Proteomic Analyses Reveal the Role of Alpha-2-Macroglobulin in Canine Osteosarcoma Cell Migration
Abstract
:1. Introduction
2. Results
2.1. Proteins Differently Expressed in Canine OSA Cells in Comparison to Osteoblasts
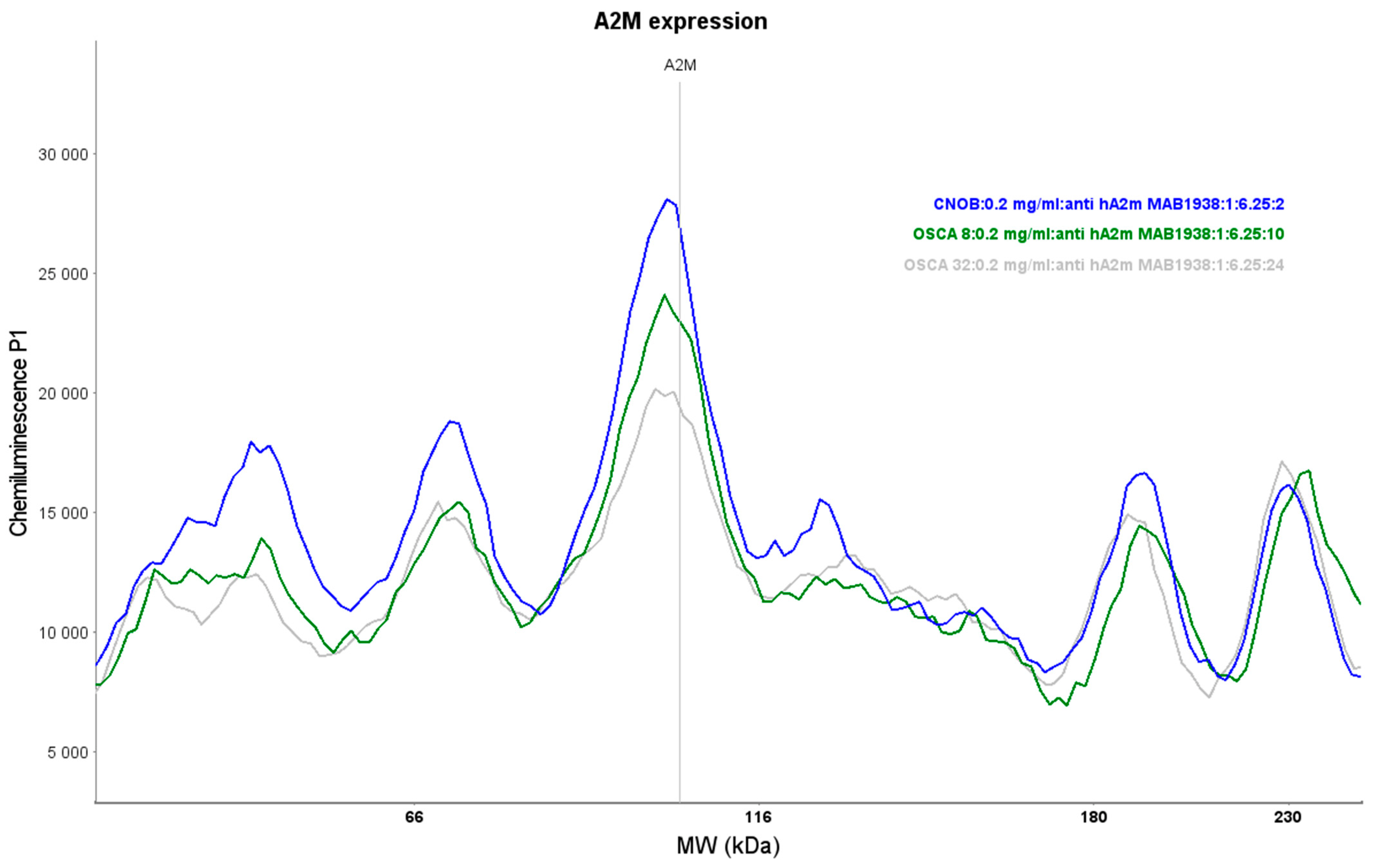
2.2. Expression of A2M Protein in Canine OSA Cell Lines and CnOb
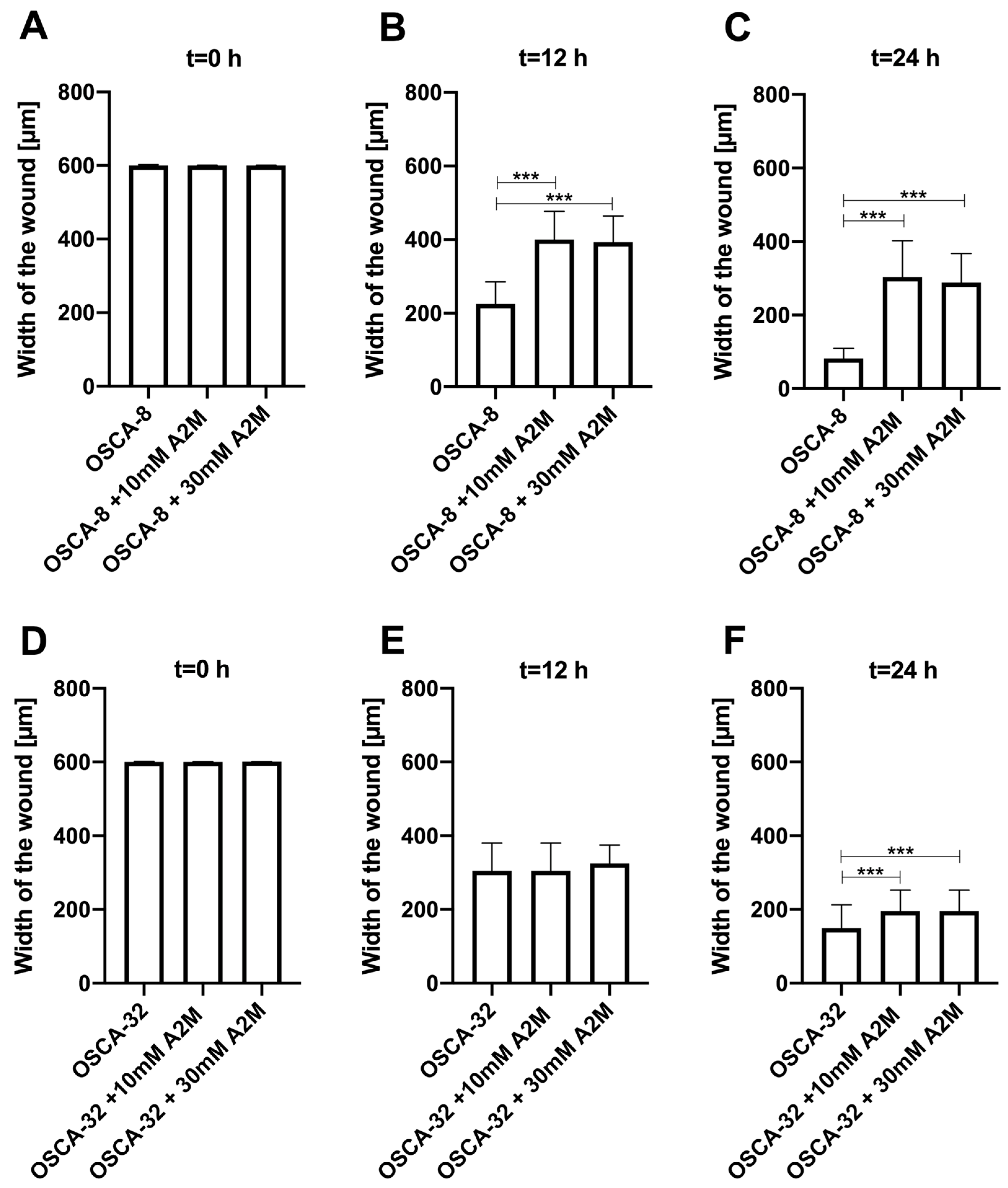
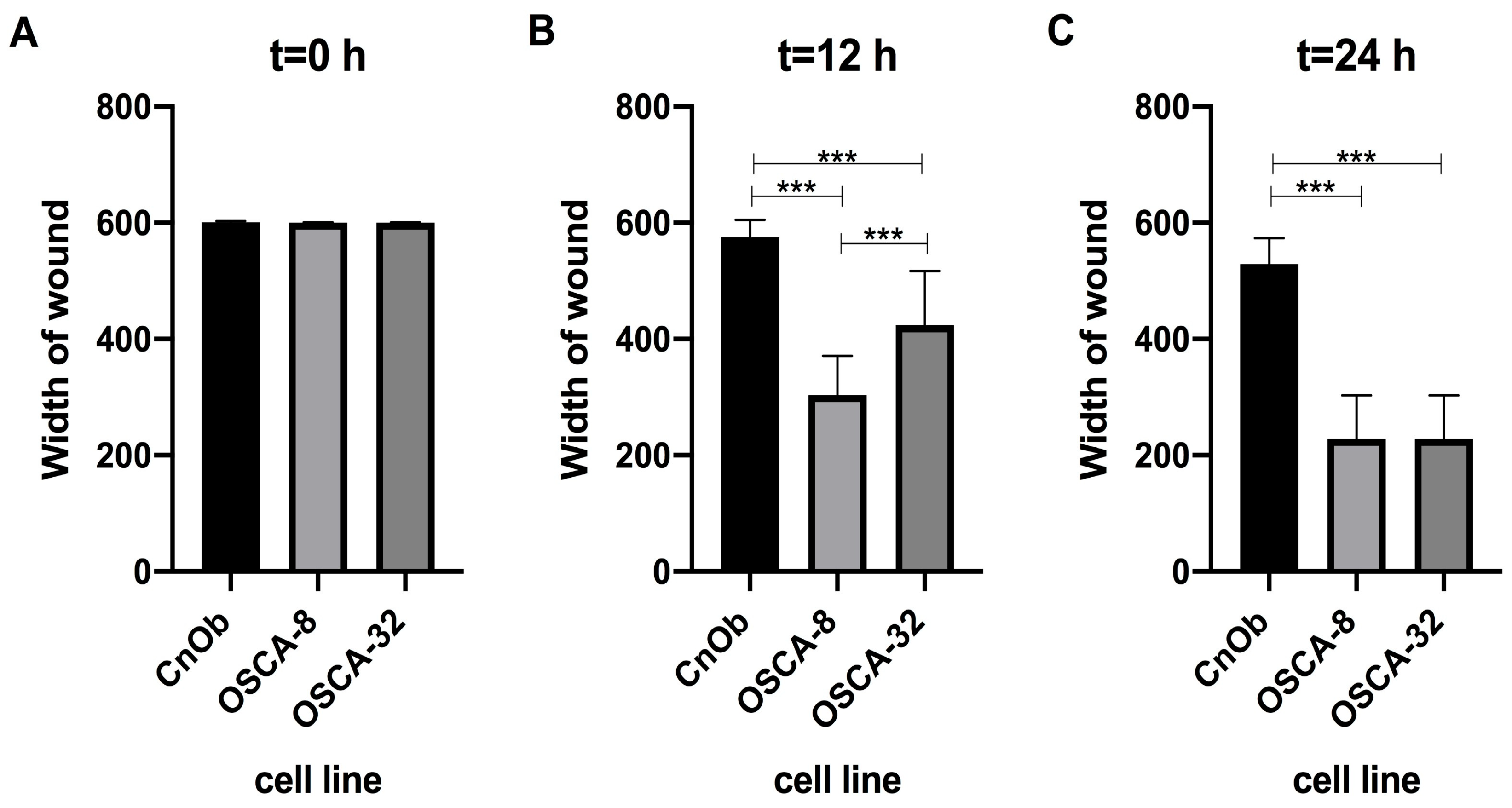
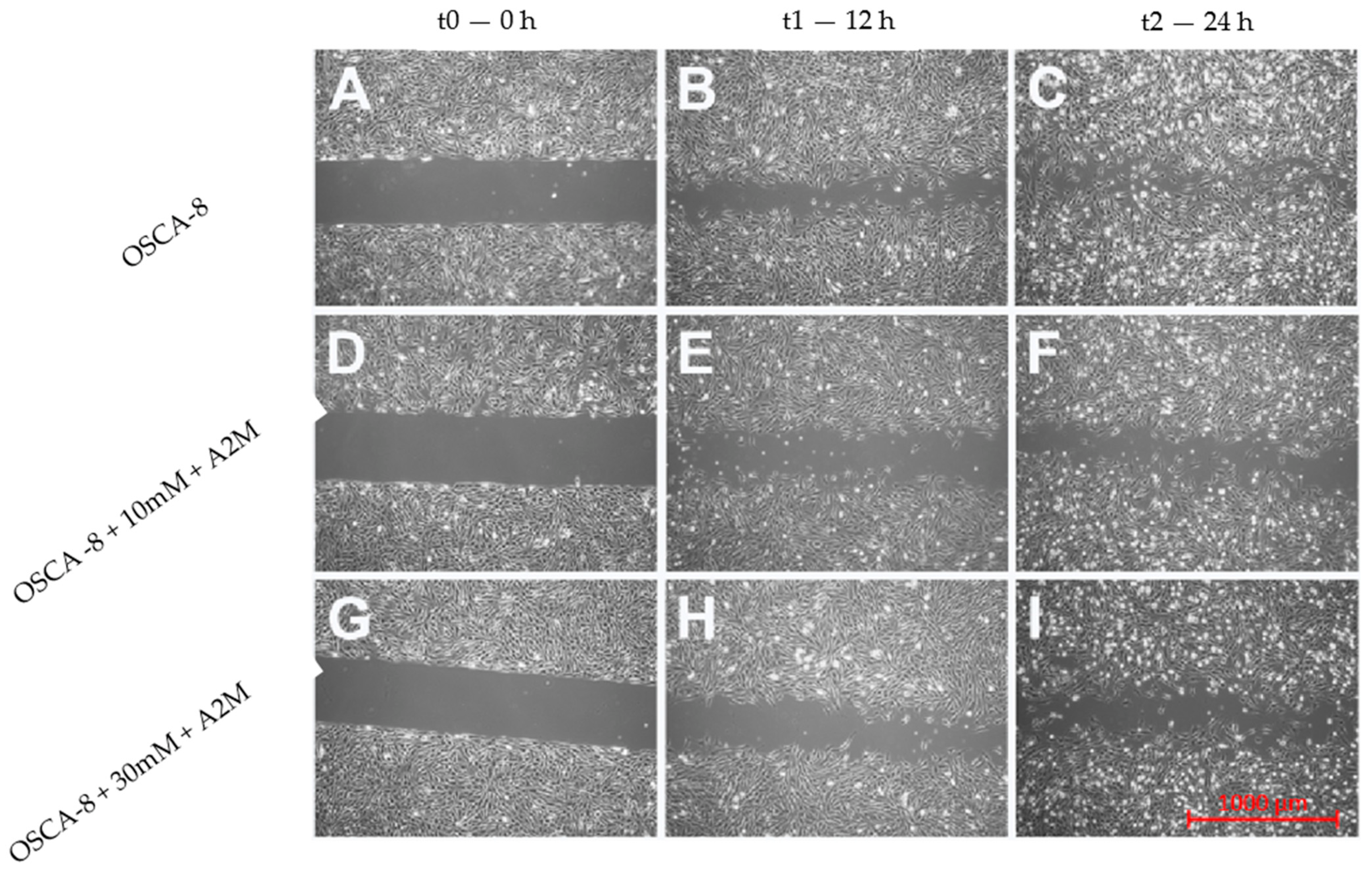
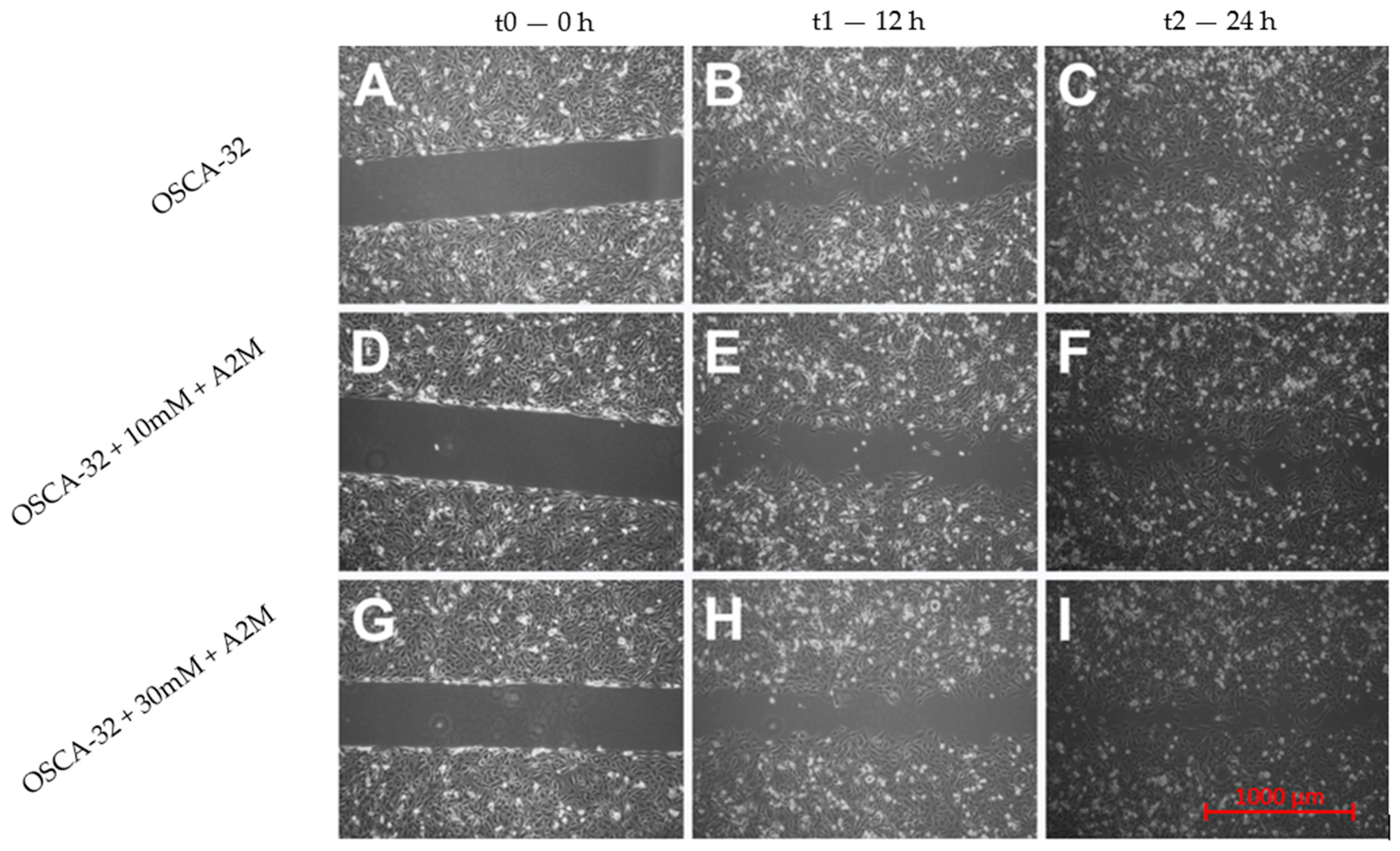
2.3. The Influence of A2M on the Migration Rate of OSCA-8 and OSCA-32 Cell Lines
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Cell Culture
4.3. 2DE Electrophoresis and MALDI-TOF/TOF MS Analysis
4.3.1. Protein Isolation, Cleaning, and Precipitation
4.3.2. Two-Dimensional Gel Electrophoresis (2DE)
4.3.3. MALDI-TOF/TOF MS Analysis
4.4. Validation of Protein Expression Using Simple Western
4.4.1. Protein Isolation, Cleaning, and Precipitation
4.4.2. Qualitative Protein Expression Analyses
Linear Range of Proteins and Primary Antibody Saturation Assessment
4.5. Migration Rate Assessment in Canine OSA Cell Lines Treated with A2M
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| 2DE | Two-dimensional gel electrophoresis |
| A2M | Alpha-2-macroglobulin |
| ANXA3 | Annexin A3 |
| ANXA5 | Annexin A5 |
| CFAP298 | Cilia- and flagella-associated protein 298 |
| CnOb | Canine osteoblasts |
| EDARADD | Ectodysplasin-A receptor-associated adapter protein |
| GTF2I | General transcription factor II-I |
| HCCP | Hibernating common carp plasma |
| ICC | Intrahepatic cholangiocarcinoma |
| LRRC72 | Leucine-rich repeat-containing protein 72 |
| MALDI-TOF/TOF MS | Matrix-assisted laser desorption/ionisation-time of flight mass spectrometry |
| MIPOL-1 | Mirror-image polydactyly gene 1 protein |
| OSA | Osteosarcoma |
| PGAM1 | Phosphoglycerate mutase 1 |
| TGFβ | Transforming growth factor beta |
| UB2L6 | Ubiquitin/ISG15-conjugating enzyme E2 |
References
- Beck, J.; Ren, L.; Huang, S.; Berger, E.; Bardales, K.; Mannheimer, J.; Mazcko, C.; LeBlanc, A. Canine and Murine Models of Osteosarcoma. Vet. Pathol. 2022, 59, 399–414. [Google Scholar] [CrossRef] [PubMed]
- Selvarajah, G.T.; Kirpensteijn, J. Prognostic and Predictive Biomarkers of Canine Osteosarcoma. Vet. J. 2010, 185, 28–35. [Google Scholar] [CrossRef]
- Szewczyk, M.; Lechowski, R.; Zabielska, K. What Do We Know about Canine Osteosarcoma Treatment?—Review. Vet. Res. Commun. 2015, 39, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Hapach, L.A.; Mosier, J.A.; Wang, W.; Reinhart-King, C.A. Engineered Models to Parse Apart the Metastatic Cascade. NPJ Precis. Oncol. 2019, 3, 20. [Google Scholar] [CrossRef]
- Wilk, S.S.; Zabielska-Koczywas, K.A. Molecular Mechanisms of Canine Osteosarcoma Metastasis. Int. J. Mol. Sci. 2021, 22, 3639. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Edfors, F.; Danielsson, F.; Hallström, B.M.; Käll, L.; Lundberg, E.; Pontén, F.; Forsström, B.; Uhlén, M. Gene-specific Correlation of RNA and Protein Levels in Human Cells and Tissues. Mol. Syst. Biol. 2016, 12, 883. [Google Scholar] [CrossRef]
- Hillenkamp, F.; Karas, M.; Beavis, R.C.; Chait, B.T. Matrix-Assisted Laser Desorption/Lonization Mass Spectrometry of Biopolymers. Anal. Chem. 1991, 63, 1193A–1203A. [Google Scholar] [CrossRef]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for Identification of Environmental Bacteria: A Review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Klose, P.; Weise, C.; Bondzio, A.; Multhaup, G.; Einspanier, R.; Gruber, A.D. Proteome of Metastatic Canine Mammary Carcinomas: Similarities to and Differences from Human Breast Cancer. J. Proteome Res. 2010, 9, 6380–6391. [Google Scholar] [CrossRef]
- Zabielska-Koczywąs, K.; Michalak, K.; Wojtalewicz, A.; Winiarczyk, M.; Adaszek, Ł.; Winiarczyk, S.; Lechowski, R. Proteomic Differences in Feline Fibrosarcomas Grown Using Doxorubicin-Sensitive and -Resistant Cell Lines in the Chick Embryo Model. Int. J. Mol. Sci. 2018, 19, 576. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, C.; Miller, I.; Hummel, K.; Neschi née Ondrovics, M.; Schlosser, S.; Walter, I. Comparative Proteome Analysis of Monolayer and Spheroid Culture of Canine Osteosarcoma Cells. J. Proteom. 2018, 177, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef] [PubMed]
- Robert, M.G.; Cornet, M.; Hennebique, A.; Rasamoelina, T.; Caspar, Y.; Pondérand, L.; Bidart, M.; Durand, H.; Jacquet, M.; Garnaud, C.; et al. Maldi-Tof Ms in a Medical Mycology Laboratory: On Stage and Backstage. Microorganisms 2021, 9, 1283. [Google Scholar] [CrossRef] [PubMed]
- Maurer, K.; Eschrich, K.; Schellenberger, W.; Bertolini, J.; Rupf, S.; Remmerbach, T.W. Oral Brush Biopsy Analysis by MALDI-ToF Mass Spectrometry for Early Cancer Diagnosis. Oral Oncol. 2013, 49, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yu, T.; Xie, Y.; Yang, Y.; Li, Y.; Zhou, X.; Tsung, S.; Loo, R.R.; Loo, J.A.; Wong, D.T. Discovery of Oral Fluid Biomarkers for Human Oral Cancer by Mass Spectrometry. Cancer Genom. Proteom. 2007, 4, 55–64. [Google Scholar]
- Zambonin, C.; Aresta, A. MALDI-TOF/MS Analysis of Non-Invasive Human Urine and Saliva Samples for the Identification of New Cancer Biomarkers. Molecules 2022, 27, 1925. [Google Scholar] [CrossRef]
- Han, Z.; Peng, C.; Yi, J.; Wang, Y.; Liu, Q.; Yang, Y.; Long, S.; Qiao, L.; Shen, Y. Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Profiling of Plasma Exosomes Evaluates Osteosarcoma Metastasis. iScience 2021, 24, 102906. [Google Scholar] [CrossRef]
- Ploypetch, S.; Jaresitthikunchai, J.; Phaonakrop, N.; Sakcamduang, W.; Manee-in, S.; Suriyaphol, P.; Roytrakul, S.; Suriyaphol, G. Utilizing MALDI-TOF MS and LC-MS/MS to Access Serum Peptidome-Based Biomarkers in Canine Oral Tumors. Sci. Rep. 2022, 12, 21641. [Google Scholar] [CrossRef]
- McCaw, D.L.; Chan, A.S.; Stegner, A.L.; Mooney, B.; Bryan, J.N.; Turnquist, S.E.; Henry, C.J.; Alexander, H.; Alexander, S. Proteomics of Canine Lymphoma Identifies Potential Cancer-Specific Protein Markers. Clin. Cancer Res. 2007, 13, 2496–2503. [Google Scholar] [CrossRef]
- Marsilio, S.; Newman, S.J.; Estep, J.S.; Giaretta, P.R.; Lidbury, J.A.; Warry, E.; Flory, A.; Morley, P.S.; Smoot, K.; Seeley, E.H.; et al. Differentiation of Lymphocytic-Plasmacytic Enteropathy and Small Cell Lymphoma in Cats Using Histology-Guided Mass Spectrometry. J. Vet. Intern. Med. 2020, 34, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Merlos Rodrigo, M.A.; Zitka, O.; Krizkova, S.; Moulick, A.; Adam, V.; Kizek, R. MALDI-TOF MS as Evolving Cancer Diagnostic Tool: A Review. J. Pharm. Biomed. Anal. 2014, 95, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, M.; Xu, B.; Steen, H.; Steen, J.A.J. Libfbi: A C++ Implementation for Fast Box Intersection and Application to Sparse Mass Spectrometry Data. Bioinformatics 2011, 27, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Sormunen, A.; Koivulehto, E.; Alitalo, K.; Saksela, K.; Laham-Karam, N.; Ylä-Herttuala, S. Comparison of Automated and Traditional Western Blotting Methods. Methods Protoc. 2023, 6, 43. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, X.; Sun, Z.; Feng, X.; Wang, H.; Hao, J.; Zhang, X. A2M Is a Potential Core Gene in Intrahepatic Cholangiocarcinoma. BMC Cancer 2022, 22, 5. [Google Scholar] [CrossRef] [PubMed]
- Sottrup-Jensen, L.; Stepanik, T.M.; Kristensen, T. Primary Structure of Human A2-Macroglobulin. V. The Complete Structure. J. Biol. Chem. 1984, 259, 8318–8327. [Google Scholar] [CrossRef]
- Wong, S.G.; Dessen, A. Structure of a Bacterial A2-Macroglobulin Reveals Mimicry of Eukaryotic Innate Immunity. Nat. Commun. 2014, 5, 4917. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.C.; Hall, P.K.; Nikolai, T.F.; McKenzie, A.K. Reduced Trypsin Binding Capacity of A2-Macroglobulin in Diabetes. Clin. Chim. Acta 1986, 154, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Kurz, S.; Thieme, R.; Amberg, R.; Groth, M.; Jahnke, H.G.; Pieroh, P.; Horn, L.C.; Kolb, M.; Huse, K.; Platzer, M.; et al. The Anti-Tumorigenic Activity of A2M—A Lesson from the Naked Mole-Rat. PLoS ONE 2017, 12, e0189514. [Google Scholar] [CrossRef]
- Lindner, I.; Hemdan, N.Y.A.; Buchold, M.; Huse, K.; Bigl, M.; Oerlecke, I.; Ricken, A.; Gaunitz, F.; Sack, U.; Naumann, A.; et al. A2-Macroglobulin Inhibits the Malignant Properties of Astrocytoma Cells By Impeding Β-Catenin Signaling. Cancer Res. 2010, 70, 277–287. [Google Scholar] [CrossRef]
- Newton, C.S.; Loukinova, E.; Mikhailenko, I.; Ranganathan, S.; Gao, Y.; Haudenschild, C.; Strickland, D.K. Platelet-Derived Growth Factor Receptor-β (PDGFR-β) Activation Promotes Its Association with the Low Density Lipoprotein Receptor-Related Protein (LRP): Evidence for Co-Receptor Function. J. Biol. Chem. 2005, 280, 27872–27878. [Google Scholar] [CrossRef]
- Lagrange, J.; Lecompte, T.; Knopp, T.; Lacolley, P.; Regnault, V. Alpha-2-Macroglobulin in Hemostasis and Thrombosis: An Underestimated Old Double-Edged Sword. J. Thromb. Haemost. 2022, 20, 806–815. [Google Scholar] [CrossRef]
- Cuéllar, J.M.; Cuéllar, V.G.; Scuderi, G.J. A2-Macroglobulin: Autologous Protease Inhibition Technology. Phys. Med. Rehabil. Clin. N. Am. 2016, 27, 909–918. [Google Scholar] [CrossRef]
- Amini, E.; Rahgozar, S.; Golpich, M.; Kefayat, A.; Fesharaki, M. Potential Anti-Cancer Effects of Hibernating Common Carp (Cyprinus Carpio) Plasma on B16-F10 Murine Melanoma: In Vitro and in Vivo Studies. Int. J. Biol. Macromol. 2023, 238, 124058. [Google Scholar] [CrossRef]
- Lee, H.C.; Chang, C.Y.; Huang, Y.C.; Wu, K.L.; Chiang, H.H.; Chang, Y.Y.; Liu, L.X.; Hung, J.Y.; Hsu, Y.L.; Wu, Y.Y.; et al. Downregulated ADAMTS1 Incorporating A2M Contributes to Tumorigenesis and Alters Tumor Immune Microenvironment in Lung Adenocarcinoma. Biology 2022, 11, 760. [Google Scholar] [CrossRef]
- Liu, J.; Wu, S.; Xie, X.; Wang, Z.; Lei, Q. Identification of Potential Crucial Genes and Key Pathways in Osteosarcoma. Hereditas 2020, 157, 29. [Google Scholar] [CrossRef]
- Niu, F.; Zhao, S.; Xu, C.Y.; Chen, L.; Ye, L.; Bi, G.B.; Tian, G.; Gong, P.; Nie, T.H. Identification and Functional Analysis of Differentially Expressed Genes Related to Metastatic Osteosarcoma. Asian Pac. J. Cancer Prev. 2014, 15, 10797–10801. [Google Scholar] [CrossRef]
- Scott, M.C.; Sarver, A.L.; Gavin, K.J.; Thayanithy, V.; Getzy, D.M.; Newman, R.A.; Cutter, G.R.; Lindblad-Toh, K.; Kisseberth, W.C.; Hunter, L.E.; et al. Molecular Subtypes of Osteosarcoma Identified by Reducing Tumor Heterogeneity through an Interspecies Comparative Approach. Bone 2011, 49, 356–367. [Google Scholar] [CrossRef]
- Scott, M.C.; Temiz, N.A.; Sarver, A.E.; Larue, R.S.; Susan, K.; Varshney, J.; Wolf, N.K.; Moriarity, B.S.; Timothy, D.; Brien, O.; et al. Comparative Transcriptome Analysis Quantifies Immune Cell Transcript Levels, Metastatic Progression and Survival in Osteosarcoma. Cancer Res. 2018, 78, 326–337. [Google Scholar] [CrossRef]
- Fenger, J.M.; Roberts, R.D.; Iwenofu, O.H.; Bear, M.D.; Zhang, X.; Couto, J.I.; Modiano, J.F.; Kisseberth, W.C.; London, C.A. MiR-9 Is Overexpressed in Spontaneous Canine Osteosarcoma and Promotes a Metastatic Phenotype Including Invasion and Migration in Osteoblasts and Osteosarcoma Cell Lines. BMC Cancer 2016, 16, 784. [Google Scholar] [CrossRef]
- Varankar, S.S.; Bapat, S.A. Migratory Metrics of Wound Healing: A Quantification Approach for in Vitro Scratch Assays. Front. Oncol. 2018, 8, 633. [Google Scholar] [CrossRef] [PubMed]
- Walewska, M.; Małek, A.; Taciak, B.; Wojtalewicz, A.; Wilk, S.; Wojtkowska, A.; Zabielska-Koczywąs, K.; Lechowski, R. PEG-Liposomal Doxorubicin as a Potential Agent for Canine Metastatic Osteosarcoma—In Vitro and Ex Ovo Studies. J. Vet. Res. 2023, 67, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.G.; Wu, X.; Guan, J.L. Wound-Healing Assay. Methods Mol. Biol. 2005, 294, 23–29. [Google Scholar] [CrossRef] [PubMed]
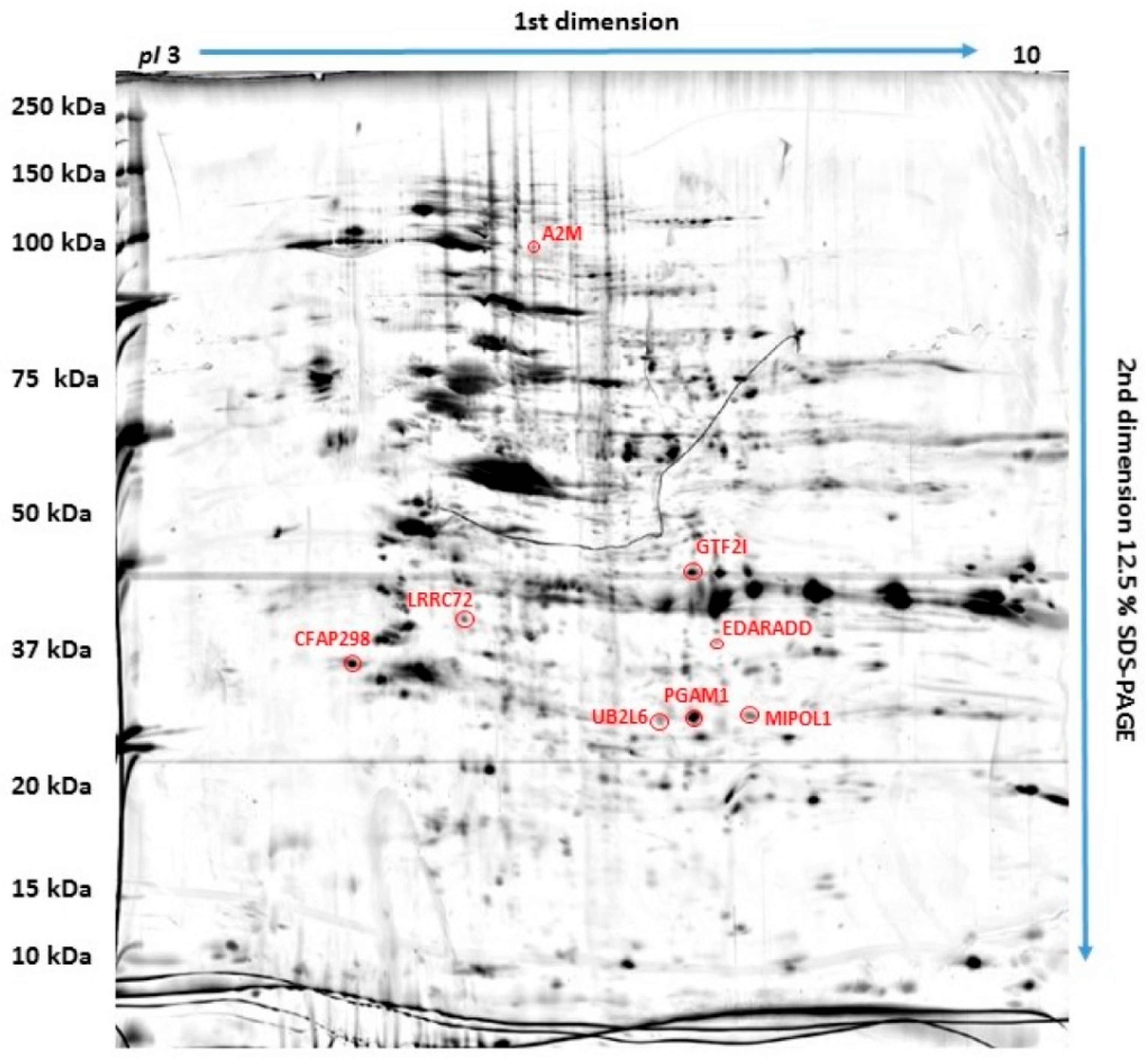
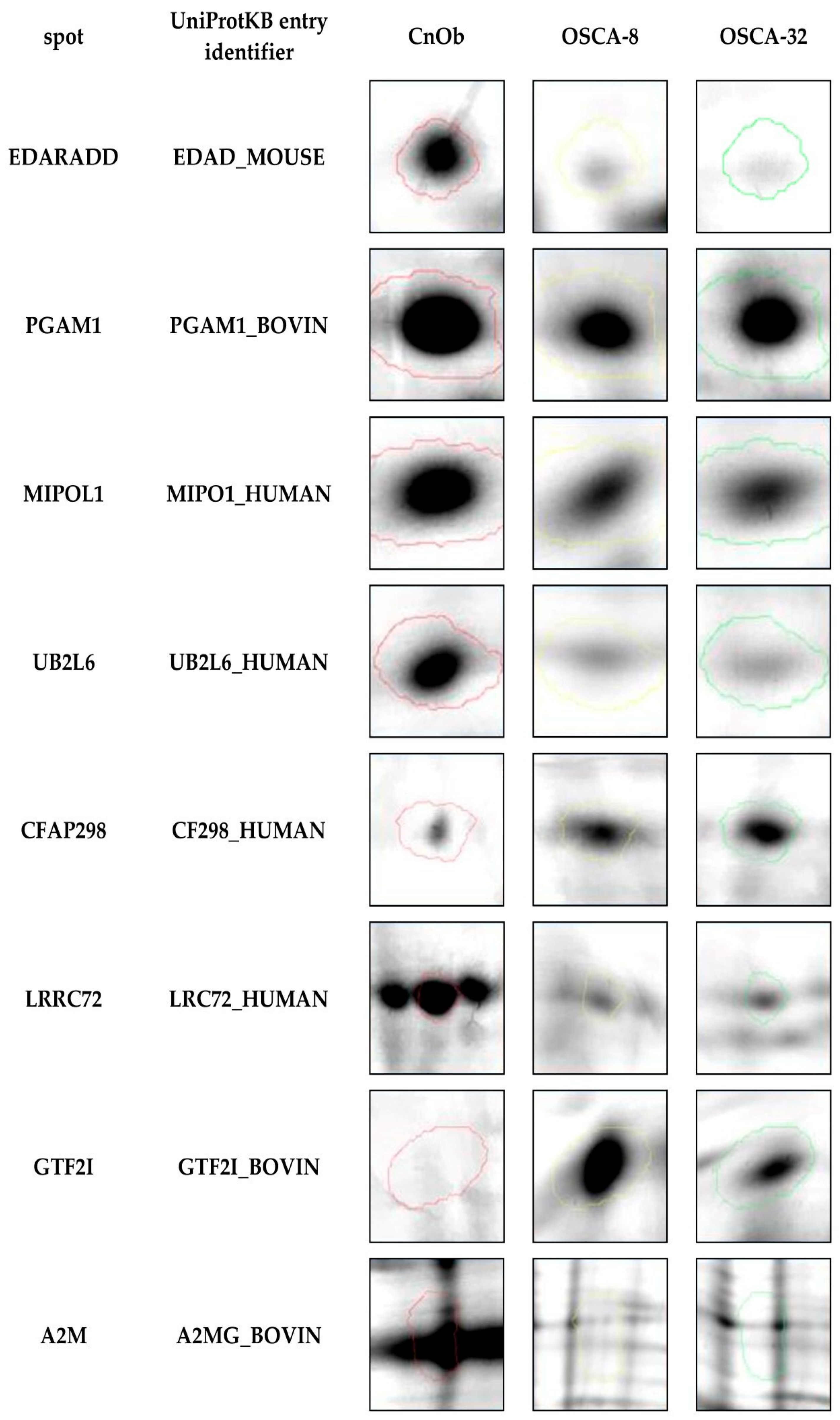
| ID | Protein | Accession Number (UniProtKB) | Score | Match | MW (Da) ** | pI * | Modif. | Seq. Cov (%) | Rt CnOb/OSCA-32 | Rt CnOb/OSCA-8 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ubiquitin/ISG15-conjugating enzyme E2 | O14933 | 95 | 6 | 17,757 | 7.71 | C, Ac, Ox | 26 | 0.664 | 0.383 |
| 2 | Leucine-rich-repeat-containing protein 72 | A6NJI9 | 150 | 14 | 33,863 | 8.91 | C, Ac, Ox | 36 | 0.471 | 0.287 |
| 3 | General transcription factor II-I | A7MB80 | 84 | 13 | 110,569 | 7.13 | C, Ac, Ox | 14 | 4.14 | 8.65 |
| 4 | Cilia- and flagella-associated protein 298 | P57076 | 68 | 10 | 33,374 | 6.99 | C, Ac | 27 | 3.78 | 6.46 |
| 5 | Mirror-image polydactyly 1 protein | Q8TD10 | 105 | 11 | 51,847 | 5.55 | C, Ac, Ox | 17 | 0.56 | 0.60 |
| 6 | Alpha-2-macroglobulin | Q7SIH1 | 158 | 14 | 168,953 | 5.71 | C, Ac, Ox | 17 | 0.19 | 0.48 |
| 7 | Ectodysplasin-A receptor-associated adapter protein | Q8VHX2 | 74 | 5 | 24,251 | 5.04 | C, Ac, Ox | 33 | 0.59 | 0.23 |
| 8 | Phosphoglycerate mutase 1 | Q3SZ62 | 77 | 10 | 28,852 | 6.67 | C, Ac, Ox | 37 | 0.71 | 0.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilk, S.S.; Michalak, K.; Owczarek, E.P.; Winiarczyk, S.; Zabielska-Koczywąs, K.A. Proteomic Analyses Reveal the Role of Alpha-2-Macroglobulin in Canine Osteosarcoma Cell Migration. Int. J. Mol. Sci. 2024, 25, 3989. https://doi.org/10.3390/ijms25073989
Wilk SS, Michalak K, Owczarek EP, Winiarczyk S, Zabielska-Koczywąs KA. Proteomic Analyses Reveal the Role of Alpha-2-Macroglobulin in Canine Osteosarcoma Cell Migration. International Journal of Molecular Sciences. 2024; 25(7):3989. https://doi.org/10.3390/ijms25073989
Chicago/Turabian StyleWilk, Sylwia S., Katarzyna Michalak, Ewelina P. Owczarek, Stanisław Winiarczyk, and Katarzyna A. Zabielska-Koczywąs. 2024. "Proteomic Analyses Reveal the Role of Alpha-2-Macroglobulin in Canine Osteosarcoma Cell Migration" International Journal of Molecular Sciences 25, no. 7: 3989. https://doi.org/10.3390/ijms25073989
APA StyleWilk, S. S., Michalak, K., Owczarek, E. P., Winiarczyk, S., & Zabielska-Koczywąs, K. A. (2024). Proteomic Analyses Reveal the Role of Alpha-2-Macroglobulin in Canine Osteosarcoma Cell Migration. International Journal of Molecular Sciences, 25(7), 3989. https://doi.org/10.3390/ijms25073989






