Functional Analysis of β-Carotene Oxygenase 2 (BCO2) Gene in Yesso Scallop (Patinopecten yessoensis)
Abstract
1. Introduction
2. Results and Discussion
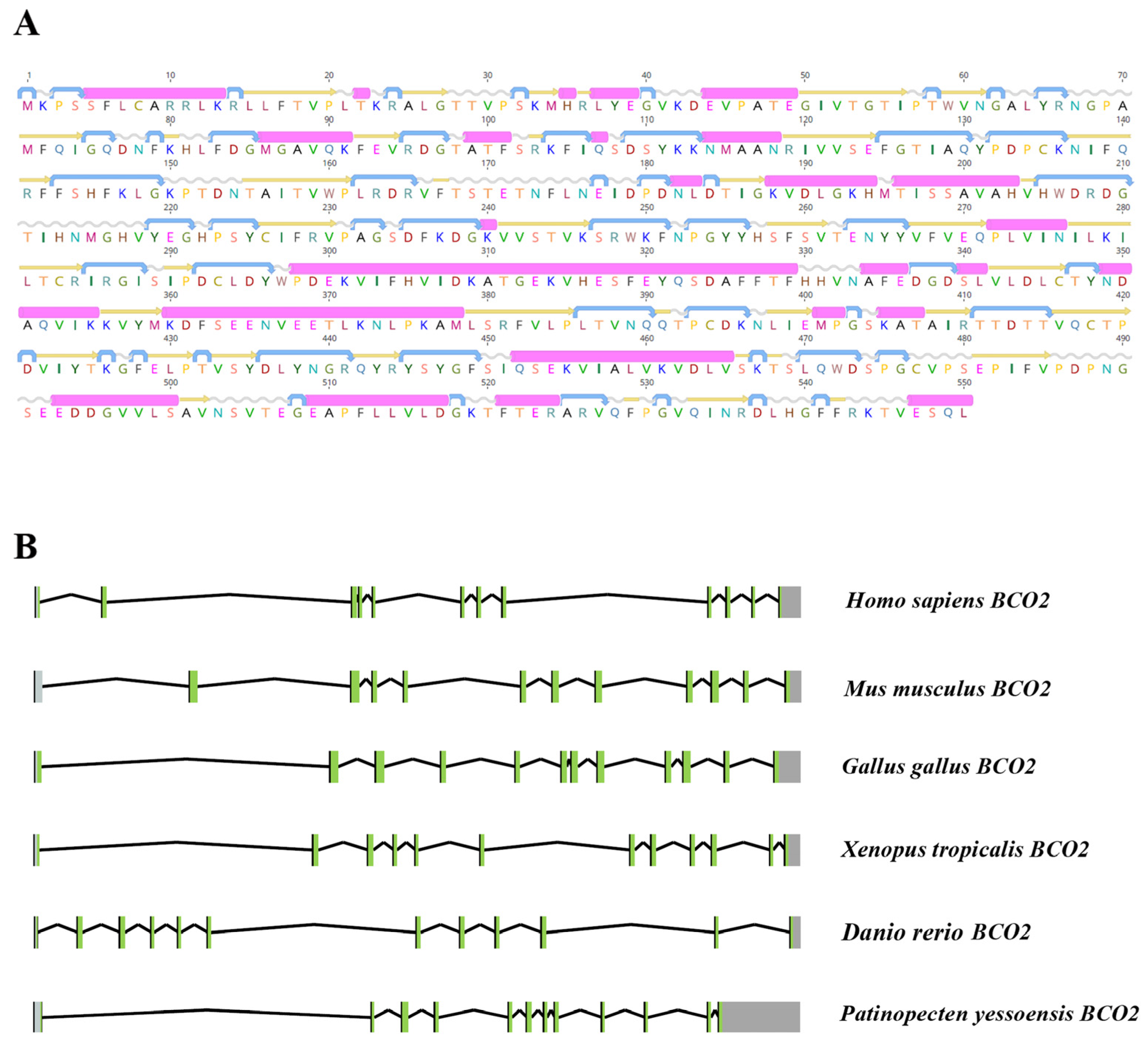
2.1. Characterization of PyBCO2 Gene Sequence
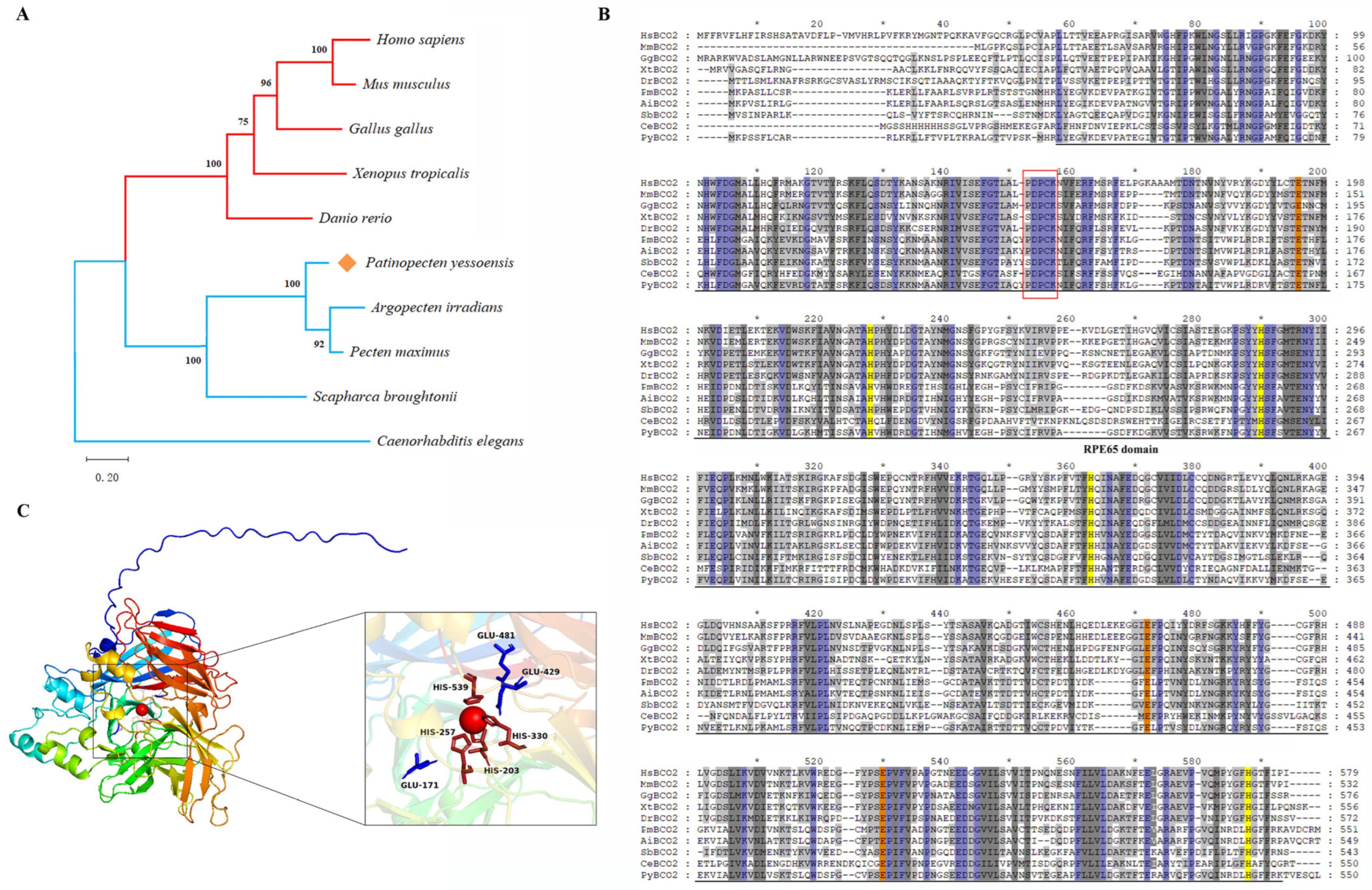
2.2. Homology and Phylogenetic Analysis
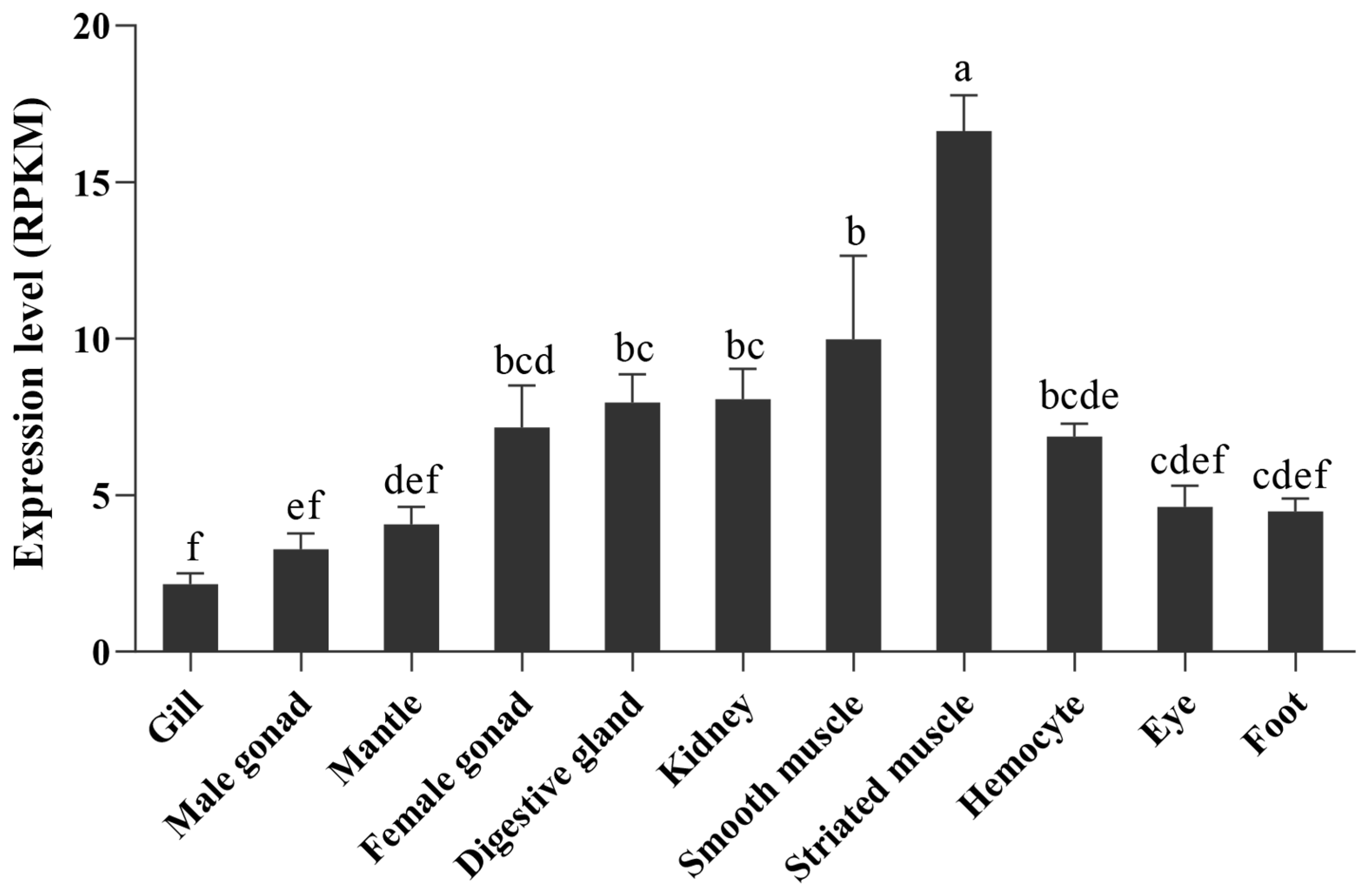
2.3. Expression Pattern of PyBCO2 in Adult Organs/Tissues
2.4. Functional Analysis of PyBCO2
3. Materials and Methods
3.1. Biological Materials
3.2. RNA Isolation and cDNA Synthesis
3.3. Cloning of PyBCO2 Gene and Sequence Analysis
3.4. Expression Analysis of PyBCO2 in Different Adult Organs/Tissues
3.5. Inhibiting PyBCO2 Expression
3.6. Quantitative Real-Time PCR (qRT-PCR)
3.7. High-Performance Liquid Chromatography (HPLC) Analysis
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.A. Carotenoids in health and disease: Recent scientific evaluations, research recommendations and the consumer. J. Nutr. 2004, 134, 221S–224S. [Google Scholar] [CrossRef] [PubMed]
- Maoka, T. Carotenoids as natural functional pigments. J. Nat. Med. 2020, 74, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.V.; Rao, L.G. Carotenoids and human health. Pharmacol. Res. 2007, 55, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.A.O.; Perez-Galvez, A. Carotenoids as a source of antioxidants in the diet. Sub-Cell. Biochem. 2016, 79, 359–375. [Google Scholar]
- da Costa, D.P.; Campos Miranda-Filho, K. The use of carotenoid pigments as food additives for aquatic organisms and their functional roles. Rev. Aquac. 2020, 12, 1567–1578. [Google Scholar] [CrossRef]
- Langi, P.; Kiokias, S.; Varzakas, T.; Proestos, C. Carotenoids: From plants to food and feed industries. Methods Mol. Biol. 2018, 1852, 57–71. [Google Scholar] [PubMed]
- Jensen, S.K.; Jensen, C.; Jakobsen, K.; Engberg, R.M.; Andersen, J.O.; Lauridsen, C.; Sorensen, P.; Skibsted, L.H.; Bertelsen, G. Supplementation of broiler diets with retinol acetate, β-carotene or canthaxanthin: Effect on vitamin status and oxidative status of broilers in vivo and on meat stability. Acta Agric. Scand. Sect. A-Anim. Sci. 1998, 48, 28–37. [Google Scholar] [CrossRef]
- Muramoto, T.; Nakanishi, N.; Shibata, M.; Aikawa, K. Effect of dietary β-carotene supplementation on beef color stability during display of two muscles from Japanese black steers. Meat Sci. 2003, 63, 39–42. [Google Scholar] [CrossRef]
- Hubbard, J.K.; Uy, J.A.C.; Hauber, M.E.; Hoekstra, H.E.; Safran, R.J. Vertebrate pigmentation: From underlying genes to adaptive function. Trends Genet. 2010, 26, 231–239. [Google Scholar] [CrossRef]
- Kiefer, C.; Hessel, S.; Lampert, J.M.; Vogt, K.; Lederer, M.O.; Breithaupt, D.E.; von Lintig, J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 2001, 276, 14110–14116. [Google Scholar] [CrossRef]
- Amengual, J.; Lobo, G.P.; Golczak, M.; Li, H.N.M.; Klimova, T.; Hoppel, C.L.; Wyss, A.; Palczewski, K.; von Lintig, J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. Faseb J. 2011, 25, 948–959. [Google Scholar] [CrossRef]
- Amengual, J.; Widjaja-Adhi, M.A.K.; Rodriguez-Santiago, S.; Hessel, S.; Golczak, M.; Palczewski, K.; von Lintig, J. Two carotenoid oxygenases contribute to mammalian provitamin a metabolism. J. Biol. Chem. 2013, 288, 34081–34096. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Kiser, P.D.; von Lintig, J.; Palczewski, K. Structural basis of carotenoid cleavage: From bacteria to mammals. Arch. Biochem. Biophys. 2013, 539, 203–213. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, M.; Yan, C.; Yang, H.; Wu, Z.; Liu, Y.; Su, N.; Hou, J.; Zhang, J.; Yang, F.; et al. CRISPR/Cas9-mediated deletion of β, β-carotene 9′, 10′-oxygenase gene (EcBCO2) from Exopalaemon carinicauda. Int. J. Biol. Macromol. 2020, 151, 168–177. [Google Scholar] [CrossRef]
- Berry, S.D.; Davis, S.R.; Beattie, E.M.; Thomas, N.L.; Burrett, A.K.; Ward, H.E.; Stanfield, A.M.; Biswas, M.; Ankersmit-Udy, A.E.; Oxley, P.E.; et al. Mutation in bovine β-carotene oxygenase 2 affects milk color. Genetics 2009, 182, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Pitchford, W.S.; Morris, C.A.; Cullen, N.G.; Bottema, C.D.K. Genetic variation in the β, β-carotene-9′,10′-dioxygenase gene and association with fat colour in bovine adipose tissue and milk. Anim. Genet. 2010, 41, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Vage, D.I.; Boman, I.A. A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genet. 2010, 11, 10. [Google Scholar] [CrossRef]
- Fallahshahroudi, A.; Sorato, E.; Altimiras, J.; Jensen, P. The domestic BCO2 allele buffers low-carotenoid diets in chickens: Possible citness increase through species hybridization. Genetics 2019, 212, 1445–1452. [Google Scholar] [CrossRef]
- Lehnert, S.J.; Christensen, K.A.; Vandersteen, W.E.; Sakhrani, D.; Pitcher, T.E.; Heath, J.W.; Koop, B.F.; Heath, D.D.; Devlin, R.H. Carotenoid pigmentation in salmon: Variation in expression at BCO2-l locus controls a key fitness trait affecting red coloration. Proc. R. Soc. B-Biol. Sci. 2019, 286, 20191588. [Google Scholar] [CrossRef]
- Wade, N.M.; Gabaudan, J.; Glencross, B.D. A review of carotenoid utilisation and function in crustacean aquaculture. Rev. Aquac. 2017, 9, 141–156. [Google Scholar] [CrossRef]
- Tan, K.; Zhang, H.; Zheng, H. Carotenoid content and composition: A special focus on commercially important fish and shellfish. Crit. Rev. Food Sci. Nutr. 2024, 64, 544–561. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Hu, J.; Wang, S.; Cheng, J.; Hu, X.; Lu, Z.; Lin, Z.; Zhu, W.; Bao, Z. Isolation and identification of the main carotenoid pigment from the rare orange muscle of the Yesso scallop. Food Chem. 2010, 118, 616–619. [Google Scholar] [CrossRef]
- Li, N. What Accounting for Orange Color of Yesso Scallop (Patinopecten yessoensis) Muscle and Its Application in Breeding. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2009. [Google Scholar]
- Li, X.; Wang, S.; Xun, X.; Zhang, M.; Wang, S.; Li, H.; Zhao, L.; Fu, Q.; Wang, H.; Li, T.; et al. A carotenoid oxygenase is responsible for muscle coloration in scallop. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2019, 1864, 966–975. [Google Scholar] [CrossRef]
- Palczewski, G.; Amengual, J.; Hoppel, C.L.; von Lintig, J. Evidence for compartmentalization of mammalian carotenoid metabolism. Faseb J. 2014, 28, 4457–4469. [Google Scholar] [CrossRef] [PubMed]
- Uppal, S.; Rogozin, I.B.; Redmond, T.M.; Poliakov, E. Palmitoylation of metazoan carotenoid oxygenases. Molecules 2020, 25, 1942. [Google Scholar] [CrossRef]
- Poliakov, E.; Uppal, S.; Rogozin, I.B.; Gentleman, S.; Redmond, T.M. Evolutionary aspects and enzymology of metazoan carotenoid cleavage oxygenases. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2020, 1865, 158665. [Google Scholar] [CrossRef]
- Wu, S.; Zhao, L.; Huang, J.; Li, Y.; Liu, Z.; Zhang, D. miR-330 targeting BCO2 is involved in carotenoid metabolism to regulate skin pigmentation in rainbow trout (Oncorhynchus mykiss). BMC Genom. 2023, 24, 124. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, A.; He, Y.-G.; Andersson, S. Cell type-specific expression of β-carotene 9′, 10′-monooxygenase in human tissues. J. Histochem. Cytochem. 2005, 53, 1403–1412. [Google Scholar] [CrossRef]
- Wu, L.; Guo, X.; Hartson, S.D.; Davis, M.A.; He, H.; Medeiros, D.M.; Wang, W.; Clarke, S.L.; Lucas, E.A.; Smith, B.J.; et al. Lack of β, β-carotene-9′, 10′-oxygenase 2 leads to hepatic mitochondrial dysfunction and cellular oxidative stress in mice. Mol. Nutr. Food Res. 2017, 61, 1600576. [Google Scholar] [CrossRef]
- Gong, M.; Bassi, A. Carotenoids from microalgae: A review of recent developments. Biotechnol. Adv. 2016, 34, 1396–1412. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Zhou, C.; Chu, R.; Chang, T.; Xu, J.; Ruan, R.; Chen, P.; Yan, X. Effect of microalgae diet and culture system on the rearing of bivalve mollusks: Nutritional properties and potential cost improvements. Algal Res. 2020, 51, 102076. [Google Scholar] [CrossRef]
- Shahidi, F.; Metusalach; Brown, J.A. Carotenoid pigments in seafoods and aquaculture. Crit. Rev. Food Sci. Nutr. 1998, 38, 1–67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, X.; Li, N.; Wang, S.; Li, X.; Sun, G.; Hu, X.; Bao, Z. Corralation analysis of carotenoid concentration and the growth traits of ‘Haida golden scallop’. Period. Ocean Univ. China/Zhongguo Haiyang Daxue Xuebao 2018, 48, 41–47. [Google Scholar]
- Li, X.; Li, N.; Zhao, L.; Shi, J.; Wang, S.; Ning, X.; Li, Y.; Hu, X. Tissue distribution and seasonal accumulation of carotenoids in Yesso scallop (Mizuhopecten yessoensis) with orange adductor muscle. Food Chem. 2022, 367, 130701. [Google Scholar] [CrossRef]
- Matsuno, T.; Hiraoka, K.; Maoka, T. Carotenoids in the gonad of scallops [Pactinopecten yessoensis]. Bull. Jap. Soc. Sci. Fish. 1981, 47, 385–390. [Google Scholar] [CrossRef]
- Narita, M.; Maoka, T.; Ebitani, K.; Nishino, H. Characteristics of chemical constituents and red pigment of orange adductor muscle of scallop Mizuhopecten yessoensis in the Okhotsk Sea and anti-oxidative activity of the pigment. Nippon. Suisan Gakkaishi 2013, 79, 48–54. [Google Scholar] [CrossRef]
- Maoka, T.; Kuwahara, T.; Narita, M. Carotenoids of sea angels Clione limacina and Paedoclione doliiformis from the perspective of the food chain. Mar. Drugs 2014, 12, 1460–1470. [Google Scholar] [CrossRef] [PubMed]
- Konishi, I.; Hosokawa, M.; Sashima, T.; Maoka, T.; Miyashita, K. Suppressive effects of alloxanthin and diatoxanthin from Halocynthia roretzi on LPS-induced expression of pro-inflammatory genes in RAW264.7 cells. J. Oleo. Sci. 2008, 57, 181–189. [Google Scholar] [CrossRef]
- Tsushima, M.; Maoka, T.; Katsuyama, M.; Kozuka, M.; Matsuno, T.; Tokuda, H.; Nishino, H.; Iwashima, A. Inhibitory effect of natural carotenoids on Epstein-Barr virus activation activity of a tumor promoter in Raji cells. A screening study for anti-tumor promoters. Biol. Pharm. Bull. 1995, 18, 227–233. [Google Scholar] [CrossRef]
- Eriksson, J.; Larson, G.; Gunnarsson, U.; Bed’hom, B.; Tixier-Boichard, M.; Strömstedt, L.; Wright, D.; Jungerius, A.; Vereijken, A.; Randi, E.; et al. Identification of the Yellow Skin gene reveals a hybrid origin of the domestic chicken. PLOS Genet. 2008, 4, e1000010. [Google Scholar] [CrossRef] [PubMed]
- Pointer, M.A.; Mundy, N.I. Chicken skin sheds light on carotenoid genetics. Heredity 2008, 101, 393–394. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Vachali, P.P.; Gorusupudi, A.; Shen, Z.; Sharifzadeh, H.; Besch, B.M.; Nelson, K.; Horvath, M.M.; Frederick, J.M.; Baehr, W.; et al. Inactivity of human beta,beta-carotene-9′, 10′-dioxygenase (BCO2) underlies retinal accumulation of the human macular carotenoid pigment. Proc. Natl. Acad. Sci. USA 2014, 111, 10173–10178. [Google Scholar] [CrossRef] [PubMed]
- Gazda, M.A.; Toomey, M.B.; Araujo, P.M.; Lopes, R.J.; Afonso, S.; Myers, C.A.; Serres, K.; Kiser, P.D.; Hill, G.E.; Corbo, J.C.; et al. Genetic basis of de novo appearance of carotenoid ornamentation in bare parts of canaries. Mol. Biol. Evol. 2020, 37, 1317–1328. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Bao, Z.; Hu, J.; Shao, M.; Zhang, L.; Bi, K.; Zhan, A.; Huang, X. Cloning and characterization of tryptophan 2,3-dioxygenase gene of Zhikong scallop Chlamys farreri (Jones and Preston 1904). Aquac. Res. 2006, 37, 1187–1194. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Jiao, W.; Li, J.; Xun, X.; Sun, Y.; Guo, X.; Huan, P.; Dong, B.; Zhang, L.; et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 2017, 1, 0120. [Google Scholar] [CrossRef] [PubMed]
- Ahern, K. Primer premier. Science 1999, 286, 433. [Google Scholar] [CrossRef]
- Yin, T.; Cook, D.; Lawrence, M. ggbio: An R package for extending the grammar of graphics for genomic data. Genome Biol. 2012, 13, R77. [Google Scholar] [CrossRef] [PubMed]
- Jeanmougin, F.; Thompson, J.D.; Gouy, M.; Higgins, D.G.; Gibson, T.J. Multiple sequence alignment with Clustal x. Trends Biochem. Sci. 1998, 23, 403–405. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Popovic, B.V. Handbook of Univariate and Multivariate Data Analysis with IBM SPSS, 2nd ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2013; Volume 42, p. 2291. [Google Scholar]

| Primer Name | Sequence | Sequence Information |
|---|---|---|
| PyBCO2-Fw | ATGAAGCCATCATCATTTCT | Sequence validation |
| PyBCO2-Rv | CTATAACTGACTCTCTACAGT | Sequence validation |
| RNAi-Fw | GATCACTAATACGACTCACTATAGGGGCGACGGAACCATCCACAA | RNAi |
| RNAi-Rv | GATCACTAATACGACTCACTATAGGGCCTCACTGAAATCCTTCATATACACCT | RNAi |
| PyBCO2-qPCR-Fw | GATGCCAGGCTCTAAAGCAAC | qRT-PCR |
| PyBCO2-qPCR-Rv | CTGAACCCGTACGAGTAACGATACT | qRT-PCR |
| UBQ-Fw | TCGCTGTAGTCTCCAGGATTGC | qRT-PCR |
| UBQ-Rv | TCGCCACATACCCTCCCAC | qRT-PCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Wang, S.; Zhao, L.; Li, T.; Zhang, Y.; Wang, H.; Bao, Z.; Hu, X. Functional Analysis of β-Carotene Oxygenase 2 (BCO2) Gene in Yesso Scallop (Patinopecten yessoensis). Int. J. Mol. Sci. 2024, 25, 3947. https://doi.org/10.3390/ijms25073947
Liu S, Wang S, Zhao L, Li T, Zhang Y, Wang H, Bao Z, Hu X. Functional Analysis of β-Carotene Oxygenase 2 (BCO2) Gene in Yesso Scallop (Patinopecten yessoensis). International Journal of Molecular Sciences. 2024; 25(7):3947. https://doi.org/10.3390/ijms25073947
Chicago/Turabian StyleLiu, Shiqi, Shuyue Wang, Liang Zhao, Tingting Li, Yihan Zhang, Huizhen Wang, Zhenmin Bao, and Xiaoli Hu. 2024. "Functional Analysis of β-Carotene Oxygenase 2 (BCO2) Gene in Yesso Scallop (Patinopecten yessoensis)" International Journal of Molecular Sciences 25, no. 7: 3947. https://doi.org/10.3390/ijms25073947
APA StyleLiu, S., Wang, S., Zhao, L., Li, T., Zhang, Y., Wang, H., Bao, Z., & Hu, X. (2024). Functional Analysis of β-Carotene Oxygenase 2 (BCO2) Gene in Yesso Scallop (Patinopecten yessoensis). International Journal of Molecular Sciences, 25(7), 3947. https://doi.org/10.3390/ijms25073947






