Epstein–Barr Virus Latent Membrane Protein 2A (LMP2A) Enhances ATP Production in B Cell Tumors through mTOR and HIF-1α
Abstract
1. Introduction
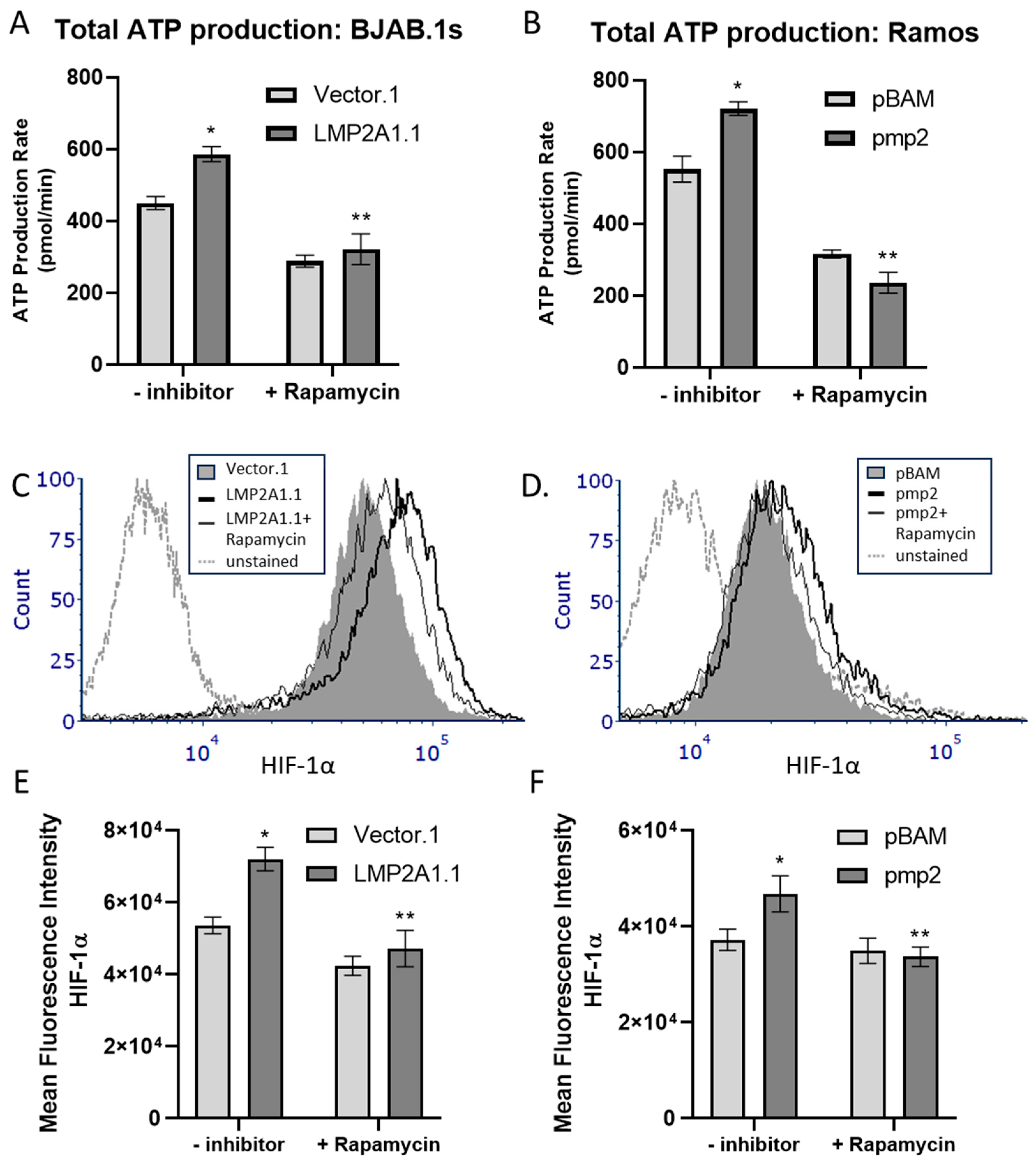
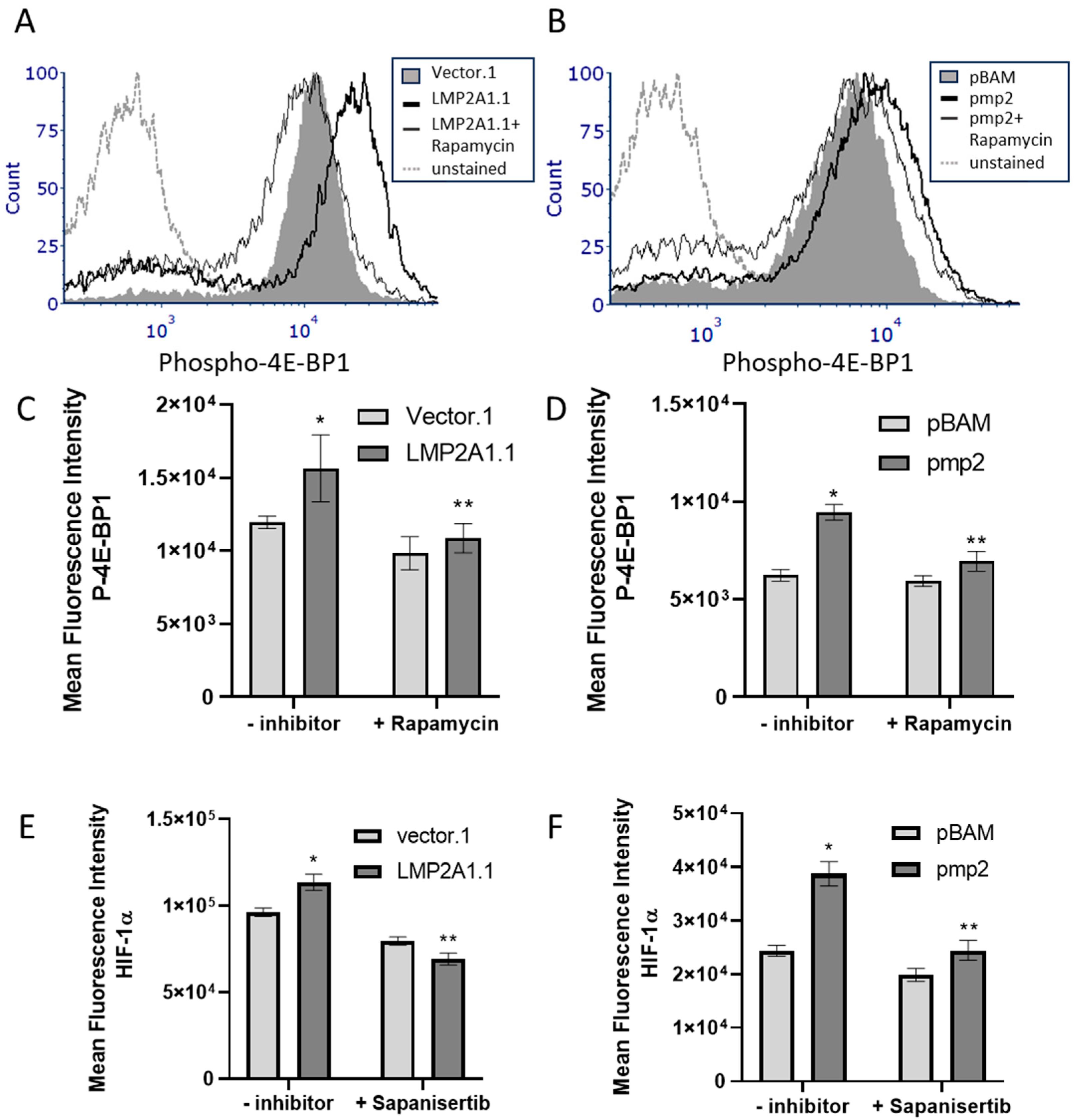
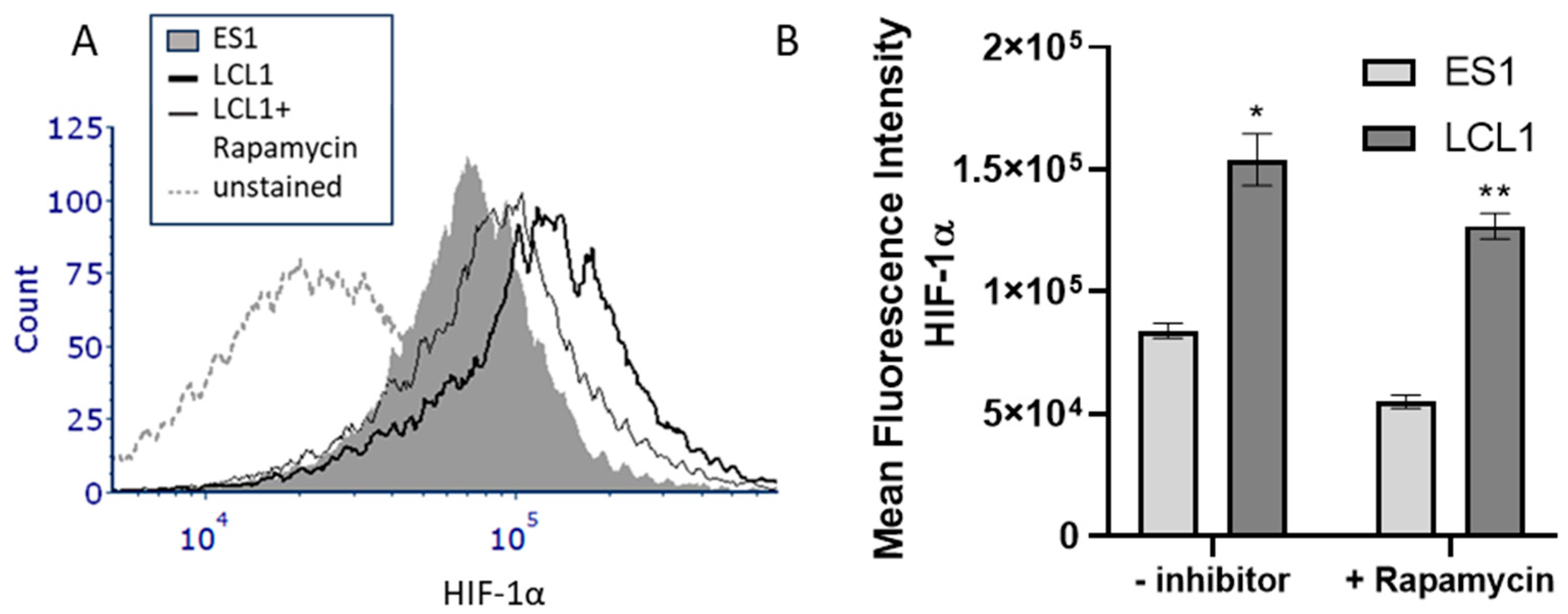
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Seahorse XF ATP Production Assay
4.3. Flow Cytometry
4.4. RT-PCR
4.5. Western Blot
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knipe, D.M.; Howley, P.M. Fields Virology, 6th ed.; Wolters Kluwer/Lippincott Williams & Wilkins Health: Philadelphia, PA, USA, 2013; 2 volumes. [Google Scholar]
- Flavell, K.J.; Murray, P.G. Hodgkin’s disease and the Epstein-Barr virus. Mol. Pathol. 2000, 53, 262–269. [Google Scholar] [CrossRef]
- Geng, L.; Wang, X. Epstein-Barr Virus-associated lymphoproliferative disorders: Experimental and clinical developments. Int. J. Clin. Exp. Med. 2015, 8, 14656–14671. [Google Scholar] [PubMed]
- Kempkes, B.; Robertson, E.S. Epstein-Barr virus latency: Current and future perspectives. Curr. Opin. Virol. 2015, 14, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Kuppers, R.; Engert, A.; Hansmann, M.L. Hodgkin lymphoma. J. Clin. Investig. 2012, 122, 3439–3447. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Allday, M.J. The curious case of the tumour virus: 50 years of Burkitt’s lymphoma. Nat. Rev. Microbiol. 2008, 6, 913–924. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Gross, A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 2004, 350, 1328–1337. [Google Scholar] [CrossRef]
- Huang, W.; Bai, L.; Tang, H. Epstein-Barr virus infection: The micro and macro worlds. Virol. J. 2023, 20, 220. [Google Scholar] [CrossRef]
- Cen, O.; Longnecker, R. Latent Membrane Protein 2 (LMP2). Curr. Top. Microbiol. Immunol. 2015, 391, 151–180. [Google Scholar] [CrossRef]
- Pang, M.F.; Lin, K.W.; Peh, S.C. The signaling pathways of Epstein-Barr virus-encoded latent membrane protein 2A (LMP2A) in latency and cancer. Cell Mol. Biol. Lett. 2009, 14, 222–247. [Google Scholar] [CrossRef]
- Fish, K.; Comoglio, F.; Shaffer, A.L., 3rd; Ji, Y.; Pan, K.T.; Scheich, S.; Oellerich, A.; Doebele, C.; Ikeda, M.; Schaller, S.J.; et al. Rewiring of B cell receptor signaling by Epstein-Barr virus LMP2A. Proc. Natl. Acad. Sci. USA 2020, 117, 26318–26327. [Google Scholar] [CrossRef]
- Merchant, M.; Caldwell, R.G.; Longnecker, R. The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. J. Virol. 2000, 74, 9115–9124. [Google Scholar] [CrossRef] [PubMed]
- Wasil, L.R.; Tomaszewski, M.J.; Hoji, A.; Rowe, D.T. The effect of Epstein-Barr virus Latent Membrane Protein 2 expression on the kinetics of early B cell infection. PLoS ONE 2013, 8, e54010. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fish, K.; Chen, J.; Longnecker, R. Epstein-Barr virus latent membrane protein 2A enhances MYC-driven cell cycle progression in a mouse model of B lymphoma. Blood 2014, 123, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Portis, T.; Longnecker, R. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene 2004, 23, 8619–8628. [Google Scholar] [CrossRef] [PubMed]
- Swart, R.; Fruehling, S.; Longnecker, R. Tyrosines 60, 64, and 101 of Epstein-Barr virus LMP2A are not essential for blocking B cell signal transduction. Virology 1999, 263, 485–495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moody, C.A.; Scott, R.S.; Amirghahari, N.; Nathan, C.O.; Young, L.S.; Dawson, C.W.; Sixbey, J.W. Modulation of the cell growth regulator mTOR by Epstein-Barr virus-encoded LMP2A. J. Virol. 2005, 79, 5499–5506. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.A.; Blenis, J. Coordinate regulation of translation by the PI 3-kinase and mTOR pathways. Adv. Cancer Res. 2002, 86, 1–39. [Google Scholar] [CrossRef]
- Vogt, P.K.; Hart, J.R. PI3K and STAT3: A new alliance. Cancer Discov. 2011, 1, 481–486. [Google Scholar] [CrossRef]
- Incrocci, R.; Barse, L.; Stone, A.; Vagvala, S.; Montesano, M.; Subramaniam, V.; Swanson-Mungerson, M. Epstein-Barr Virus Latent Membrane Protein 2A (LMP2A) enhances IL-10 production through the activation of Bruton’s tyrosine kinase and STAT3. Virology 2017, 500, 96–102. [Google Scholar] [CrossRef]
- Ricci, J.E.; Chiche, J. Metabolic Reprogramming of Non-Hodgkin’s B-Cell Lymphomas and Potential Therapeutic Strategies. Front. Oncol. 2018, 8, 556. [Google Scholar] [CrossRef]
- Duvel, K.; Yecies, J.L.; Menon, S.; Raman, P.; Lipovsky, A.I.; Souza, A.L.; Triantafellow, E.; Ma, Q.; Gorski, R.; Cleaver, S.; et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 2010, 39, 171–183. [Google Scholar] [CrossRef]
- Cantor, J.R.; Sabatini, D.M. Cancer cell metabolism: One hallmark, many faces. Cancer Discov. 2012, 2, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Agani, F.; Jiang, B.H. Oxygen-independent regulation of HIF-1: Novel involvement of PI3K/AKT/mTOR pathway in cancer. Curr. Cancer Drug Targets 2013, 13, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Dodd, K.M.; Yang, J.; Shen, M.H.; Sampson, J.R.; Tee, A.R. mTORC1 drives HIF-1alpha and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene 2015, 34, 2239–2250. [Google Scholar] [CrossRef] [PubMed]
- Doughty, C.A.; Bleiman, B.F.; Wagner, D.J.; Dufort, F.J.; Mataraza, J.M.; Roberts, M.F.; Chiles, T.C. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: Role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood 2006, 107, 4458–4465. [Google Scholar] [CrossRef] [PubMed]
- Franchina, D.G.; Grusdat, M.; Brenner, D. B-Cell Metabolic Remodeling and Cancer. Trends Cancer 2018, 4, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Daly, L.A.; Mennerich, D.; Kietzmann, T.; See, V. The Role of Hypoxia-Inducible Factor Post-Translational Modifications in Regulating Its Localisation, Stability, and Activity. Int. J. Mol. Sci. 2020, 22, 268. [Google Scholar] [CrossRef]
- Meng, X.; Grotsch, B.; Luo, Y.; Knaup, K.X.; Wiesener, M.S.; Chen, X.X.; Jantsch, J.; Fillatreau, S.; Schett, G.; Bozec, A. Hypoxia-inducible factor-1alpha is a critical transcription factor for IL-10-producing B cells in autoimmune disease. Nat. Commun. 2018, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; You, L.; Nepovimova, E.; Heger, Z.; Wu, W.; Kuca, K.; Adam, V. Hypoxia-inducible factors: Master regulators of hypoxic tumor immune escape. J. Hematol. Oncol. 2022, 15, 77. [Google Scholar] [CrossRef]
- Hua, H.; Kong, Q.; Zhang, H.; Wang, J.; Luo, T.; Jiang, Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019, 12, 71. [Google Scholar] [CrossRef]
- Ben-Sahra, I.; Howell, J.J.; Asara, J.M.; Manning, B.D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 2013, 339, 1323–1328. [Google Scholar] [CrossRef]
- Pourdehnad, M.; Truitt, M.L.; Siddiqi, I.N.; Ducker, G.S.; Shokat, K.M.; Ruggero, D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc. Natl. Acad. Sci. USA 2013, 110, 11988–11993. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A.B.; Bell, A.I. Epstein-Barr virus-associated lymphomas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160271. [Google Scholar] [CrossRef]
- Yang, T.; You, C.; Meng, S.; Lai, Z.; Ai, W.; Zhang, J. EBV Infection and Its Regulated Metabolic Reprogramming in Nasopharyngeal Tumorigenesis. Front. Cell Infect. Microbiol. 2022, 12, 935205. [Google Scholar] [CrossRef]
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964, 1, 702–703. [Google Scholar] [CrossRef]
- Xia, Y.; Jiang, L.; Zhong, T. The role of HIF-1alpha in chemo-/radioresistant tumors. Onco Targets Ther. 2018, 11, 3003–3011. [Google Scholar] [CrossRef]
- Pearce, E.L.; Pearce, E.J. Metabolic pathways in immune cell activation and quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef]
- Gaballah, A.; Bartosch, B. An Update on the Metabolic Landscape of Oncogenic Viruses. Cancers 2022, 14, 5742. [Google Scholar] [CrossRef]
- Cuninghame, S.; Jackson, R.; Zehbe, I. Hypoxia-inducible factor 1 and its role in viral carcinogenesis. Virology 2014, 456, 370–383. [Google Scholar] [CrossRef]
- Piccaluga, P.P.; Weber, A.; Ambrosio, M.R.; Ahmed, Y.; Leoncini, L. Epstein-Barr Virus-Induced Metabolic Rearrangements in Human B-Cell Lymphomas. Front. Microbiol. 2018, 9, 1233. [Google Scholar] [CrossRef]
- Cao, Y.; Xie, L.L.; Shi, F.; Tang, M.; Li, Y.S.; Hu, J.M.; Zhao, L.; Zhao, L.Q.; Yu, X.F.; Luo, X.J.; et al. Targeting the signaling in Epstein-Barr virus-associated diseases: Mechanism, regulation, and clinical study. Signal Transduct. Target. Ther. 2021, 6, 15. [Google Scholar] [CrossRef]
- Saha, A.; Robertson, E.S. Mechanisms of B-Cell Oncogenesis Induced by Epstein-Barr Virus. J. Virol. 2019, 93, e00238-19. [Google Scholar] [CrossRef]
- O’Neil, J.D.; Owen, T.J.; Wood, V.H.J.; Date, K.L.; Valentine, R.; Chukwuma, M.B.; Arrand, J.R.; Dawson, C.W.; Young, L.S. Epstein-Barr virus-encoded EBNA1 modulates the AP-1 transcription factor pathway in nasopharyngeal carcinoma cells and enhances angiogenesis in vitro. J. Gen. Virol. 2008, 89, 2833–2842. [Google Scholar] [CrossRef]
- Kondo, S.; Seo, S.Y.; Yoshizaki, T.; Wakisaka, N.; Furukawa, M.; Joab, I.; Jang, K.L.; Pagano, J.S. EBV latent membrane protein 1 up-regulates hypoxia-inducible factor 1alpha through Siah1-mediated down-regulation of prolyl hydroxylases 1 and 3 in nasopharyngeal epithelial cells. Cancer Res. 2006, 66, 9870–9877. [Google Scholar] [CrossRef]
- Darekar, S.; Georgiou, K.; Yurchenko, M.; Yenamandra, S.P.; Chachami, G.; Simos, G.; Klein, G.; Kashuba, E. Epstein-Barr virus immortalization of human B-cells leads to stabilization of hypoxia-induced factor 1 alpha, congruent with the Warburg effect. PLoS ONE 2012, 7, e42072. [Google Scholar] [CrossRef]
- Ariaans, G.; Jalving, M.; Vries, E.G.; Jong, S. Anti-tumor effects of everolimus and metformin are complementary and glucose-dependent in breast cancer cells. BMC Cancer 2017, 17, 232. [Google Scholar] [CrossRef]
- Sakamoto, T.; Weng, J.S.; Hara, T.; Yoshino, S.; Kozuka-Hata, H.; Oyama, M.; Seiki, M. Hypoxia-inducible factor 1 regulation through cross talk between mTOR and MT1-MMP. Mol. Cell Biol. 2014, 34, 30–42. [Google Scholar] [CrossRef]
- Kraus, R.J.; Yu, X.; Cordes, B.A.; Sathiamoorthi, S.; Iempridee, T.; Nawandar, D.M.; Ma, S.; Romero-Masters, J.C.; McChesney, K.G.; Lin, Z.; et al. Hypoxia-inducible factor-1alpha plays roles in Epstein-Barr virus’s natural life cycle and tumorigenesis by inducing lytic infection through direct binding to the immediate-early BZLF1 gene promoter. PLoS Pathog. 2017, 13, e1006404. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.H.; Wang, N.; Li, A.; Liao, W.T.; Pan, Z.G.; Mai, S.J.; Li, D.J.; Zeng, M.S.; Wen, J.M.; Zeng, Y.X. Hypoxia can contribute to the induction of the Epstein-Barr virus (EBV) lytic cycle. J. Clin. Virol. 2006, 37, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, R.G.; Brown, R.C.; Longnecker, R. Epstein-Barr virus LMP2A-induced B-cell survival in two unique classes of EmuLMP2A transgenic mice. J. Virol. 2000, 74, 1101–1113. [Google Scholar] [CrossRef]
- Caldwell, R.G.; Wilson, J.B.; Anderson, S.J.; Longnecker, R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 1998, 9, 405–411. [Google Scholar] [CrossRef]
- Gold, M.R.; Matsuuchi, L.; Kelly, R.B.; DeFranco, A.L. Tyrosine phosphorylation of components of the B-cell antigen receptors following receptor crosslinking. Proc. Natl. Acad. Sci. USA 1991, 88, 3436–3440. [Google Scholar] [CrossRef]
- Portis, T.; Longnecker, R. Epstein-Barr virus (EBV) LMP2A alters normal transcriptional regulation following B-cell receptor activation. Virology 2004, 318, 524–533. [Google Scholar] [CrossRef]
- Kaymaz, Y.; Oduor, C.I.; Yu, H.; Otieno, J.A.; Ong’echa, J.M.; Moormann, A.M.; Bailey, J.A. Comprehensive Transcriptome and Mutational Profiling of Endemic Burkitt Lymphoma Reveals EBV Type-Specific Differences. Mol. Cancer Res. 2017, 15, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Bieging, K.T.; Amick, A.C.; Longnecker, R. Epstein-Barr virus LMP2A bypasses p53 inactivation in a MYC model of lymphomagenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 17945–17950. [Google Scholar] [CrossRef]
- Cen, O.; Longnecker, R. Rapamycin reverses splenomegaly and inhibits tumor development in a transgenic model of Epstein-Barr virus-related Burkitt’s lymphoma. Mol. Cancer Ther. 2011, 10, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Incrocci, R.; McAloon, J.; Montesano, M.; Bardahl, J.; Vagvala, S.; Stone, A.; Swanson-Mungerson, M. Epstein-Barr virus LMP2A utilizes Syk and PI3K to activate NF-kappaB in B-cell lymphomas to increase MIP-1alpha production. J. Med. Virol. 2019, 91, 845–855. [Google Scholar] [CrossRef]
- Kung, C.P.; Meckes, D.G., Jr.; Raab-Traub, N. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCdelta. J. Virol. 2011, 85, 4399–4408. [Google Scholar] [CrossRef] [PubMed]
- Laherty, C.D.; Hu, H.M.; Opipari, A.W.; Wang, F.; Dixit, V.M. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappa B. J. Biol. Chem. 1992, 267, 24157–24160. [Google Scholar] [CrossRef]
- Ma, S.D.; Tsai, M.H.; Romero-Masters, J.C.; Ranheim, E.A.; Huebner, S.M.; Bristol, J.A.; Delecluse, H.J.; Kenney, S.C. Latent Membrane Protein 1 (LMP1) and LMP2A Collaborate To Promote Epstein-Barr Virus-Induced B Cell Lymphomas in a Cord Blood-Humanized Mouse Model but Are Not Essential. J. Virol. 2017, 91, e01928-16. [Google Scholar] [CrossRef] [PubMed]
- Wowro, S.J.; Schmitt, K.R.L.; Tong, G.; Berger, F.; Schubert, S. Effects of mTOR and calcineurin inhibitors combined therapy in Epstein-Barr virus positive and negative Burkitt lymphoma cells. Int. Immunopharmacol. 2016, 30, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Azizian, N.G.; Sullivan, D.K.; Li, Y. mTOR inhibition attenuates chemosensitivity through the induction of chemotherapy resistant persisters. Nat. Commun. 2022, 13, 7047. [Google Scholar] [CrossRef]
- Fukuda, M.; Longnecker, R. Latent membrane protein 2A inhibits transforming growth factor-beta 1-induced apoptosis through the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 2004, 78, 1697–1705. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Incrocci, R.; Monroy Del Toro, R.; Devitt, G.; Salimian, M.; Braich, K.; Swanson-Mungerson, M. Epstein–Barr Virus Latent Membrane Protein 2A (LMP2A) Enhances ATP Production in B Cell Tumors through mTOR and HIF-1α. Int. J. Mol. Sci. 2024, 25, 3944. https://doi.org/10.3390/ijms25073944
Incrocci R, Monroy Del Toro R, Devitt G, Salimian M, Braich K, Swanson-Mungerson M. Epstein–Barr Virus Latent Membrane Protein 2A (LMP2A) Enhances ATP Production in B Cell Tumors through mTOR and HIF-1α. International Journal of Molecular Sciences. 2024; 25(7):3944. https://doi.org/10.3390/ijms25073944
Chicago/Turabian StyleIncrocci, Ryan, Rosalinda Monroy Del Toro, Grace Devitt, Melody Salimian, Kamaljit Braich, and Michelle Swanson-Mungerson. 2024. "Epstein–Barr Virus Latent Membrane Protein 2A (LMP2A) Enhances ATP Production in B Cell Tumors through mTOR and HIF-1α" International Journal of Molecular Sciences 25, no. 7: 3944. https://doi.org/10.3390/ijms25073944
APA StyleIncrocci, R., Monroy Del Toro, R., Devitt, G., Salimian, M., Braich, K., & Swanson-Mungerson, M. (2024). Epstein–Barr Virus Latent Membrane Protein 2A (LMP2A) Enhances ATP Production in B Cell Tumors through mTOR and HIF-1α. International Journal of Molecular Sciences, 25(7), 3944. https://doi.org/10.3390/ijms25073944






