The Impact of SNP-Induced Amino Acid Substitutions L19P and G66R in the dRP-Lyase Domain of Human DNA Polymerase β on Enzyme Activities
Abstract
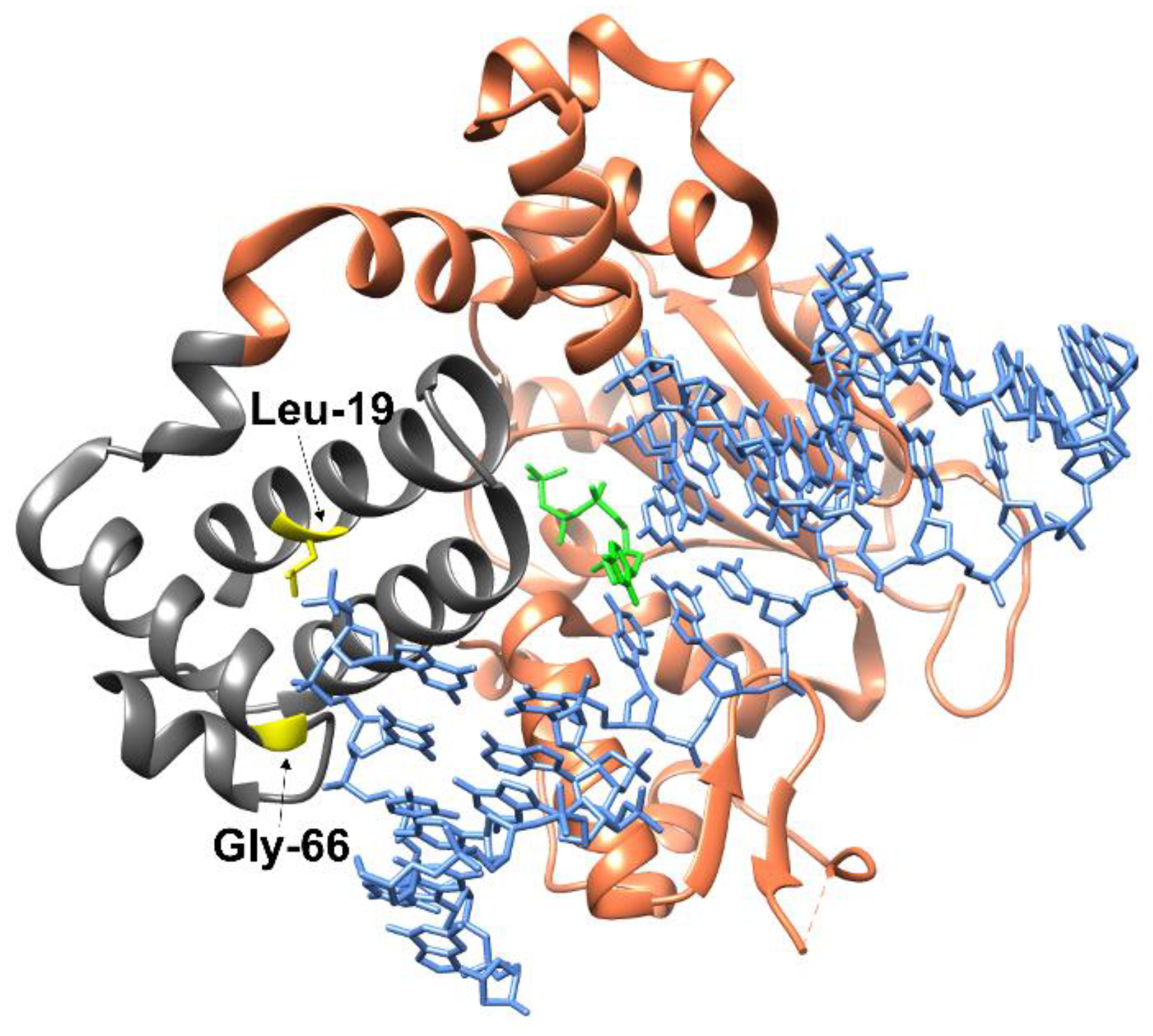
1. Introduction
2. Results and Discussion
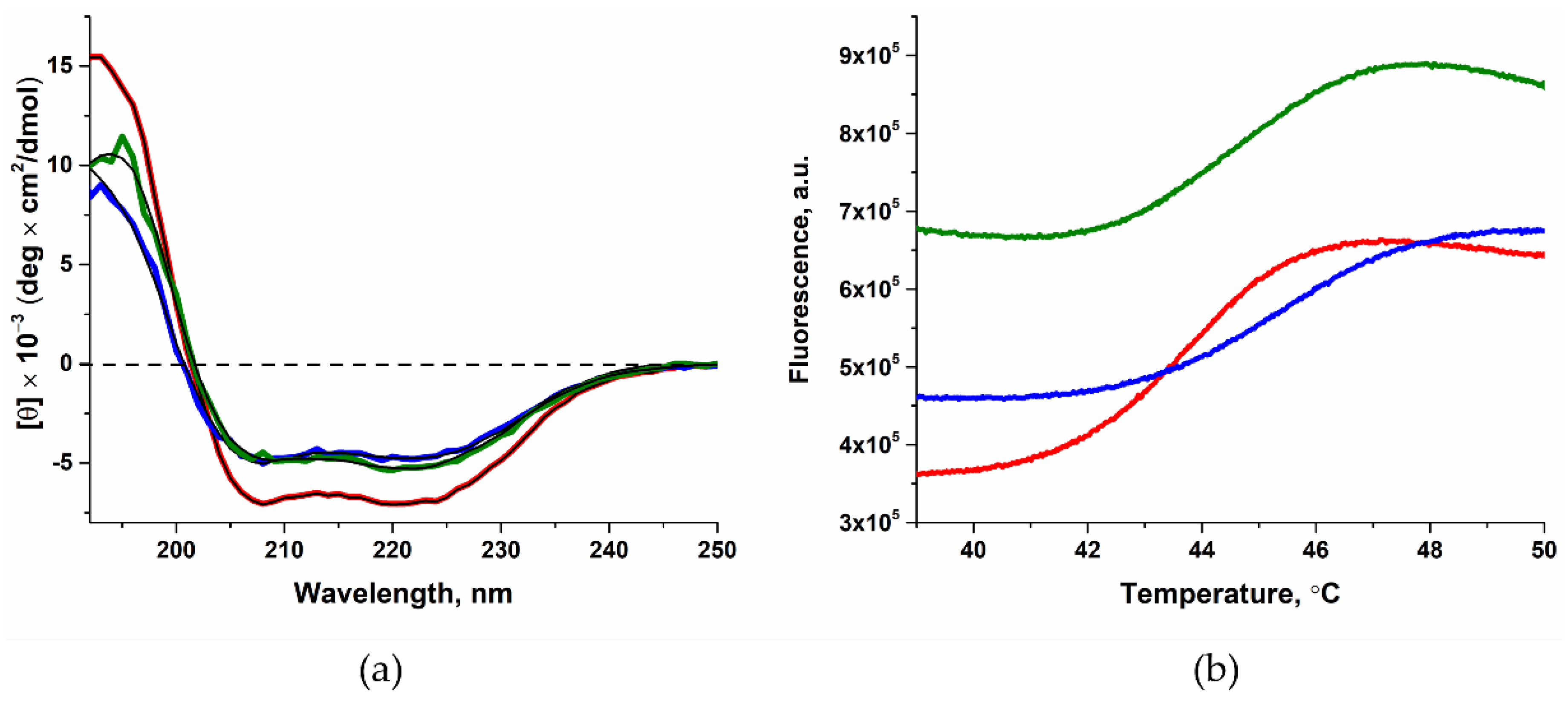
2.1. Determining the Effect of Amino Acid Substitutions on the Thermal Stability and Secondary Structure of Polβ
| WT Polβ | Polβ L19P | Polβ G66R | |
|---|---|---|---|
| Content of α-helices, % | 79 ± 16 * | 55 ± 11 | 66 ± 13 |
| Tm, °C | 44.9 ± 0.2 ** | 45.3 ± 0.1 | 44.6 ± 0.1 |
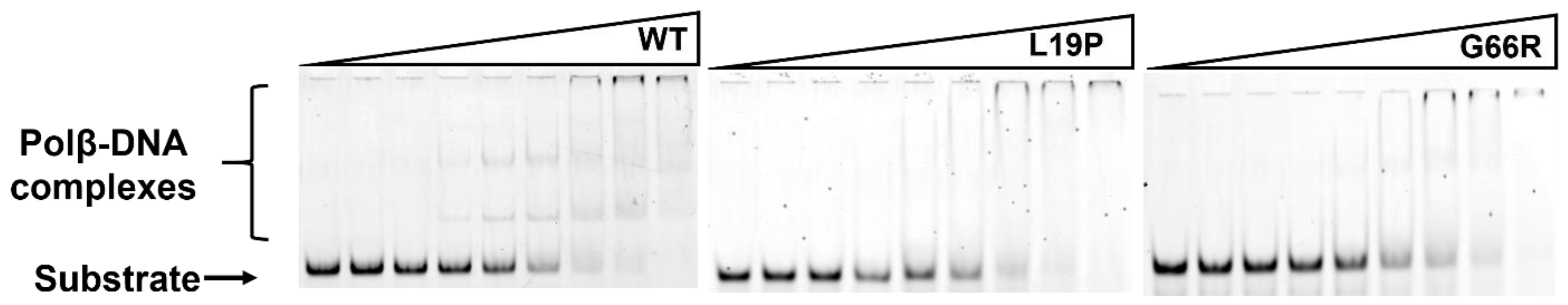
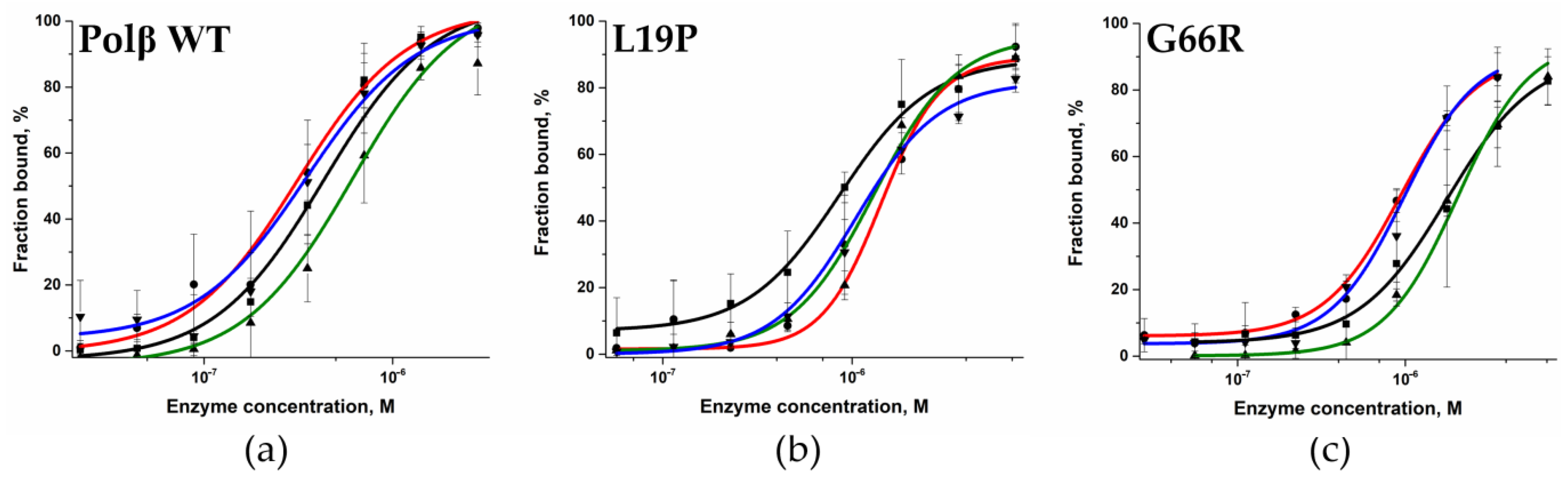
2.2. Determination of the Dissociation Constant for the Enzymes and DNA
2.3. Determination of the dRP-Lyase Activity of the Polβ Variants
2.4. An Assay of Primer Extension Efficiency of the Polβ Variants
2.5. Determination of the Gap-Filling Efficiency of the Polβ Variants
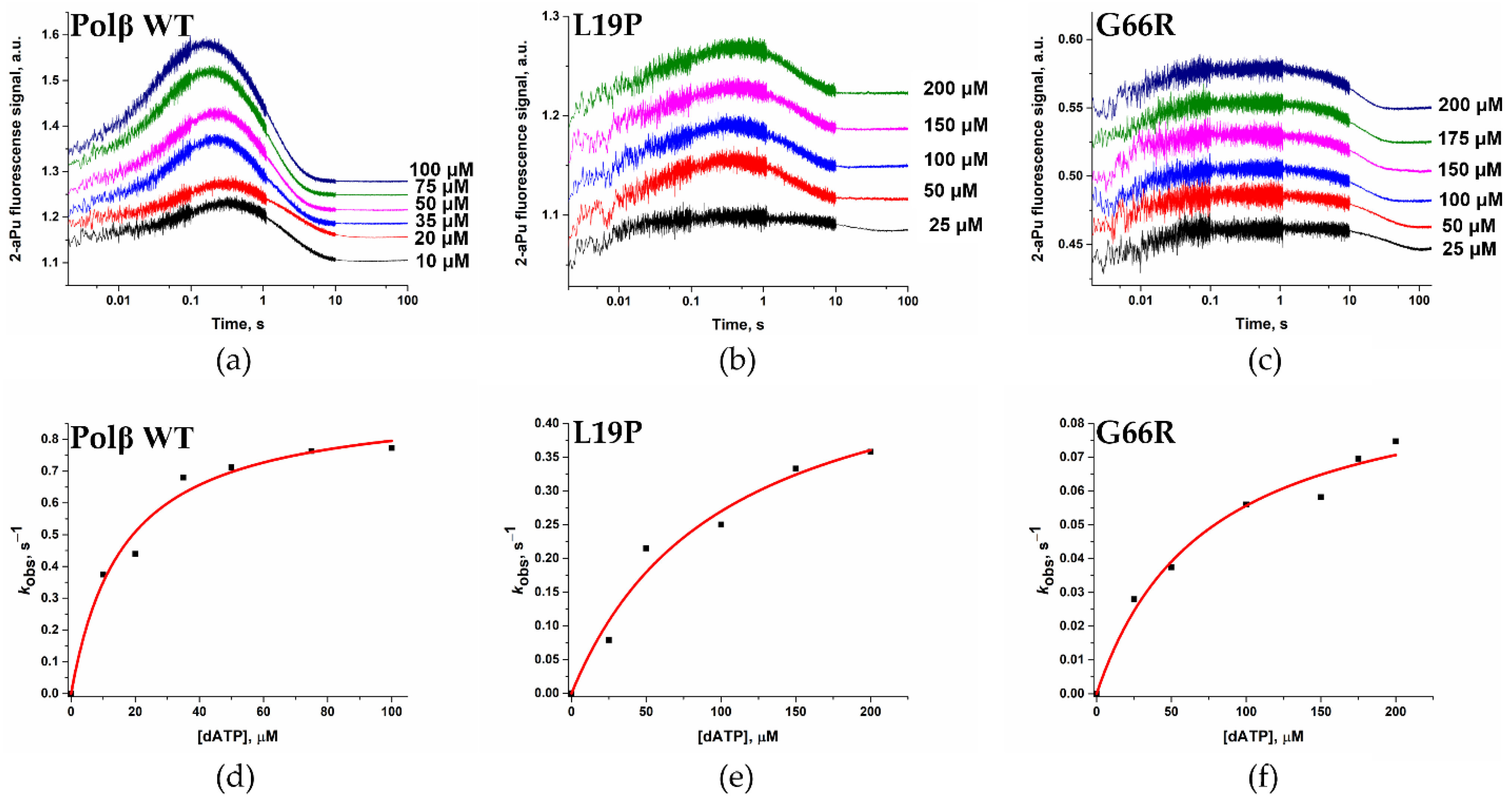
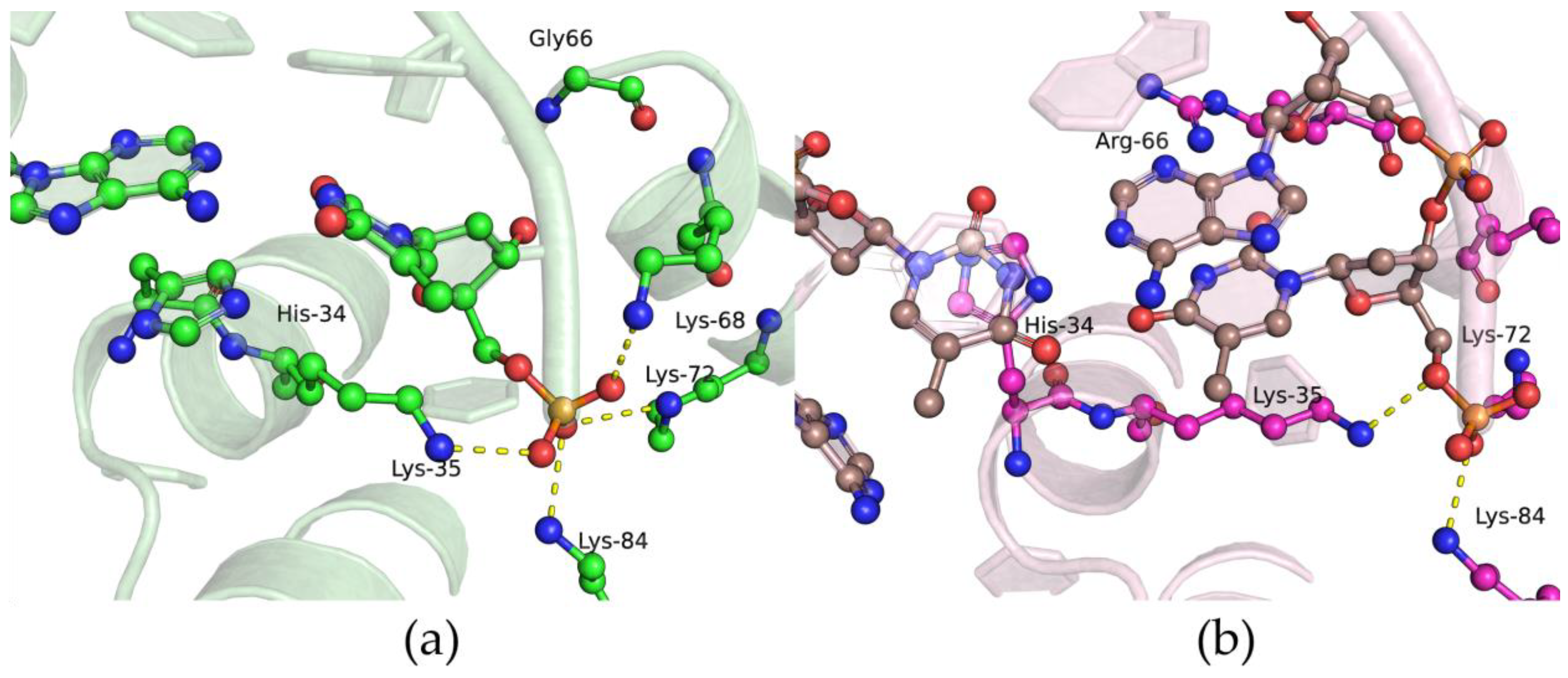
2.6. Effects of the Amino Acid Residue Substitutions on the Binding of dNTPs and Catalysis
3. Materials and Methods
3.1. Enzymes
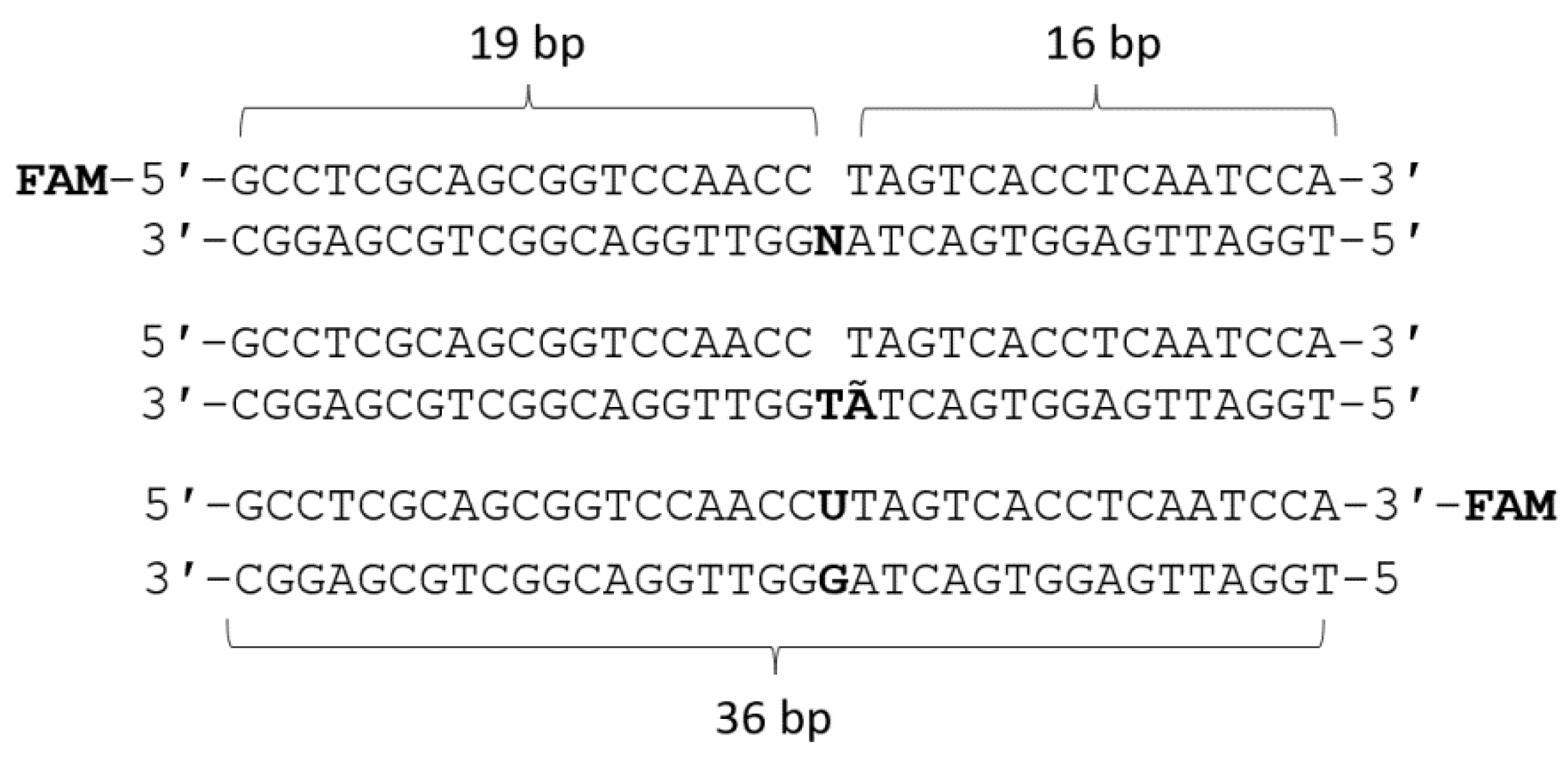
3.2. DNA Substrates
3.3. Circular Dichroism (CD) Spectroscopy
3.4. Analysis of the Melting Point of the Enzymes
3.5. DNA-Binding Analysis
3.6. Analysis of Polβ dRP-Lyase Activity
3.7. Analysis of Polβ Polymerase Activity
3.8. Registration of Conformational Changes in the DNA Substrate by the Stopped-Flow Method
3.9. Molecular Dynamics (MD) Simulations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, D.; Kreutzer, D.A.; Essigmann, J.M. Mutagenicity and Repair of Oxidative DNA Damage: Insights from Studies Using Defined Lesions. Mutat. Res. 1998, 400, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Møller, P.; Wallin, H. Adduct Formation, Mutagenesis and Nucleotide Excision Repair of DNA Damage Produced by Reactive Oxygen Species and Lipid Peroxidation Product. Mutat. Res. Rev. Mutat. Res. 1998, 410, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Izakovic, M.; Mazur, M.; Rhodes, C.J.; Telser, J. Role of Oxygen Radicals in DNA Damage and Cancer Incidence. Mol. Cell Biochem. 2004, 266, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Bjelland, S.; Seeberg, E. Mutagenicity, Toxicity and Repair of DNA Base Damage Induced by Oxidation. Mutat. Res.—Fundam. Mol. Mech. Mutagen. 2003, 531, 37–80. [Google Scholar] [CrossRef] [PubMed]
- Frederico, L.A.; Kunkel, T.A.; Shaw, B.R. A Sensitive Genetic Assay for the Detection of Cytosine Deamination: Determination of Rate Constants and the Activation Energy. Biochemistry 1990, 29, 2532–2537. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, J.; Walker, V.E.; Upton, P.B.; Chiang, S.Y.; Kow, Y.W.; Swenberg, J.A. Highly Sensitive Apurinic/Apyrimidinic Site Assay Can Detect Spontaneous and Chemically Induced Depurination under Physiological Conditions. Cancer Res. 1998, 58, 222–225. [Google Scholar] [PubMed]
- Wallace, S.S. Biological Consequences of Free Radical-Damaged DNA Bases. Free Radic. Biol. Med. 2002, 33, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Coppede, F.; Migliore, L. DNA Damage in Neurodegenerative Diseases. Mutat. Res.—Fundam. Mol. Mech. Mutagen. 2015, 776, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjørås, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- David, S.S.; Williams, S.D. Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem. Rev. 1998, 98, 1221–1262. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Biade, S.; Matsumoto, Y. Involvement of Flap Endonuclease 1 in Base Excision DNA Repair. J. Biol. Chem. 1998, 273, 8842–8848. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Nash, R.A.; Klungland, A.; Schär, P.; Barnes, D.E.; Lindahl, T. Reconstitution of DNA Base Excision-Repair with Purified Human Proteins: Interaction between DNA Polymerase β and the XRCC1 Protein. EMBO J. 1996, 15, 6662–6670. [Google Scholar] [CrossRef] [PubMed]
- Sobol, R.W.; Horton, J.K.; Kühn, R.; Gu, H.; Singhal, R.K.; Prasad, R.; Rajewsky, K.; Wilson, S.H. Requirement of Mammalian DNA Polymerase-β in Base-Excision Repair. Nature 1996, 379, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Sobol, R.W.; Wilson, S.H. Mammalian DNA Beta-Polymerase in Base Excision Repair of Alkylation Damage. Prog. Nucleic Acid Res. Mol. Biol. 2001, 68, 57–74. [Google Scholar] [PubMed]
- Beard, W.A.; Wilson, S.H. Structural Design of a Eukaryotic DNA Repair Polymerase: DNA Polymerase Beta. Mutat. Res. 2000, 460, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Menezes, M.R.; Senejani, A.; Sweasy, J.B. Cellular Roles of DNA Polymerase Beta. Yale J. Biol. Med. 2013, 86, 463–469. [Google Scholar] [PubMed]
- Sawaya, M.R.; Prasad, R.; Wilson, S.H.; Kraut, J.; Pelletier, H. Crystal Structures of Human DNA Polymerase Beta Complexed with Gapped and Nicked DNA: Evidence for an Induced Fit Mechanism. Biochemistry 1997, 36, 11205–11215. [Google Scholar] [CrossRef] [PubMed]
- Nemec, A.A.; Abriola, L.; Merkel, J.S.; De Stanchina, E.; DeVeaux, M.; Zelterman, D.; Glazer, P.M.; Sweasy, J.B. DNA Polymerase Beta Germline Variant Confers Cellular Response to Cisplatin Therapy. Mol. Cancer Res. 2017, 15, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Almeida, K.H.; Sobol, R.W. A Unified View of Base Excision Repair: Lesion-Dependent Protein Complexes Regulated by Post-Translational Modification. DNA Repair 2007, 6, 695–711. [Google Scholar] [CrossRef] [PubMed]
- Fotiadou, P.; Henegariu, O.; Sweasy, J.B. DNA Polymerase β Interacts with TRF2 and Induces Telomere Dysfunction in a Murine Mammary Cell Line. Cancer Res. 2004, 64, 3830–3837. [Google Scholar] [CrossRef] [PubMed]
- Kidane, D.; Jonason, A.S.; Gorton, T.S.; Mihaylov, I.; Pan, J.; Keeney, S.; De Rooij, D.G.; Ashley, T.; Keh, A.; Liu, Y.; et al. DNA Polymerase Β Is Critical for Mouse Meiotic Synapsis. EMBO J. 2010, 29, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.K.; Srivastava, D.K.; Zmudzka, B.Z.; Wilson, S.H. Strategic Down-Regulation of DNA Polymerase β by Antisense RNA Sensitizes Mammalian Cells to Specific DNA Damaging Agents. Nucleic Acids Res. 1995, 23, 3810–3815. [Google Scholar] [CrossRef] [PubMed]
- Brookes, A.J. The Essence of SNPs. Gene 1999, 234, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Kiyohara, C.; Takayama, K.; Nakanishi, Y. Association of Genetic Polymorphisms in the Base Excision Repair Pathway with Lung Cancer Risk: A Meta-Analysis. Lung Cancer 2006, 54, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.M.; Goode, E.L.; Ladiges, W.C.; Ulrich, C.M. Polymorphic Variation in HOGG1 and Risk of Cancer: A Review of the Functional and Epidemiologic Literature. Mol. Carcinog. 2005, 42, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zheng, B.; Li, Y.; Yuan, Y.; Xing, C. Genetic Polymorphisms of DNA Repair Pathways in Sporadic Colorectal Carcinogenesis. J. Cancer 2019, 10, 1417–1433. [Google Scholar] [CrossRef] [PubMed]
- Alnajjar, K.S.; Garcia-Barboza, B.; Negahbani, A.; Nakhjiri, M.; Kashemirov, B.; McKenna, C.; Goodman, M.F.; Sweasy, J.B. A Change in the Rate-Determining Step of Polymerization by the K289M DNA Polymerase β Cancer-Associated Variant. Biochemistry 2017, 56, 2096–2105. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.M.; Schechter, A.; Alnajjar, K.S.; Huang, J.; Towle-Weicksel, J.; Eckenroth, B.E.; Doublié, S.; Sweasy, J.B. Defective Nucleotide Release by DNA Polymerase β Mutator Variant E288K Is the Basis of Its Low Fidelity. Biochemistry 2017, 56, 5550–5559. [Google Scholar] [CrossRef] [PubMed]
- Liptak, C.; Mahmoud, M.M.; Eckenroth, B.E.; Moreno, M.V.; Kyle, E.; Alnajjar, K.S.; Huang, J.; Towle-Weicksel, J.B.; Doublie, S.; Loria, J.P.; et al. I260Q DNA Polymerase β Highlights Precatalytic Conformational Rearrangements Critical for Fidelity. Nucleic Acids Res. 2018, 46, 10740–10756. [Google Scholar] [CrossRef] [PubMed]
- Dalal, S.; Starcevic, D.; Jaeger, J.; Sweasy, J.B. The I260Q Variant of DNA Polymerase Beta Extends Mispaired Primer-Termini Due to Increased Affinity for Deoxynucleotide Triphosphate Substrates. Biochemistry 2008, 47, 12118–12125. [Google Scholar] [CrossRef] [PubMed]
- Kosa, J.L.; Sweasy, J.B. The E249K Mutator Mutant of DNA Polymerase β Extends Mispaired Termini. J. Biol. Chem. 1999, 274, 35866–35872. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.L.; Kosa, J.; Jaeger, J.; Sweasy, J.B. The Asp285 Variant of DNA Polymerase Beta Extends Mispaired Primer Termini via Increased Nucleotide Binding. Biochemistry 2008, 47, 8048–8057. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Starcevic, D.; Dalal, S.; Sweasy, J.B. Is There a Link Between DNA Polymerase Beta and Cancer? Cell Cycle 2004, 3, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Kladova, O.A.; Fedorova, O.S.; Kuznetsov, N.A. The Role of Natural Polymorphic Variants of DNA Polymerase β in DNA Repair. Int. J. Mol. Sci. 2022, 23, 2390. [Google Scholar] [CrossRef] [PubMed]
- Dalal, S.; Chikova, A.; Jaeger, J.; Sweasy, J.B. The Leu22Pro Tumor-Associated Variant of DNA Polymerase Beta Is DRP Lyase Deficient. Nucleic Acids Res. 2008, 36, 411–422. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beard, W.A.; Wilson, S.H. Structural Insights into the Origins of DNA Polymerase Fidelity. Structure 2003, 11, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Woolfson, D.N.; Williams, D.H. The Influence of Proline Residues on α-Helical Structure. FEBS Lett. 1990, 277, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.S.; Richardson, D.C. Amino Acid Preferences for Specific Locations at the Ends of Alpha Helices. Science 1988, 240, 1648–1652. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Kang, Y.K. Positional Preference of Proline in Alpha-Helices. Protein Sci. 1999, 8, 1492–1499. [Google Scholar] [CrossRef] [PubMed]
- Kirby, T.W.; Derose, E.F.; Beard, W.A.; Shock, D.D.; Wilson, S.H.; London, R.E. Substrate Rescue of DNA Polymerase β Containing a Catastrophic L22P Mutation. Biochemistry 2014, 53, 2413–2422. [Google Scholar] [CrossRef] [PubMed]
- Kladova, O.A.; Tyugashev, T.E.; Mikushina, E.S.; Soloviev, N.O.; Kuznetsov, N.A.; Novopashina, D.S.; Kuznetsova, A.A. Human Polβ Natural Polymorphic Variants G118V and R149I Affects Substate Binding and Catalysis. Int. J. Mol. Sci. 2023, 24, 5892. [Google Scholar] [CrossRef] [PubMed]
- Kladova, O.A.; Tyugashev, T.E.; Mikushina, E.S.; Kuznetsov, N.A.; Novopashina, D.S.; Kuznetsova, A.A. The Activity of Natural Polymorphic Variants of Human DNA Polymerase β Having an Amino Acid Substitution in the Transferase Domain. Cells 2023, 12, 1300. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Beard, W.A.; Strauss, P.R.; Wilson, S.H. Human DNA Polymerase β Deoxyribose Phosphate Lyase: Substrate Specificity and Catalytic Mechanism. J. Biol. Chem. 1998, 273, 15263–15270. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.A.; Crasto, C.J.; Matsumoto, Y. Deoxyribose Phosphate Excision by the N-Terminal Domain of the Polymerase β: The Mechanism Revisited. Biochemistry 1998, 37, 9605–9611. [Google Scholar] [CrossRef] [PubMed]
- Piersen, C.E.; Prasad, R.; Wilson, S.H.; Lloyd, R.S. Evidence for an Imino Intermediate in the DNA Polymerase β Deoxyribose Phosphate Excision Reaction. J. Biol. Chem. 1996, 271, 17811–17815. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Batra, V.K.; Yang, X.P.; Krahn, J.M.; Pedersen, L.C.; Beard, W.A.; Wilson, S.H. Structural Insight into the DNA Polymerase β Deoxyribose Phosphate Lyase Mechanism. DNA Repair 2005, 4, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Beard, W.A.; Chyan, J.Y.; Maciejewski, M.W.; Mullen, G.P.; Wilson, S.H. Functional Analysis of the Amino-Terminal 8-KDa Domain of DNA Polymerase β as Revealed by Site-Directed Mutagenesis: DNA Binding and 5′-Deoxyribose Phosphate Lyase Activities. J. Biol. Chem. 1998, 273, 11121–11126. [Google Scholar] [CrossRef]
- Daskalova, S.M.; Bai, X.; Hecht, S.M. Study of the Lyase Activity of Human DNA Polymerase β Using Analogues of the Intermediate Schiff Base Complex. Biochemistry 2018, 57, 2711–2722. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Kim, K. Excision of Deoxyribose Phosphate Residues by DNA Polymerase β during DNA Repair. Science 1995, 269, 699–702. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kim, K.; Katz, D.S.; Feng, J.A. Catalytic Center of DNA Polymerase β for Excision of Deoxyribose Phosphate Groups. Biochemistry 1998, 37, 6456–6464. [Google Scholar] [CrossRef] [PubMed]
- Deterding, L.J.; Prasad, R.; Mullen, G.P.; Wilson, S.H.; Tomer, K.B. Mapping of the 5′-2-Deoxyribose-5-Phosphate Lyase Active Site in DNA Polymerase β by Mass Spectrometry. J. Biol. Chem. 2000, 275, 10463–10471. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Reed, A.J.; Zahurancik, W.J.; Daskalova, S.M.; Hecht, S.M.; Suo, Z. Interlocking Activities of DNA Polymerase β in the Base Excision Repair Pathway. Proc. Natl. Acad. Sci. USA 2022, 119, e2118940119. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.C.; Neely, R.K. 2-Aminopurine as a Fluorescent Probe of DNA Conformation and the DNA–Enzyme Interface. Q. Rev. Biophys. 2015, 48, 244–279. [Google Scholar] [CrossRef] [PubMed]
- Tleugabulova, D.; Reha-Krantz, L.J. Probing DNA Polymerase—DNA Interactions: Examining the Template Strand in Exonuclease Complexes Using 2-Aminopurine Fluorescence and Acrylamide Quenching. Biochemistry 2007, 46, 6559–6569. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, C.; Bloom, L.B.; Helquist, S.A.; Kool, E.T.; Reha-Krantz, L.J. Dynamics of Nucleotide Incorporation: Snapshots Revealed by 2-Aminopurine Fluorescence Studies. Biochemistry 2006, 45, 2836–2844. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hariharan, C.; Reha-Krantz, L.J. Using 2-Aminopurine Fluorescence to Detect Bacteriophage T4 DNA Polymerase-DNA Complexes That Are Important for Primer Extension and Proofreading Reactions. Biochemistry 2005, 44, 15674–15684. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, C.A.; Tsai, M.D. Use of 2-Aminopurine and Tryptophan Fluorescence as Probes in Kinetic Analyses of DNA Polymerase β. Biochemistry 2002, 41, 11226–11235. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Patel, S.S.; Werneburg, B.G.; Tsai, M.D. DNA Polymerase β: Multiple Conformational Changes in the Mechanism of Catalysis. Biochemistry 1997, 36, 11891–11900. [Google Scholar] [CrossRef] [PubMed]
- Bakman, A.S.; Boichenko, S.S.; Kuznetsova, A.A.; Ishchenko, A.A.; Saparbaev, M.; Kuznetsov, N.A. The Impact of Human DNA Glycosylases on the Activity of DNA Polymerase β toward Various Base Excision Repair Intermediates. Int. J. Mol. Sci. 2023, 24, 9594. [Google Scholar] [CrossRef] [PubMed]
- Bakman, A.S.; Boichenko, S.S.; Kuznetsova, A.A.; Ishchenko, A.A.; Saparbaev, M.; Kuznetsov, N.A. Coordination between Human DNA Polymerase β and Apurinic/Apyrimidinic Endonuclease 1 in the Course of DNA Repair. Biochimie 2024, 216, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A Web Server for Accurate Protein Secondary Structure Prediction and Fold Recognition from the Circular Dichroism Spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef] [PubMed]
- Gridley, C.L.; Rangarajan, S.; Firbank, S.; Dalal, S.; Sweasy, J.B.; Jaeger, J. Structural Changes in the Hydrophobic Hinge Region Adversely Affect the Activity and Fidelity of the I260Q Mutator DNA Polymerase β. Biochemistry 2013, 52, 4422–4432. [Google Scholar] [CrossRef] [PubMed]
- Maciejewski, M.W.; Liu, D.; Prasad, R.; Wilson, S.H.; Mullen, G.P. Backbone Dynamics and Refined Solution Structure of the N-Terminal Domain of DNA Polymerase Beta. Correlation with DNA Binding and DRP Lyase Activity. J. Mol. Biol. 2000, 296, 229–253. [Google Scholar] [CrossRef] [PubMed]
- Freudenthal, B.D.; Beard, W.A.; Shock, D.D.; Wilson, S.H. Observing a DNA Polymerase Choose Right from Wrong. Cell 2013, 154, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. In Protein Structure Prediction; Methods Molecular Biology; Springer: Berlin, Germany, 2014; Volume 1137. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.M.; SØndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical PKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1998, 79, 926–935. [Google Scholar] [CrossRef]
- Joung, I.S.; Cheatham, T.E. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, H.; Tan, T. Rational Design of Methodology-Independent Metal Parameters Using a Nonbonded Dummy Model. J. Chem. Theory Comput. 2016, 12, 3250–3260. [Google Scholar] [CrossRef] [PubMed]
- Bayly, C.I.; Merz, K.M.; Ferguson, D.M.; Cornell, W.D.; Fox, T.; Caldwell, J.W.; Kollman, P.A.; Cieplak, P.; Gould, I.R.; Spellmeyer, D.C. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef]
- Zgarbová, M.; Šponer, J.; Otyepka, M.; Cheatham, T.E.; Galindo-Murillo, R.; Jurečka, P. Refinement of the Sugar-Phosphate Backbone Torsion Beta for AMBER Force Fields Improves the Description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Meagher, K.L.; Redman, L.T.; Carlson, H.A. Development of Polyphosphate Parameters for Use with the AMBER Force Field. J. Comput. Chem. 2003, 24, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.Y.R.E.D. Server: A Web Service for Deriving RESP and ESP Charges and Building Force Field Libraries for New Molecules and Molecular Fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed]
- Sousa Da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser InterfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Wennberg, C.L.; Murtola, T.; Hess, B.; Lindahl, E. Lennard-Jones Lattice Summation in Bilayer Simulations Has Critical Effects on Surface Tension and Lipid Properties. J. Chem. Theory Comput. 2013, 9, 3527–3537. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Bernetti, M.; Bussi, G. Pressure Control Using Stochastic Cell Rescaling. J. Chem. Phys. 2020, 153, 114107. [Google Scholar] [CrossRef] [PubMed]

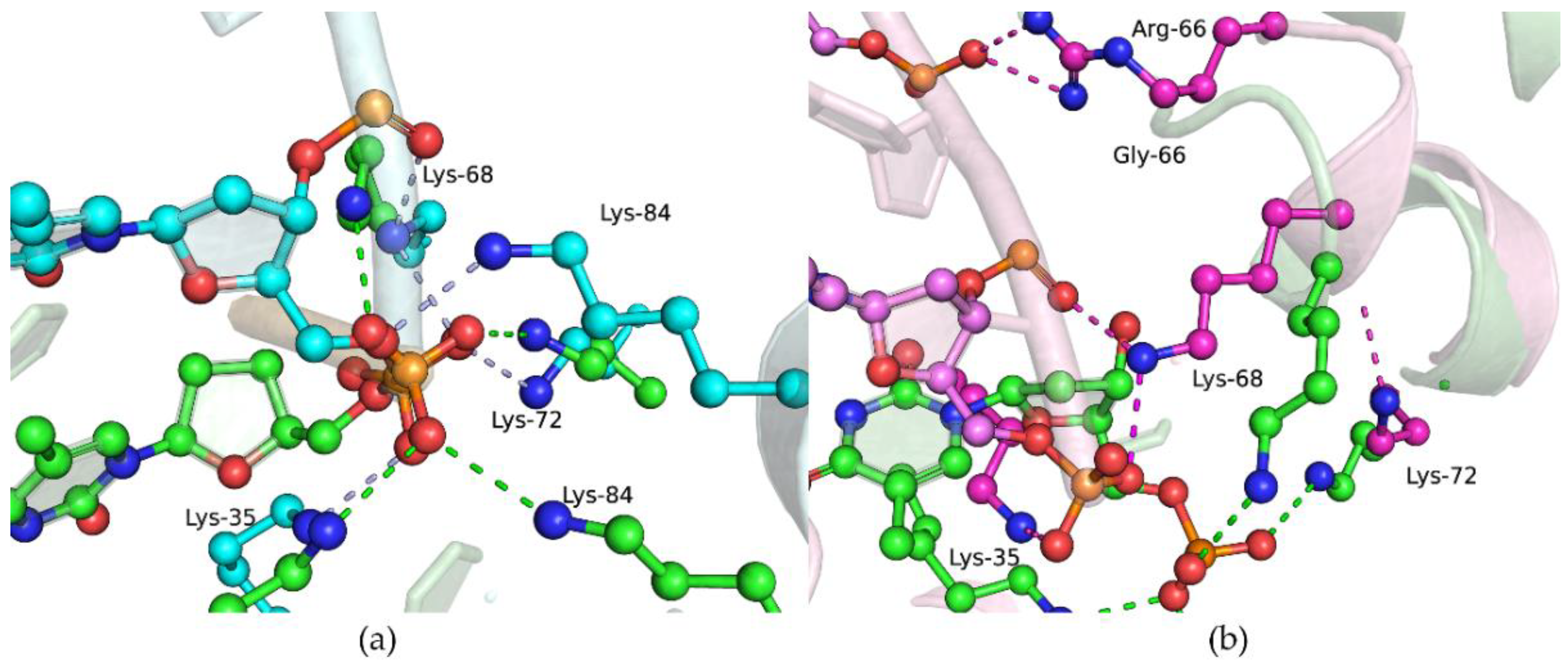


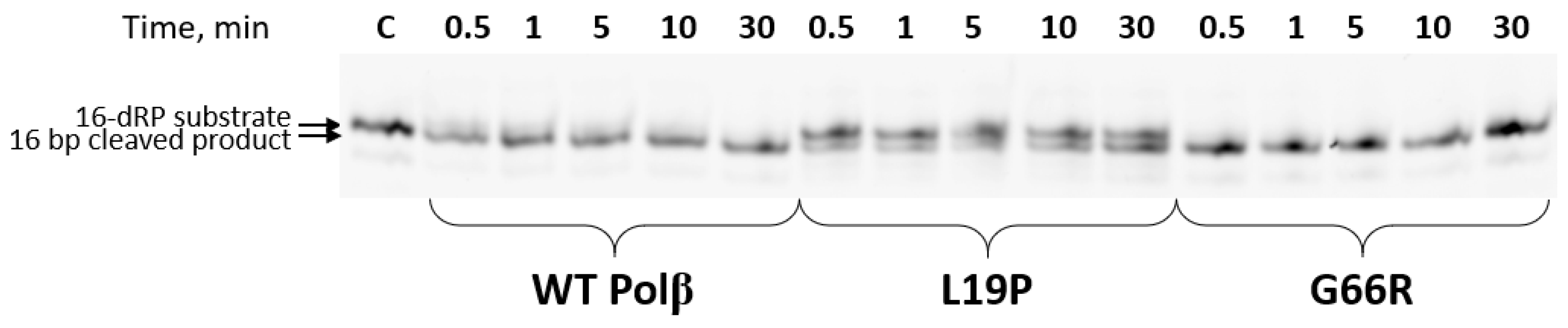
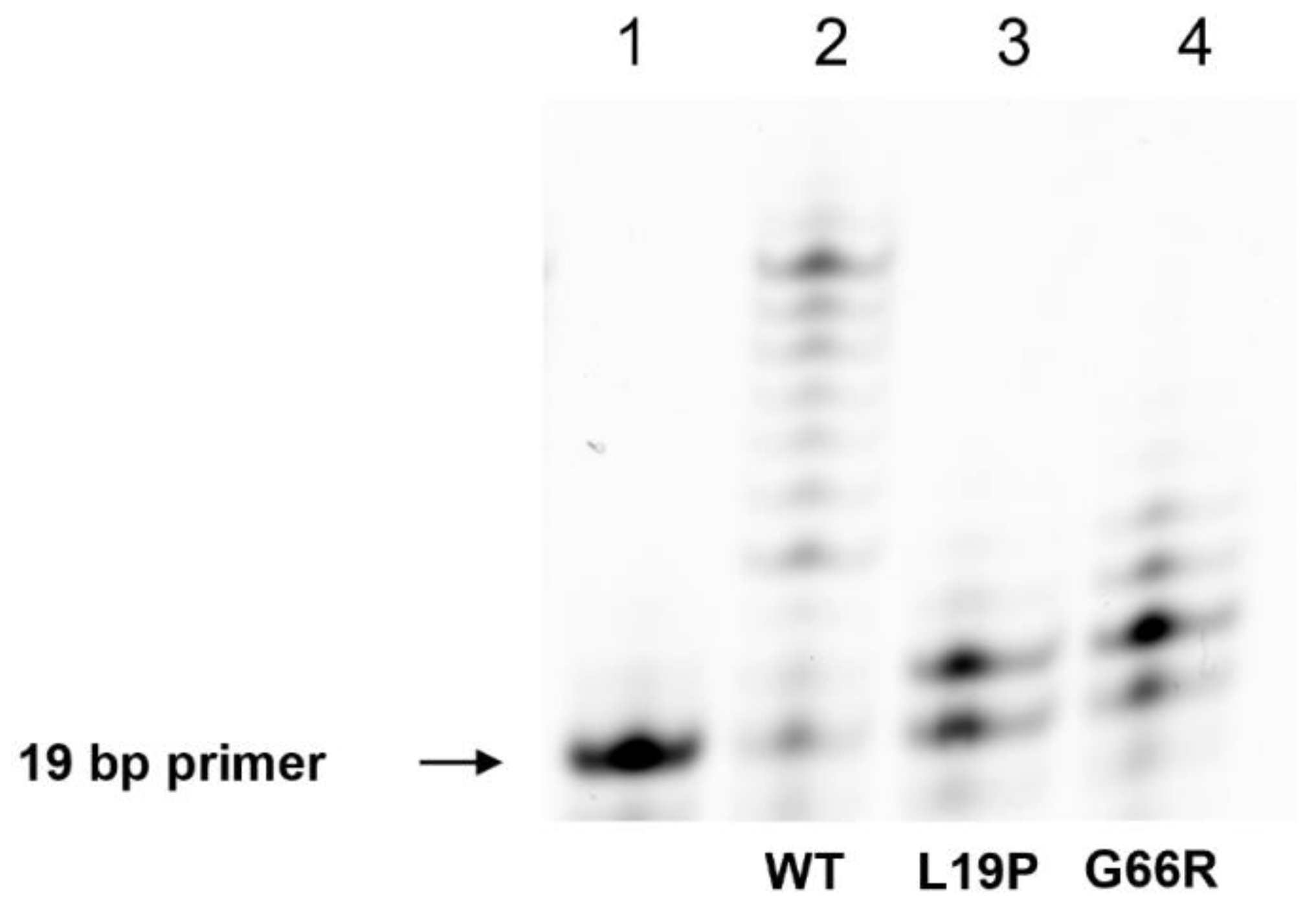
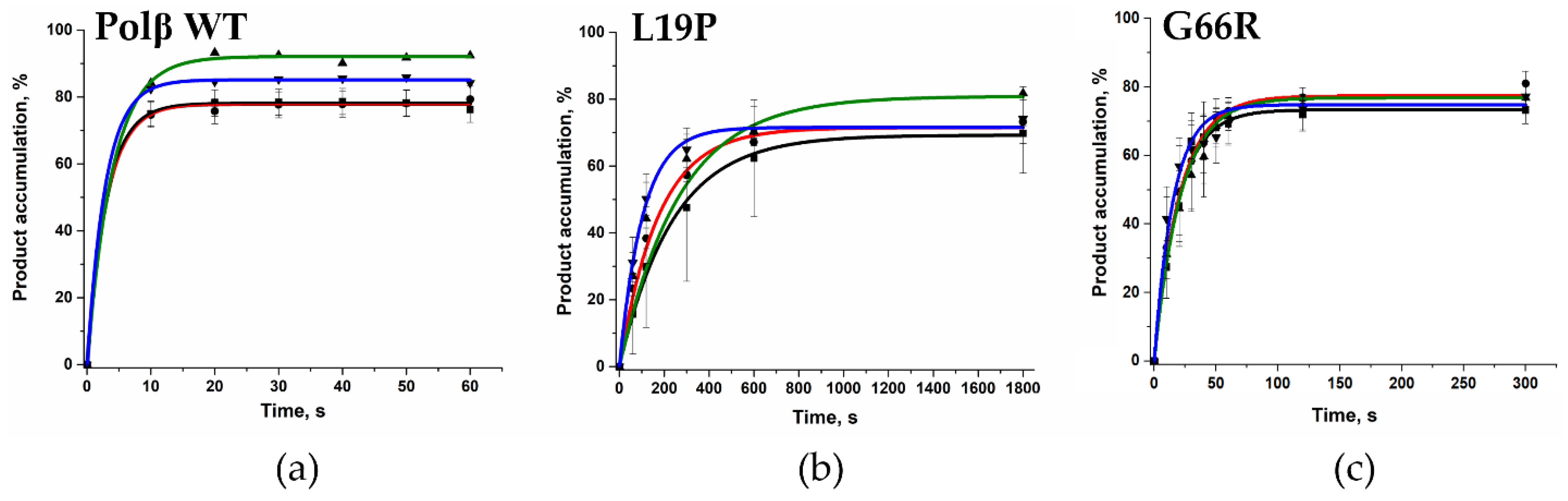
| Amino Acid Substitution | COSMIC (https://cancer.sanger.ac.uk, accessed on 21 February 2021) | cBioportal (https://www.cbioportal.org, accessed on 21 February 2021) | Hivebiochemistry (https://hive.biochemistry.gwu.edu, accessed on 21 February 2021) |
|---|---|---|---|
| M1V | Liver cancer | ||
| S2I | Blastoma | ||
| S2R | Liver cancer | ||
| R4L | Choriocarcinoma | ||
| A6S | Small cell lung carcinoma | ||
| Q8R | Non–small cell lung carcinoma | ||
| E9D | Uterine endometrioid carcinoma | Uterine cancer | |
| L11F | Glioblastoma | ||
| L11V | Colon adenocarcinoma | Uterine cancer | |
| G13V | Astrocytoma | Melanoma | |
| D17N | Thyroid carcinoma | Papillary thyroid cancer | Malignant glioma |
| M18I | Colon adenocarcinoma, rectal adenocarcinoma | Rectal adenocarcinoma | Colorectal cancer, thyroid carcinoma |
| L22F | Bile duct adenocarcinoma | ||
| A23T | Melanoma | ||
| A23V | Squamous cell lung carcinoma | Lung squamous cell carcinoma | Kidney cancer |
| K27R | Lung adenocarcinoma | Lung adenocarcinoma | Lung cancer |
| N37H | Liver carcinoma | Blastoma, lung cancer | |
| A38S | Liver cancer | ||
| S44C | Stomach adenocarcinoma | Tubular stomach adenocarcinoma | Liver cancer |
| S44F | Stomach cancer | ||
| Y49S | Prostate adenocarcinoma | Colorectal cancer | |
| P50L | Prostate carcinoma | Prostate cancer | |
| A59S | Prostate cancer | ||
| L62F | Thyroid carcinoma | Liver cancer | |
| E71K | Endometrioid carcinoma | Uterine serous carcinoma | Thyroid carcinoma |
| F76C | Endometrioid carcinoma | Uterine endometrioid carcinoma | Uterine cancer |
| G80R | Serous carcinoma | Uterine cancer | |
| R83C | Endometrioid carcinoma, lung adenocarcinoma, stomach adenocarcinoma | Uterine endometrioid carcinoma, lung adenocarcinoma, tubular stomach adenocarcinoma | Germ cell cancer, lung cancer, gastric cancer |
| R83H | Cecum adenocarcinoma | ||
| L85V | Esophageal squamous cell carcinoma | ||
| E86G | Colon adenocarcinoma | Uterine cancer |
| Substrate | WT Polβ [24] | Polβ L19P | Polβ G66R |
|---|---|---|---|
| Gap_A | 0.38 ± 0.02 | 0.86 ± 0.04 | 1.7 ± 0.2 |
| Gap_T | 0.33 ± 0.03 | 2.0 ± 0.4 | 0.9 ± 0.1 |
| Gap_G | 0.59 ± 0.07 | 1.3 ± 0.1 | 2.0 ± 0.3 |
| Gap_C | 0.38 ± 0.03 | 1.1 ± 0.1 | 1.0 ± 0.1 |
| kobs, s−1 | WT Polβ [24] | Polβ L19P | Polβ G66R |
|---|---|---|---|
| Gap_A | 0.33 ± 0.03 | 0.005 ± 0.001 | 0.052 ± 0.004 |
| Gap_T | 0.32 ± 0.04 | 0.009 ± 0.001 | 0.048 ± 0.006 |
| Gap_G | 0.25 ± 0.02 | 0.008 ± 0.001 | 0.047 ± 0.002 |
| Gap_C | 0.34 ± 0.02 | 0.011 ± 0.001 | 0.065 ± 0.008 |
| Enzyme | WT Polβ [41] | Polβ L19P | Polβ G66R |
|---|---|---|---|
| kpol, s−1 | 0.93 ± 0.05 | 0.54 ± 0.22 | 0.10 ± 0.01 |
| Kd, app(dATP), μM | 16 ± 3 | 101 ± 36 | 74 ± 19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kladova, O.A.; Tyugashev, T.E.; Yakimov, D.V.; Mikushina, E.S.; Novopashina, D.S.; Kuznetsov, N.A.; Kuznetsova, A.A. The Impact of SNP-Induced Amino Acid Substitutions L19P and G66R in the dRP-Lyase Domain of Human DNA Polymerase β on Enzyme Activities. Int. J. Mol. Sci. 2024, 25, 4182. https://doi.org/10.3390/ijms25084182
Kladova OA, Tyugashev TE, Yakimov DV, Mikushina ES, Novopashina DS, Kuznetsov NA, Kuznetsova AA. The Impact of SNP-Induced Amino Acid Substitutions L19P and G66R in the dRP-Lyase Domain of Human DNA Polymerase β on Enzyme Activities. International Journal of Molecular Sciences. 2024; 25(8):4182. https://doi.org/10.3390/ijms25084182
Chicago/Turabian StyleKladova, Olga A., Timofey E. Tyugashev, Denis V. Yakimov, Elena S. Mikushina, Daria S. Novopashina, Nikita A. Kuznetsov, and Aleksandra A. Kuznetsova. 2024. "The Impact of SNP-Induced Amino Acid Substitutions L19P and G66R in the dRP-Lyase Domain of Human DNA Polymerase β on Enzyme Activities" International Journal of Molecular Sciences 25, no. 8: 4182. https://doi.org/10.3390/ijms25084182
APA StyleKladova, O. A., Tyugashev, T. E., Yakimov, D. V., Mikushina, E. S., Novopashina, D. S., Kuznetsov, N. A., & Kuznetsova, A. A. (2024). The Impact of SNP-Induced Amino Acid Substitutions L19P and G66R in the dRP-Lyase Domain of Human DNA Polymerase β on Enzyme Activities. International Journal of Molecular Sciences, 25(8), 4182. https://doi.org/10.3390/ijms25084182







