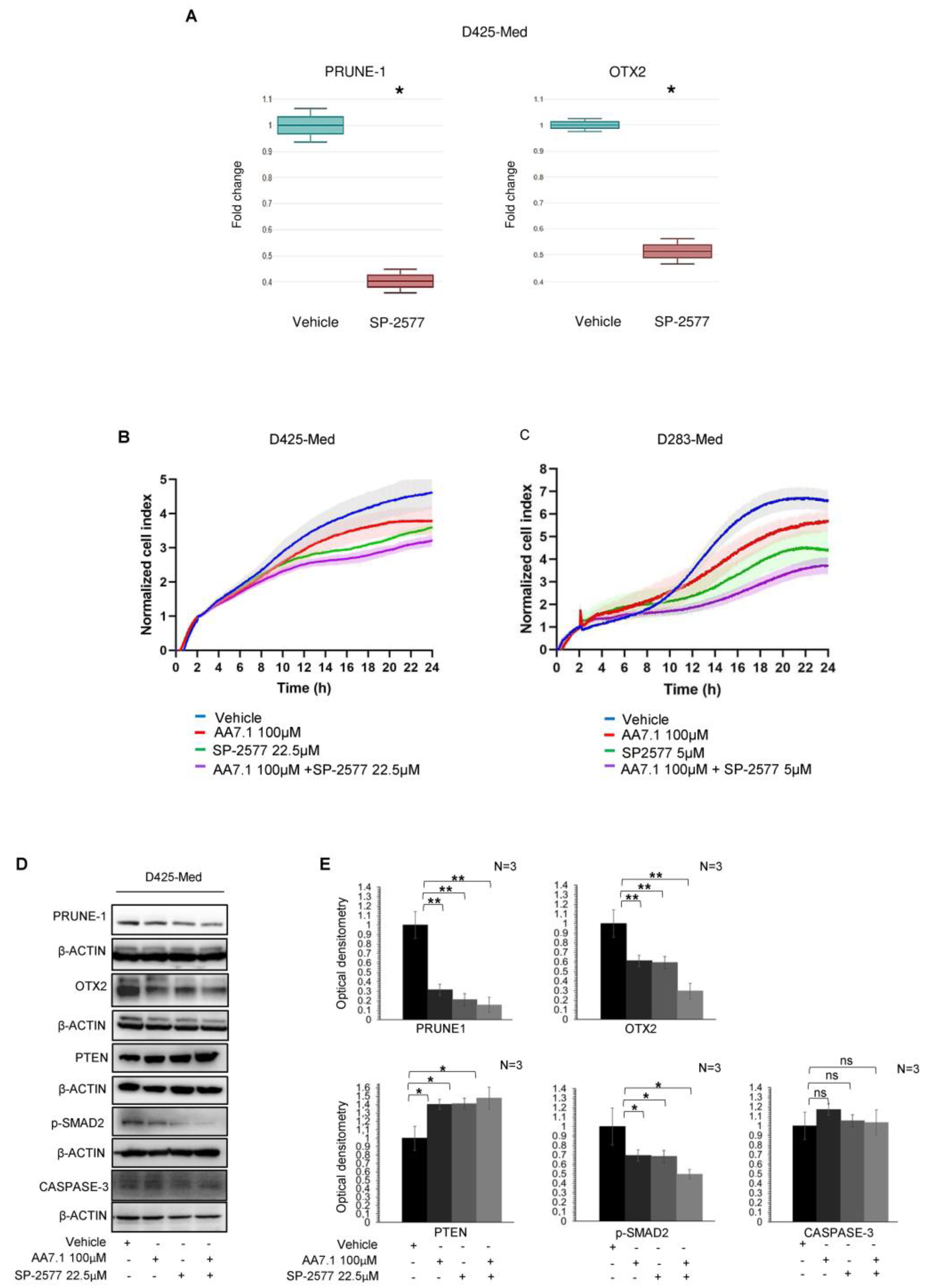
2.2. The Combination of the Prune-1 Inhibitor AA7.1 and the Selective LSD1/KDM1A Inhibitor SP-2577 impairs Cell Proliferation and TGF-β Signalling in D425-Med Gr3 MB Cells
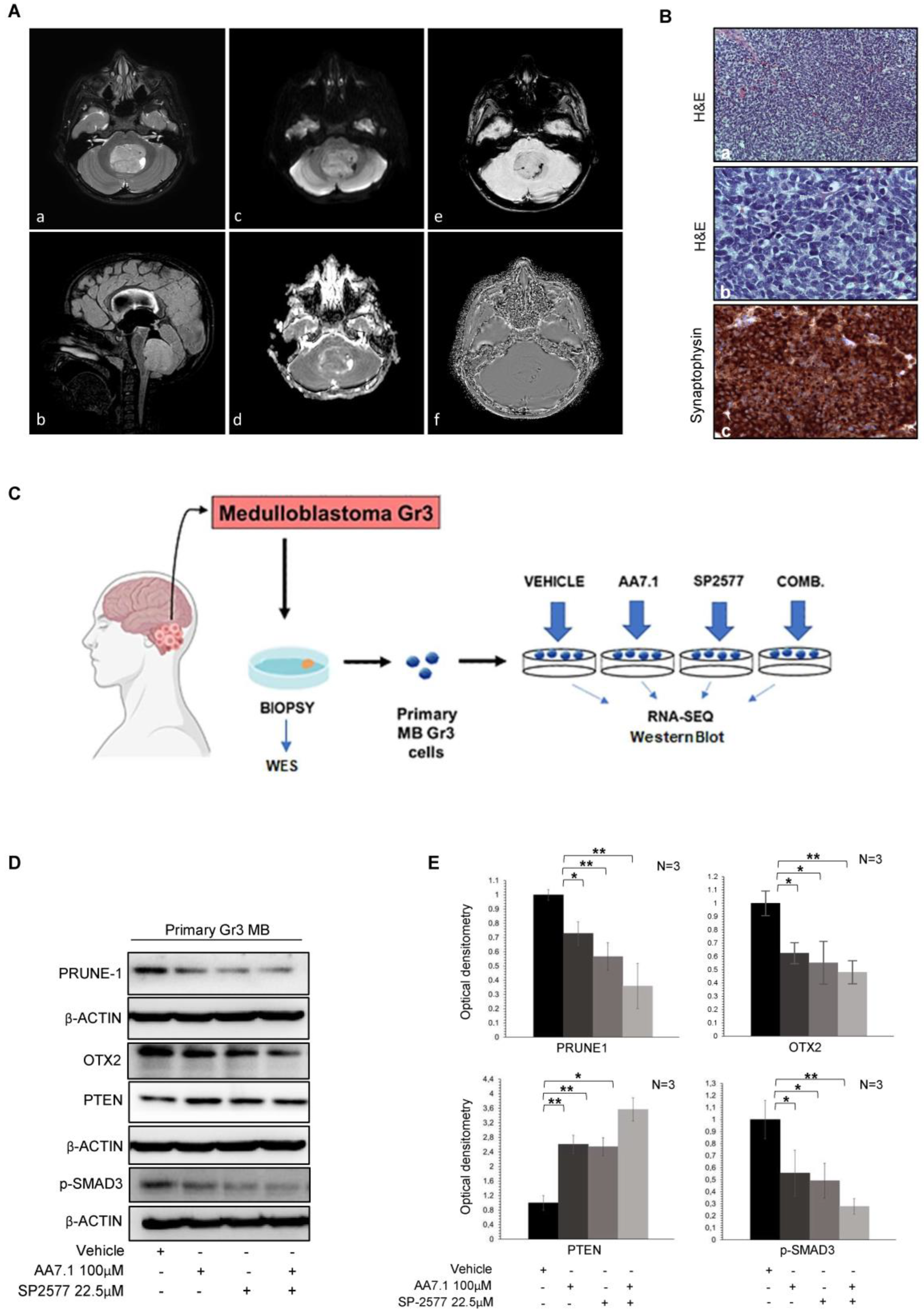
2.3. Treatment of Primary Gr3 MB Cells with the Combination of AA7.1 and SP-2577 Confirms its Efficacy on Targets
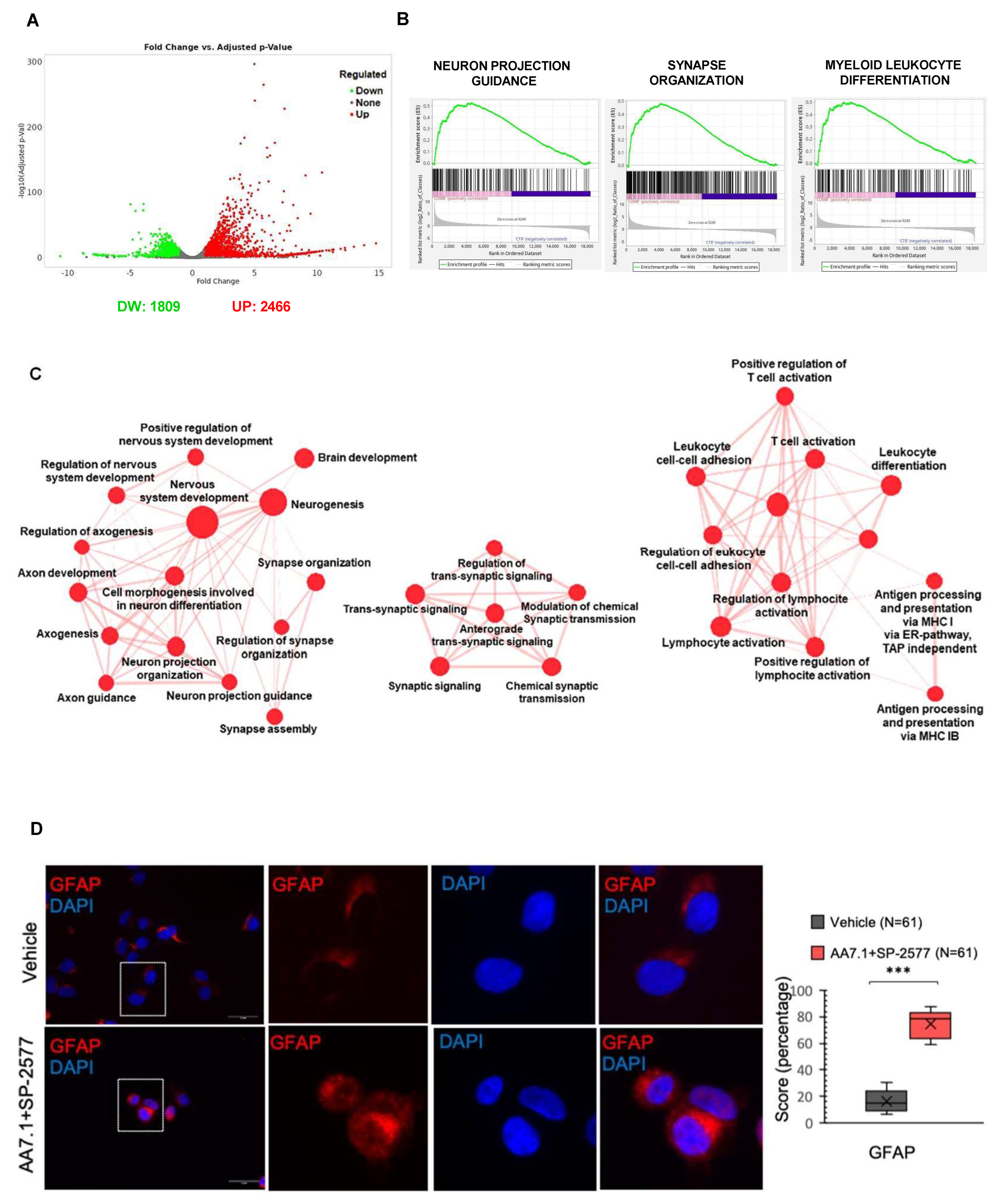
2.4. The Combination of AA7.1 and SP-2577 Affects Neuronal Commitment
2.5. The Combination of AA7.1 and SP-2577 Affects Leukocyte Differentiation
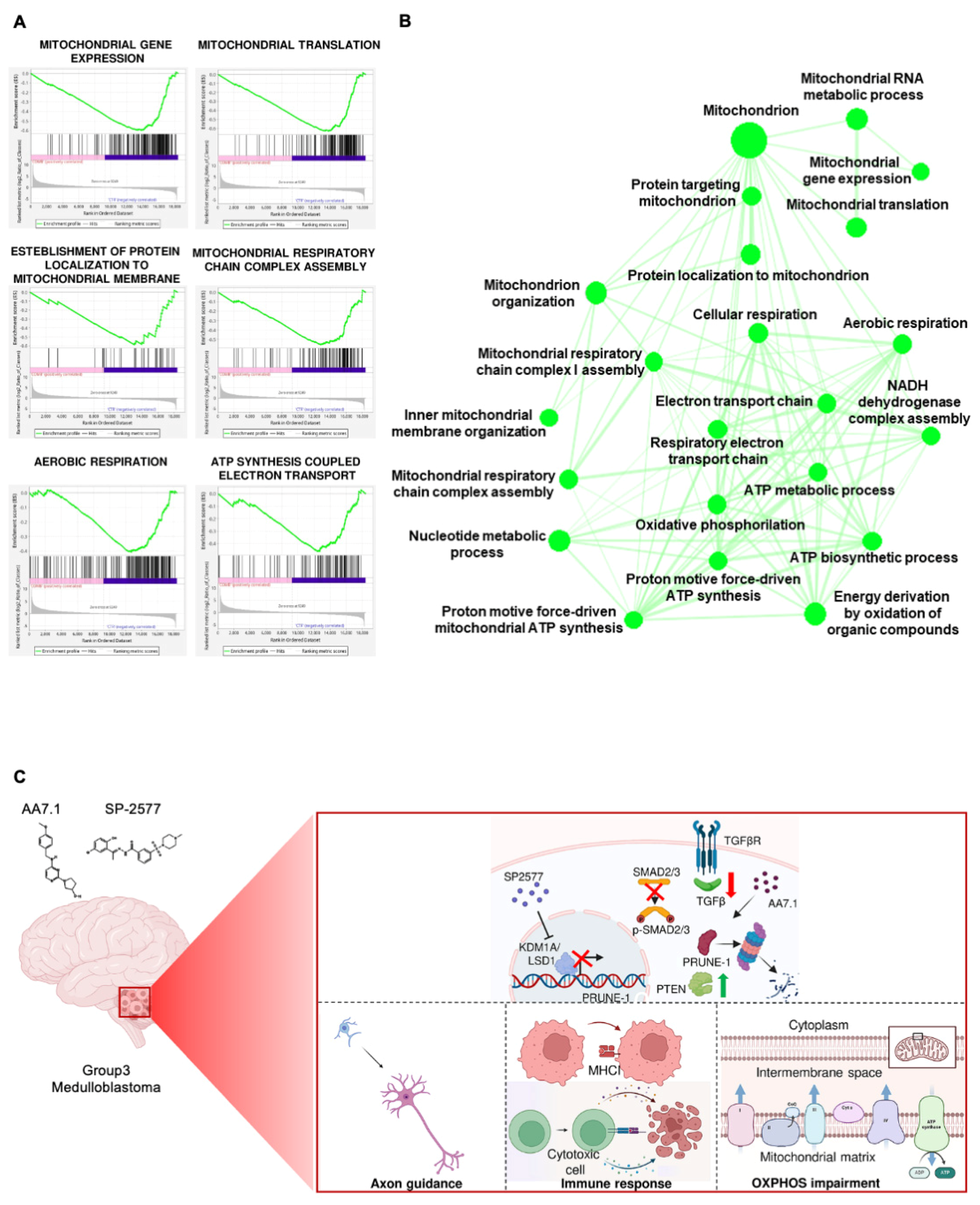
2.6. The Combination of AA7.1 and SP-2577 Affects Mitochondrial Metabolism
3. Discussion
4. Materials and Methods
4.1. Data Source
4.2. Cell Culture
4.3. In Vitro Treatments
4.4. Cell Proliferation Assays (Cell Index)
4.5. Immunoblotting
4.6. Brain MRI
4.7. Immunohistochemistry
4.8. Immunofluorescence
4.9. Genomic DNA Extraction
4.10. Whole-Exome Sequencing
4.11. Analysis of Variants
4.12. RNA Extraction and qPCR Assays
4.13. RNA Sequencing (RNA-Seq)
4.14. Clustering of Primary Gr3 MB Cells
4.15. Analysis of Differentially Expressed Genes
4.16. Statistical Analysis
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gajjar, A.; Pfister, S.M.; Taylor, M.D.; Gilbertson, R.J. Molecular Insights into Pediatric Brain Tumors Have the Potential to Transform Therapy. Clin. Cancer Res. 2014, 20, 5630–5640. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Koster, J.; Bunt, J.; Hasselt, N.E.; Lakeman, A.; van Sluis, P.; Troost, D.; Meeteren, N.S.; Caron, H.N.; Cloos, J.; et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE 2008, 3, e3088. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.J.; Tsherniak, A.; Tamayo, P.; Santagata, S.; Ligon, A.; Greulich, H.; Berhoukim, R.; Amani, V.; Goumnerova, L.; Eberhart, C.G.; et al. Integrative Genomic Analysis of Medulloblastoma Identifies a Molecular Subgroup That Drives Poor Clinical Outcome. J. Clin. Oncol. 2011, 29, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Korshunov, A.; Witt, H.; Hielscher, T.; Eberhart, C.G.; Mack, S.; Bouffet, E.; Clifford, S.C.; Hawkins, C.E.; French, P.; et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 2011, 29, 1408–1414. [Google Scholar] [CrossRef] [PubMed]
- Remke, M.; Hielscher, T.; Northcott, P.A.; Witt, H.; Ryzhova, M.; Wittmann, A.; Benner, A.; von Deimling, A.; Scheurlen, W.; Perry, A.; et al. Adult medulloblastoma comprises three major molecular variants. J. Clin. Oncol. 2011, 29, 2717–2723. [Google Scholar] [CrossRef] [PubMed]
- Rausch, T.; Jones, D.T.; Zapatka, M.; Stutz, A.M.; Zichner, T.; Weischenfeldt, J.; Jager, N.; Remke, M.; Shih, D.; Northcott, P.A.; et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 2012, 148, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.E6. [Google Scholar] [CrossRef]
- Hovestadt, V.; Smith, K.S.; Bihannic, L.; Filbin, M.G.; Shaw, M.L.; Baumgartner, A.; DeWitt, J.C.; Groves, A.; Mayr, L.; Weisman, H.R.; et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 2019, 572, 74–79. [Google Scholar] [CrossRef]
- Robinson, G.W.; Orr, B.A.; Wu, G.; Gururangan, S.; Lin, T.; Qaddoumi, I.; Packer, R.J.; Goldman, S.; Prados, M.D.; Desjardins, A.; et al. Vismodegib Exerts Targeted Efficacy against Recurrent Sonic Hedgehog-Subgroup Medulloblastoma: Results from Phase II Pediatric Brain Tumor Consortium Studies PBTC-025B and PBTC-032. J. Clin. Oncol. 2015, 33, 2646–2654. [Google Scholar] [CrossRef]
- Goschzik, T.; Schwalbe, E.C.; Hicks, D.; Smith, A.; Zur Muehlen, A.; Figarella-Branger, D.; Doz, F.; Rutkowski, S.; Lannering, B.; Pietsch, T.; et al. Prognostic effect of whole chromosomal aberration signatures in standard-risk, non-WNT/non-SHH medulloblastoma: A retrospective, molecular analysis of the HIT-SIOP PNET 4 trial. Lancet Oncol. 2018, 19, 1602–1616. [Google Scholar] [CrossRef]
- Lannering, B.; Rutkowski, S.; Doz, F.; Pizer, B.; Gustafsson, G.; Navajas, A.; Massimino, M.; Reddingius, R.; Benesch, M.; Carrie, C.; et al. Hyperfractionated versus conventional radiotherapy followed by chemotherapy in standard-risk medulloblastoma: Results from the randomized multicenter HIT-SIOP PNET 4 trial. J. Clin. Oncol. 2012, 30, 3187–3193. [Google Scholar] [CrossRef] [PubMed]
- Merchant, T.E.; Kun, L.E.; Krasin, M.J.; Wallace, D.; Chintagumpala, M.M.; Woo, S.Y.; Ashley, D.M.; Sexton, M.; Kellie, S.J.; Ahern, V.; et al. Multi-institution prospective trial of reduced-dose craniospinal irradiation (23.4 Gy) followed by conformal posterior fossa (36 Gy) and primary site irradiation (55.8 Gy) and dose-intensive chemotherapy for average-risk medulloblastoma. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Mynarek, M.; Milde, T.; Padovani, L.; Janssens, G.O.; Kwiecien, R.; Mosseri, V.; Clifford, S.C.; Doz, F.; Rutkowski, S. SIOP PNET5 MB Trial: History and Concept of a Molecularly Stratified Clinical Trial of Risk-Adapted Therapies for Standard-Risk Medulloblastoma. Cancers 2021, 13, 6077. [Google Scholar] [CrossRef] [PubMed]
- Michalski, J.M.; Janss, A.J.; Vezina, L.G.; Smith, K.S.; Billups, C.A.; Burger, P.C.; Embry, L.M.; Cullen, P.L.; Hardy, K.K.; Pomeroy, S.L.; et al. Children’s Oncology Group Phase III Trial of Reduced-Dose and Reduced-Volume Radiotherapy with Chemotherapy for Newly Diagnosed Average-Risk Medulloblastoma. J. Clin. Oncol. 2021, 39, 2685–2697. [Google Scholar] [CrossRef]
- Ferrucci, V.; de Antonellis, P.; Pennino, F.P.; Asadzadeh, F.; Virgilio, A.; Montanaro, D.; Galeone, A.; Boffa, I.; Pisano, I.; Scognamiglio, I.; et al. Metastatic group 3 medulloblastoma is driven by PRUNE1 targeting NME1-TGF-β-OTX2-SNAIL via PTEN inhibition. Brain 2018, 141, 1300–1319. [Google Scholar] [CrossRef] [PubMed]
- Hendrikse, L.D.; Haldipur, P.; Saulnier, O.; Millman, J.; Sjoboen, A.H.; Erickson, A.W.; Ong, W.; Gordon, V.; Coudiere-Morrison, L.; Mercier, A.L.; et al. Author Correction: Failure of human rhombic lip differentiation underlies medulloblastoma formation. Nature 2022, 612, E12. [Google Scholar] [CrossRef] [PubMed]
- Tammenkoski, M.; Koivula, K.; Cusanelli, E.; Zollo, M.; Steegborn, C.; Baykov, A.A.; Lahti, R. Human metastasis regulator protein H-prune is a short-chain exopolyphosphatase. Biochemistry 2008, 47, 9707–9713. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Simard, L.R.; Ding, H. Generation of Conditional Knockout Alleles for PRUNE-1. Cells 2023, 12, 524. [Google Scholar] [CrossRef] [PubMed]
- Bibbo, F.; Sorice, C.; Ferrucci, V.; Zollo, M. Functional Genomics of PRUNE1 in Neurodevelopmental Disorders (NDDs) Tied to Medulloblastoma (MB) and Other Tumors. Front. Oncol. 2021, 11, 758146. [Google Scholar] [CrossRef]
- Zollo, M.; Ahmed, M.; Ferrucci, V.; Salpietro, V.; Asadzadeh, F.; Carotenuto, M.; Maroofian, R.; Al-Amri, A.; Singh, R.; Scognamiglio, I.; et al. PRUNE is crucial for normal brain development and mutated in microcephaly with neurodevelopmental impairment. Brain 2017, 140, 940–952. [Google Scholar] [CrossRef]
- Scorrano, G.; Battaglia, L.; Spiaggia, R.; Basile, A.; Palmucci, S.; Foti, P.V.; David, E.; Marinangeli, F.; Mascilini, I.; Corsello, A.; et al. Corrigendum: Neuroimaging in PRUNE1 syndrome: A mini-review of the literature. Front. Neurol. 2024, 15, 1360347. [Google Scholar] [CrossRef] [PubMed]
- Forus, A.; D’Angelo, A.; Henriksen, J.; Merla, G.; Maelandsmo, G.M.; Florenes, V.A.; Olivieri, S.; Bjerkehagen, B.; Meza-Zepeda, L.A.; del Vecchio Blanco, F.; et al. Amplification and overexpression of PRUNE in human sarcomas and breast carcinomas-a possible mechanism for altering the nm23-H1 activity. Oncogene 2001, 20, 6881–6890. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, A.; Garzia, L.; Andre, A.; Carotenuto, P.; Aglio, V.; Guardiola, O.; Arrigoni, G.; Cossu, A.; Palmieri, G.; Aravind, L.; et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell 2004, 5, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, M.; De Antonellis, P.; Liguori, L.; Benvenuto, G.; Magliulo, D.; Alonzi, A.; Turino, C.; Attanasio, C.; Damiani, V.; Bello, A.M.; et al. H-Prune through GSK-3beta interaction sustains canonical WNT/beta-catenin signaling enhancing cancer progression in NSCLC. Oncotarget 2014, 5, 5736–5749. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, V.; Lomada, S.; Wieland, T.; Zollo, M. PRUNE1 and NME/NDPK family proteins influence energy metabolism and signaling in cancer metastases. Cancer Metastasis Rev. 2024. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ and immune evasion in metastatic cancer. Cancer Res. 2021, 81. [Google Scholar] [CrossRef]
- Pickup, M.; Novitskiy, S.; Moses, H.L. The roles of TGFbeta in the tumour microenvironment. Nat. Rev. Cancer 2013, 13, 788–799. [Google Scholar] [CrossRef]
- Ferrucci, V.; Asadzadeh, F.; Collina, F.; Siciliano, R.; Boccia, A.; Marrone, L.; Spano, D.; Carotenuto, M.; Chiarolla, C.M.; De Martino, D.; et al. Prune-1 drives polarization of tumor-associated macrophages (TAMs) within the lung metastatic niche in triple-negative breast cancer. Iscience 2021, 24, 101938. [Google Scholar] [CrossRef]
- Pugh, T.J.; Weeraratne, S.D.; Archer, T.C.; Pomeranz Krummel, D.A.; Auclair, D.; Bochicchio, J.; Carneiro, M.O.; Carter, S.L.; Cibulskis, K.; Erlich, R.L.; et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012, 488, 106–110. [Google Scholar] [CrossRef]
- Northcott, P.A.; Robinson, G.W.; Kratz, C.P.; Mabbott, D.J.; Pomeroy, S.L.; Clifford, S.C.; Rutkowski, S.; Ellison, D.W.; Malkin, D.; Taylor, M.D.; et al. Medulloblastoma. Nat. Rev. Dis. Primers 2019, 5, 11. [Google Scholar] [CrossRef]
- Shi, Y.J.; Matson, C.; Lan, F.; Iwase, S.; Baba, T.; Shi, Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 2005, 19, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.G.; Wynder, C.; Cooch, N.; Shiekhattar, R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005, 437, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Chen, Y.P.; Sun, Y.M.; Yang, F.; Yu, W.H.; Liang, J.; Sun, L.Y.; Yang, X.H.; Shi, L.; et al. LSD1 Is a Subunit of the NuRD Complex and Targets the Metastasis Programs in Breast Cancer. Cell 2009, 138, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Wissmann, M.; Yin, N.; Muller, J.M.; Schneider, R.; Peters, A.H.; Gunther, T.; Buettner, R.; Schule, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Telese, F.; Tan, Y.; Li, W.; Jin, C.; He, X.; Basnet, H.; Ma, Q.; Merkurjev, D.; Zhu, X.; et al. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat. Neurosci. 2015, 1265, 64. [Google Scholar] [CrossRef]
- Shoaib, M.; Chen, Q.M.; Shi, X.Y.; Nair, N.; Prasanna, C.; Yang, R.L.; Walter, D.; Frederiksen, K.S.; Einarsson, H.; Svensson, J.P.; et al. Histone H4 lysine 20 mono-methylation directly facilitates chromatin openness and promotes transcription of housekeeping genes. Nat. Commun. 2021, 12, 4800. [Google Scholar] [CrossRef]
- Karakaidos, P.; Verigos, J.; Magklara, A. LSD1/KDM1A, a Gate-Keeper of Cancer Stemness and a Promising Therapeutic Target. Cancers 2019, 11, 1821. [Google Scholar] [CrossRef] [PubMed]
- Vinyard, M.E.; Su, C.; Siegenfeld, A.P.; Waterbury, A.L.; Freedy, A.M.; Gosavi, P.M.; Park, Y.; Kwan, E.E.; Senzer, B.D.; Doench, J.G.; et al. CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML. Nat. Chem. Biol. 2019, 15, 529–539. [Google Scholar] [CrossRef]
- Ferrari-Amorotti, G.; Fragliasso, V.; Esteki, R.; Prudente, Z.; Soliera, A.R.; Cattelani, S.; Manzotti, G.; Grisendi, G.; Dominici, M.; Pieraccioli, M.; et al. Inhibiting Interactions of Lysine Demethylase LSD1 with Snail/Slug Blocks Cancer Cell Invasion. Cancer Res. 2013, 73, 235–245. [Google Scholar] [CrossRef]
- Pajtler, K.W.; Weingarten, C.; Thor, T.; Künkele, A.; Heukamp, L.C.; Büttner, R.; Suzuki, T.; Miyata, N.; Grotzer, M.; Rieb, A.; et al. The KDM1A histone demethylase is a promising new target for the epigenetic therapy of medulloblastoma. Acta Neuropathol. Com. 2013, 1, 19. [Google Scholar] [CrossRef]
- Callegari, K.; Maegawa, S.; Bravo-Alegria, J.; Gopalakrishnan, V. Pharmacological inhibition of LSD1 activity blocks REST-dependent medulloblastoma cell migration. Cell Commun. Signal 2018, 16, 60. [Google Scholar] [CrossRef]
- Lee, C.; Rudneva, V.A.; Erkek, S.; Zapatka, M.; Chau, L.Q.; Tacheva-Grigorova, S.K.; Garancher, A.; Rusert, J.M.; Aksoy, O.; Lea, R.; et al. Lsd1 as a therapeutic target in Gfi1-activated medulloblastoma. Nat. Commun. 2019, 10, 332. [Google Scholar] [CrossRef]
- Sacilotto, N.; Dessanti, P.; Lufino, M.M.P.; Ortega, A.; Rodriguez-Gimeno, A.; Salas, J.; Maes, T.; Buesa, C.; Mascaro, C.; Soliva, R. Comprehensive In Vitro Characterization of the LSD1 Small Molecule Inhibitor Class in Oncology. ACS Pharmacol. Transl. Sci. 2021, 4, 1818–1834. [Google Scholar] [CrossRef]
- Fang, Y.; Liao, G.C.; Yu, B. LSD1/KDM1A inhibitors in clinical trials: Advances and prospects. J. Hematol. Oncol. 2019, 12, 129. [Google Scholar] [CrossRef]
- Dai, X.J.; Liu, Y.; Xiong, X.P.; Xue, L.P.; Zheng, Y.C.; Liu, H.M. Tranylcypromine Based Lysine-Specific Demethylase 1 Inhibitor: Summary and Perspective. J. Med. Chem. 2020, 63, 14197–14215. [Google Scholar] [CrossRef]
- Mould, D.P.; McGonagle, A.E.; Wiseman, D.H.; Williams, E.L.; Jordan, A.M. Reversible Inhibitors of LSD1 as Therapeutic Agents in Acute Myeloid Leukemia: Clinical Significance and Progress to Date. Med. Res. Rev. 2015, 35, 586–618. [Google Scholar] [CrossRef]
- Laurent, B.; Ruitu, L.; Murn, J.; Hempel, K.; Ferrao, R.; Xiang, Y.; Liu, S.C.; Garcia, B.A.; Wu, H.; Wu, F.Z.; et al. A Specific LSD1/KDM1A Isoform Regulates Neuronal Differentiation through H3K9 Demethylation. Mol. Cell 2015, 57, 957–970. [Google Scholar] [CrossRef]
- Yang, M.; Culhane, J.C.; Szewczuk, L.M.; Jalili, P.; Ball, H.L.; Machius, M.; Cole, P.A.; Yu, H. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry 2007, 46, 8058–8065. [Google Scholar] [CrossRef]
- Stazi, G.; Zwergel, C.; Valente, S.; Mai, A. LSD1 inhibitors: A patent review (2010–2015). Expert. Opin. Ther. Pat. 2016, 26, 565–580. [Google Scholar] [CrossRef]
- Saez, J.E.; Gomez, A.V.; Barrios, A.P.; Parada, G.E.; Galdames, L.; Gonzalez, M.; Andres, M.E. Decreased Expression of CoREST1 and CoREST2 Together with LSD1 and HDAC1/2 during Neuronal Differentiation. PLoS ONE 2015, 10, e0131760. [Google Scholar] [CrossRef]
- Di Giovannantonio, L.G.; Di Salvio, M.; Omodei, D.; Prakash, N.; Wurst, W.; Pierani, A.; Acampora, D.; Simeone, A. Otx2 cell-autonomously determines dorsal mesencephalon versus cerebellum fate independently of isthmic organizing activity. Development 2014, 141, 377–388. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell 2017, 31, 326–341. [Google Scholar] [CrossRef]
- Klemm, F.; Maas, R.R.; Bowman, R.L.; Kornete, M.; Soukup, K.; Nassiri, S.; Brouland, J.P.; Iacobuzio-Donahue, C.A.; Brennan, C.; Tabar, V.; et al. Interrogation of the Microenvironmental Landscape in Brain Tumors Reveals Disease-Specific Alterations of Immune Cells. Cell 2020, 181, 1643–1660.e17. [Google Scholar] [CrossRef]
- Bockmayr, M.; Mohme, M.; Klauschen, F.; Winkler, B.; Budczies, J.; Rutkowski, S.; Schüller, U. Subgroup-specific immune and stromal microenvironment in medulloblastoma. OncoImmunology 2018, 7, e1462430. [Google Scholar] [CrossRef]
- Shen, D.D.; Pang, J.R.; Bi, Y.P.; Zhao, L.F.; Li, Y.R.; Zhao, L.J.; Gao, Y.; Wang, B.; Wang, N.; Wei, L.; et al. LSD1 deletion decreases exosomal PD-L1 and restores T-cell response in gastric cancer. Mol. Cancer 2022, 21, 75. [Google Scholar] [CrossRef]
- Qin, Y.; Vasilatos, S.N.; Chen, L.; Wu, H.; Cao, Z.; Fu, Y.; Huang, M.; Vlad, A.M.; Lu, B.; Oesterreich, S.; et al. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 2019, 38, 390–405. [Google Scholar] [CrossRef]
- Soldi, R.; Ghosh Halder, T.; Weston, A.; Thode, T.; Drenner, K.; Lewis, R.; Kaadige, M.R.; Srivastava, S.; Daniel Ampanattu, S.; Rodriguez Del Villar, R.; et al. The novel reversible LSD1 inhibitor SP-2577 promotes anti-tumor immunity in SWItch/Sucrose-NonFermentable (SWI/SNF) complex mutated ovarian cancer. PLoS ONE 2020, 15, e0235705. [Google Scholar] [CrossRef]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 2019, 37, 457–495. [Google Scholar] [CrossRef]
- Blank, C.U.; Haining, W.N.; Held, W.; Hogan, P.G.; Kallies, A.; Lugli, E.; Lynn, R.C.; Philip, M.; Rao, A.; Restifo, N.P.; et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019, 19, 665–674. [Google Scholar] [CrossRef]
- Tu, W.J.; McCuaig, R.D.; Tan, A.H.Y.; Hardy, K.; Seddiki, N.; Ali, S.; Dahlstrom, J.E.; Bean, E.G.; Dunn, J.; Forwood, J.; et al. Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch. Front. Immunol. 2020, 11, 1228. [Google Scholar] [CrossRef]
- Lee, D.Y.; Salahuddin, T.; Iqbal, J. Lysine-Specific Demethylase 1 (LSD1)-Mediated Epigenetic Modification of Immunogenicity and Immunomodulatory Effects in Breast Cancers. Curr. Oncol. 2023, 30, 2127–2143. [Google Scholar] [CrossRef]
- Hong, Y.; Li, X.; Zhu, J. LSD1-mediated stabilization of SEPT6 protein activates the TGF-beta1 pathway and regulates non-small-cell lung cancer metastasis. Cancer Gene Ther. 2022, 29, 189–201. [Google Scholar] [CrossRef]
- Li, M.O.; Wan, Y.Y.; Sanjabi, S.; Robertson, A.K.; Flavell, R.A. Transforming growth factor-beta regulation of immune responses. Annu. Rev. Immunol. 2006, 24, 99–146. [Google Scholar] [CrossRef]
- Zuber, P.; Kuppner, M.C.; De Tribolet, N. Transforming growth factor-beta 2 down-regulates HLA-DR antigen expression on human malignant glioma cells. Eur. J. Immunol. 1988, 18, 1623–1626. [Google Scholar] [CrossRef]
- Rook, A.H.; Kehrl, J.H.; Wakefield, L.M.; Roberts, A.B.; Sporn, M.B.; Burlington, D.B.; Lane, H.C.; Fauci, A.S. Effects of Transforming Growth-Factor-Beta on the Functions of Natural-Killer-Cells—Depressed Cytolytic Activity and Blunting of Interferon Responsiveness. J. Immunol. 1986, 136, 3916–3920. [Google Scholar] [CrossRef]
- Weller, M.; Fontana, A. The failure of current immunotherapy for malignant glioma. Tumor-derived TGF-beta, T-cell apoptosis, and the immune privilege of the brain. Brain Res. Brain Res. Rev. 1995, 21, 128–151. [Google Scholar] [CrossRef]
- Gamble, J.R.; Vadas, M.A. Endothelial-Cell Adhesiveness for Human Lymphocytes-T Is Inhibited by Transforming Growth-Factor-Beta. J. Immunol. 1991, 146, 1149–1154. [Google Scholar] [CrossRef]
- Platten, M.; Wick, W.; Weller, M. Malignant glioma biology: Role for TGF-beta in growth, motility, angiogenesis, and immune escape. Microsc. Res. Tech. 2001, 52, 401–410. [Google Scholar] [CrossRef]
- Hess, C.; Means, T.K.; Autissier, P.; Woodberry, T.; Altfeld, M.; Addo, M.M.; Frahm, N.; Brander, C.; Walker, B.D.; Luster, A.D. IL-8 responsiveness defines a subset of CD8 T cells poised to kill. Blood 2004, 104, 3463–3471. [Google Scholar] [CrossRef]
- Li, E.; Yang, X.; Du, Y.; Wang, G.; Chan, D.W.; Wu, D.; Xu, P.; Ni, P.; Xu, D.; Hu, Y. CXCL8 Associated Dendritic Cell Activation Marker Expression and Recruitment as Indicators of Favorable Outcomes in Colorectal Cancer. Front. Immunol. 2021, 12, 667177. [Google Scholar] [CrossRef]
- Kallingal, A.; Olszewski, M.; Maciejewska, N.; Brankiewicz, W.; Baginski, M. Cancer immune escape: The role of antigen presentation machinery. J. Cancer Res. Clin. Oncol. 2023, 149, 8131–8141. [Google Scholar] [CrossRef]
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New insights into cancer immunoediting and its three component phases elimination, equilibrium and escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef]
- Reeves, E.; James, E. Antigen processing and immune regulation in the response to tumours. Immunology 2017, 150, 16–24. [Google Scholar] [CrossRef]
- Kulkarni, B.; Kirave, P.; Gondaliya, P.; Jash, K.; Jain, A.; Tekade, R.K.; Kalia, K. Exosomal miRNA in chemoresistance, immune evasion, metastasis and progression of cancer. Drug Discov. Today 2019, 24, 2058–2067. [Google Scholar] [CrossRef]
- Reiman, J.M.; Kmieciak, M.; Manjili, M.H.; Knutson, K.L. Tumor immunoediting and immunosculpting pathways to cancer progression. Semin. Cancer Biol. 2007, 17, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ren, Y.; Weng, S.; Xu, H.; Li, L.; Han, X. A New Trend in Cancer Treatment: The Combination of Epigenetics and Immunotherapy. Front. Immunol. 2022, 13, 809761. [Google Scholar] [CrossRef]
- Gryder, B.E.; Sodji, Q.H.; Oyelere, A.K. Targeted cancer therapy: Giving histone deacetylase inhibitors all they need to succeed. Future Med. Chem. 2012, 4, 505–524. [Google Scholar] [CrossRef]
- Doroshow, D.B.; Bhalla, S.; Beasley, M.B.; Sholl, L.M.; Kerr, K.M.; Gnjatic, S.; Wistuba, I.I.; Rimm, D.L.; Tsao, M.S.; Hirsch, F.R. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2021, 18, 345–362. [Google Scholar] [CrossRef]
- Warburg, O. On respiratory impairment in cancer cells. Science 1956, 124, 269–270. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, W.; Bian, Y.; Li, Y.; Cong, L. Multifaceted roles of aerobic glycolysis and oxidative phosphorylation in hepatocellular carcinoma. PeerJ. 2023, 11, e14797. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.T.; Li, Y.; Zhang, Z.Y. Crosstalk between oxidative phosphorylation and immune escape in cancer: A new concept of therapeutic targets selection. Cell Oncol. 2023, 46, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Reznik, E.; Miller, M.L.; Senbabaoglu, Y.; Riaz, N.; Sarungbam, J.; Tickoo, S.K.; Al-Ahmadie, H.A.; Lee, W.; Seshan, V.E.; Hakimi, A.A.; et al. Mitochondrial DNA copy number variation across human cancers. eLife 2016, 5, e10769. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.S.; Baty, J.W.; Dong, L.F.; Bezawork-Geleta, A.; Endaya, B.; Goodwin, J.; Bajzikova, M.; Kovarova, J.; Peterka, M.; Yan, B.; et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015, 21, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, S.E.; Chandel, N.S. Targeting mitochondria metabolism for Cancer therapy. Nat. Chem. Biol. 2015, 11, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Van den Ameele, J.; Brand, A.H. Neural stem cell temporal patterning and brain tumour growth rely on oxidative phosphorylation. eLife 2019, 8, e47887. [Google Scholar] [CrossRef] [PubMed]
- Kuramoto, K.; Yamamoto, M.; Suzuki, S.; Sanomachi, T.; Togashi, K.; Seino, S.; Kitanaka, C.; Okada, M. Verteporfin inhibits oxidative phosphorylation and induces cell death specifically in glioma stem cells. FEBS J. 2020, 287, 2023–2036. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef]
- Martirosian, V.; Deshpande, K.; Zhou, H.; Shen, K.; Smith, K.; Northcott, P.; Lin, M.; Stepanosyan, V.; Das, D.; Remsik, J.; et al. Medulloblastoma uses GABA transaminase to survive in the cerebrospinal fluid microenvironment and promote leptomeningeal dissemination. Cell Rep. 2021, 35, 109302. [Google Scholar] [CrossRef]
- Gui, D.Y.; Sullivan, L.B.; Luengo, A.; Hosios, A.M.; Bush, L.N.; Gitego, N.; Davidson, S.M.; Freinkman, E.; Thomas, C.J.; Vander Heiden, M.G. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab 2016, 24, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.F.; Lim, S.K.; Liang, Q.R.; Iyer, S.V.; Wang, H.Y.; Wang, Z.L.; Xie, X.H.; Sun, D.C.; Chen, Y.J.; Tabar, V.; et al. Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature 2019, 567, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Tang, L.; Du, K.; Paraiso, K.; Sun, Q.; Liu, Z.; Ye, X.; Fang, Y.; Yuan, F.; Chen, H.; et al. Anterograde regulation of mitochondrial genes and FGF21 signaling by hepatic LSD1. JCI Insight 2021, 6, e147692. [Google Scholar] [CrossRef] [PubMed]
- Duteil, D.; Metzger, E.; Willmann, D.; Karagianni, P.; Friedrichs, N.; Greschik, H.; Günther, T.; Buettner, R.; Talianidis, I.; Metzger, D.; et al. LSD1 promotes oxidative metabolism of white adipose tissue. Nat. Commun. 2014, 5, 4093. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, E.; Aschar-Sobbi, R.; Campanella, M.; Turner, R.J.; Gomez-Garcia, M.R.; Abramov, A.Y. Inorganic polyphosphate and energy metabolism in mammalian cells. J. Biol. Chem. 2010, 285, 9420–9428. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jia, Y.; Tang, B.; Yang, H.; Yang, Q.; Luo, X.; Pan, Y. Mitochondrial subtype MB-G3 contains potential novel biomarkers and therapeutic targets associated with prognosis of medulloblastoma. Biomarkers 2023, 28, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Linke, F.; Aldighieri, M.; Lourdusamy, A.; Grabowska, A.M.; Stolnik, S.; Kerr, I.D.; Merry, C.L.; Coyle, B. 3D hydrogels reveal medulloblastoma subgroup diferences and identify extracellular matrix subtypes that predict patient outcome. J. Pathol. 2021, 253, 326–338. [Google Scholar] [CrossRef]
- Linke, F.; Johnson, J.E.C.; Kern, S.; Bennett, C.D.; Lourdusamy, A.; Lea, D.; Clifford, S.C.; Merry, C.L.R.; Stolnik, S.; Alexander, M.R.; et al. Identifying new biomarkers of aggressive Group 3 and SHH medulloblastoma using 3D hydrogel models, single cell RNA sequencing and 3D OrbiSIMS imaging. Acta Neuropathol. Commun. 2023, 11, 6. [Google Scholar] [CrossRef]
- Contenti, J.; Guo, Y.; Mazzu, A.; Irondelle, M.; Rouleau, M.; Lago, C.; Leva, G.; Tiberi, L.; Ben-Sahra, I.; Bost, F.; et al. The mitochondrial NADH shuttle system is a targetable vulnerability for Group 3 medulloblastoma in a hypoxic microenvironment. Cell Death Dis. 2023, 14, 784. [Google Scholar] [CrossRef]
- Martell, E.; Kuzmychova, H.; Kaul, E.; Senthil, H.; Chowdhury, S.R.; Morrison, L.C.; Fresnoza, A.; Zagozewski, J.; Venugopal, C.; Anderson, C.M.; et al. Metabolism-based targeting of MYC via MPC-SOD2 axis-mediated oxidation promotes cellular differentiation in group 3 medulloblastoma. Nat. Commun. 2023, 14, 2502. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Talbot, J.; Piris, P.; Grand, M.L.; Montero, M.P.; Matteudi, M.; Agavnian-Couquiaud, E.; Appay, R.; Keime, C.; Williamson, D.; et al. Beta-blockers disrupt mitochondrial bioenergetics and increase radiotherapy efficacy independently of beta-adrenergic receptors in medulloblastoma. eBioMedicine 2022, 82, 104149. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.; Ohkuni, A.; Kitamura, T.; Abe, K.; Naganuma, T.; Ohno, Y.; Zoeller, R.A.; Kihara, A. The Sjögren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol. Cell. 2012, 46, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, H.; Gerstl, M.P.; Ruckerbauer, D.; Hanscho, M.; Himmelbauer, H.; Clarke, C.; Barron, N.; Zanghellini, J.; Borth, N. Genetic and Epigenetic Variation across Genes Involved in Energy Metabolism and Mitochondria of Chinese Hamster Ovary Cell Lines. Biotechnol. J. 2019, 14, e1800681. [Google Scholar] [CrossRef] [PubMed]
- Sivanand, S.; Viney, I.; Wellen, K.E. Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem. Sci. 2018, 43, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wei, G.; Ma, J.; Cheng, S.; Jia, L.; Song, X.; Zhang, M.; Ju, M.; Wang, L.; Zhao, L.; et al. Identification of the prognostic value of ferroptosis-related gene signature in breast cancer patients. BMC Cancer 2021, 21, 645. [Google Scholar] [CrossRef] [PubMed]
- Parsazad, E.; Esrafili, F.; Yazdani, B.; Ghafarzadeh, S.; Razmavar, N.; Sirous, H. Integrative bioinformatics analysis of ACS enzymes as candidate prognostic and diagnostic biomarkers in colon adenocarcinoma. Res. Pharm. Sci. 2023, 18, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Zeuner, S.; Vollmer, J.; Sigaud, R.; Oppermann, S.; Peterziel, H.; ElHarouni, D.; Oehme, I.; Witt, O.; Milde, T.; Ecker, J. Combination drug screen identifies synergistic drug interaction of BCL-XL and class I histone deacetylase inhibitors in MYC-amplified medulloblastoma cells. J. Neuro-Oncol. 2024, 166, 99–112. [Google Scholar] [CrossRef]
- 109. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015, 17, 405–424. [CrossRef]
| Gene Symbol | ID Transcript | Exon | DNA Base Positions | Amino Acid Changes | Variant Function | Gene Description |
|---|---|---|---|---|---|---|
| DAPK1 | NM_001288729 | exon11 | c.C938A | p.S313X | stopgain | Death-associated protein kinase 1 is a positive mediator of gamma-interferon-induced programed cell death. It is a tumor suppressor candidate. |
| HSPH1 | NM_001286503 | exon2 | c.G124T | p.G42X | stopgain | This gene encodes a member of the heat shock protein 70 family of proteins. An elevated expression of this protein has been observed in numerous human cancers. |
| LRP1B | NM_018557 | exon25 | c.G4168T | p.G1390X | stopgain | This gene encodes a member of the low density lipoprotein (LDL) receptor family. The disruption of this gene has been reported in several types of cancer. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bibbò, F.; Asadzadeh, F.; Boccia, A.; Sorice, C.; Bianco, O.; Saccà, C.D.; Majello, B.; Donofrio, V.; Bifano, D.; De Martino, L.; et al. Targeting Group 3 Medulloblastoma by the Anti-PRUNE-1 and Anti-LSD1/KDM1A Epigenetic Molecules. Int. J. Mol. Sci. 2024, 25, 3917. https://doi.org/10.3390/ijms25073917
Bibbò F, Asadzadeh F, Boccia A, Sorice C, Bianco O, Saccà CD, Majello B, Donofrio V, Bifano D, De Martino L, et al. Targeting Group 3 Medulloblastoma by the Anti-PRUNE-1 and Anti-LSD1/KDM1A Epigenetic Molecules. International Journal of Molecular Sciences. 2024; 25(7):3917. https://doi.org/10.3390/ijms25073917
Chicago/Turabian StyleBibbò, Francesca, Fatemeh Asadzadeh, Angelo Boccia, Carmen Sorice, Orazio Bianco, Carmen Daniela Saccà, Barbara Majello, Vittoria Donofrio, Delfina Bifano, Lucia De Martino, and et al. 2024. "Targeting Group 3 Medulloblastoma by the Anti-PRUNE-1 and Anti-LSD1/KDM1A Epigenetic Molecules" International Journal of Molecular Sciences 25, no. 7: 3917. https://doi.org/10.3390/ijms25073917
APA StyleBibbò, F., Asadzadeh, F., Boccia, A., Sorice, C., Bianco, O., Saccà, C. D., Majello, B., Donofrio, V., Bifano, D., De Martino, L., Quaglietta, L., Cristofano, A., Covelli, E. M., Cinalli, G., Ferrucci, V., De Antonellis, P., & Zollo, M. (2024). Targeting Group 3 Medulloblastoma by the Anti-PRUNE-1 and Anti-LSD1/KDM1A Epigenetic Molecules. International Journal of Molecular Sciences, 25(7), 3917. https://doi.org/10.3390/ijms25073917








