Circulating Blood Prognostic Biomarker Signatures for Hemorrhagic Cerebral Cavernous Malformations (CCMs)
Abstract
1. Introduction
2. Results
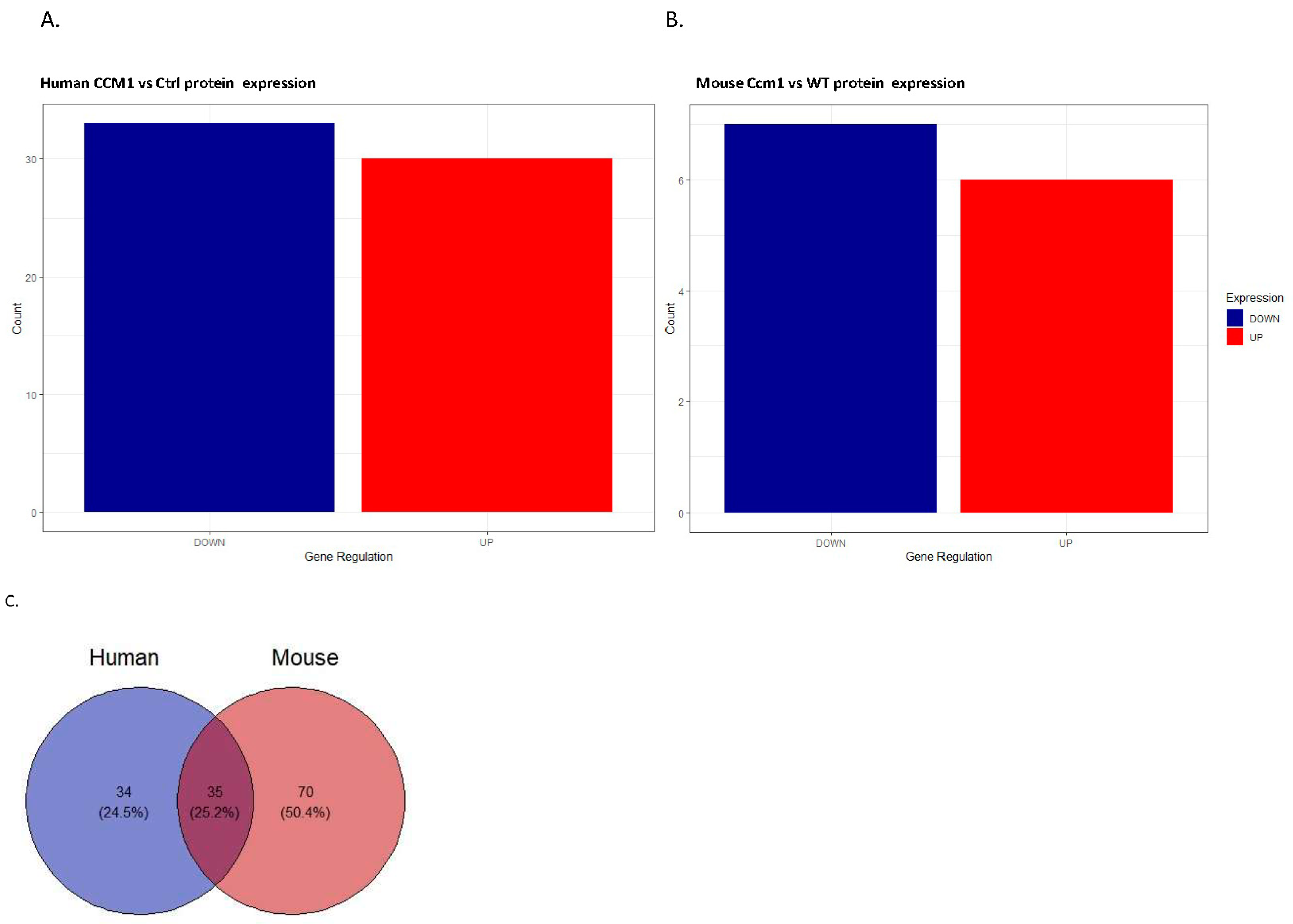
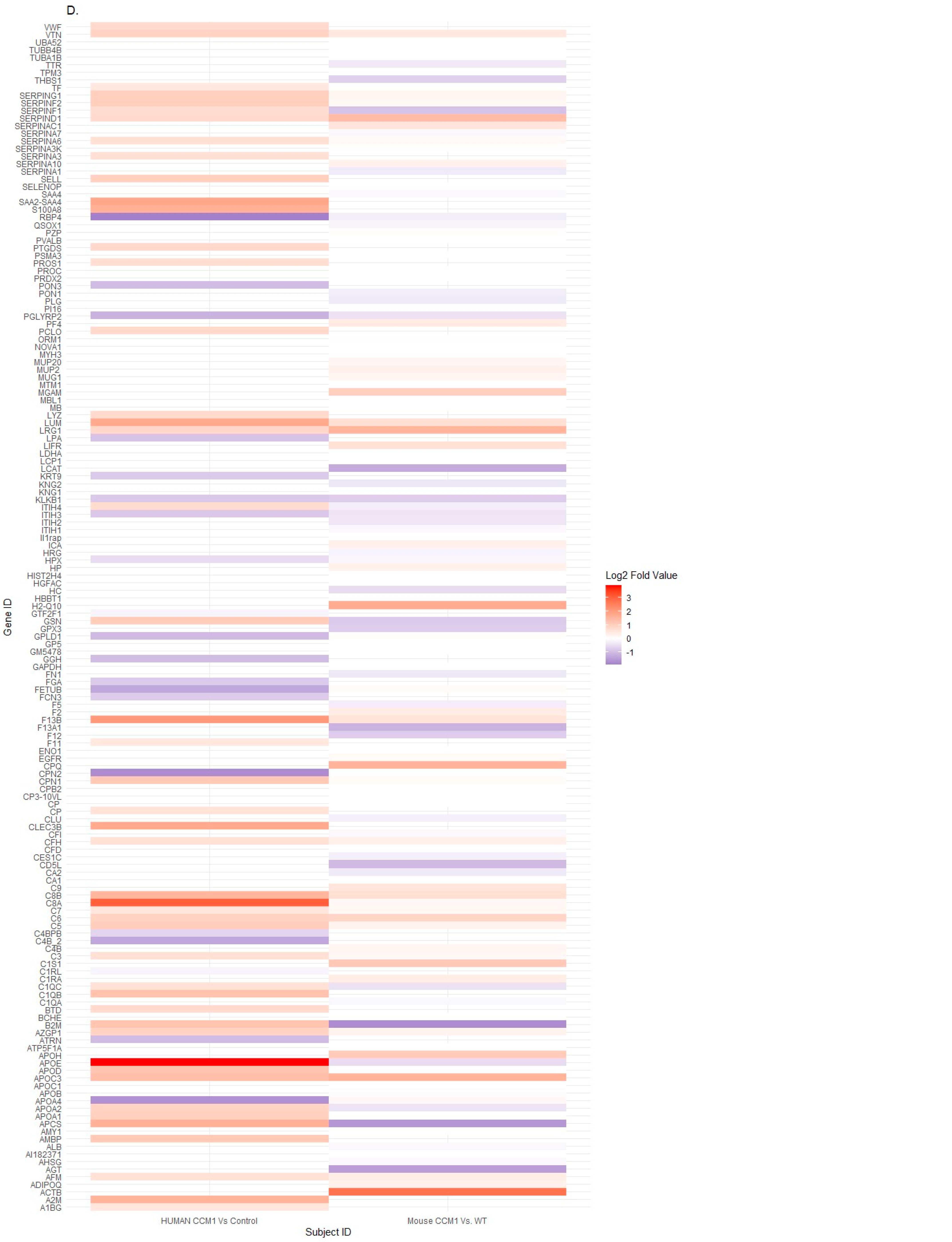
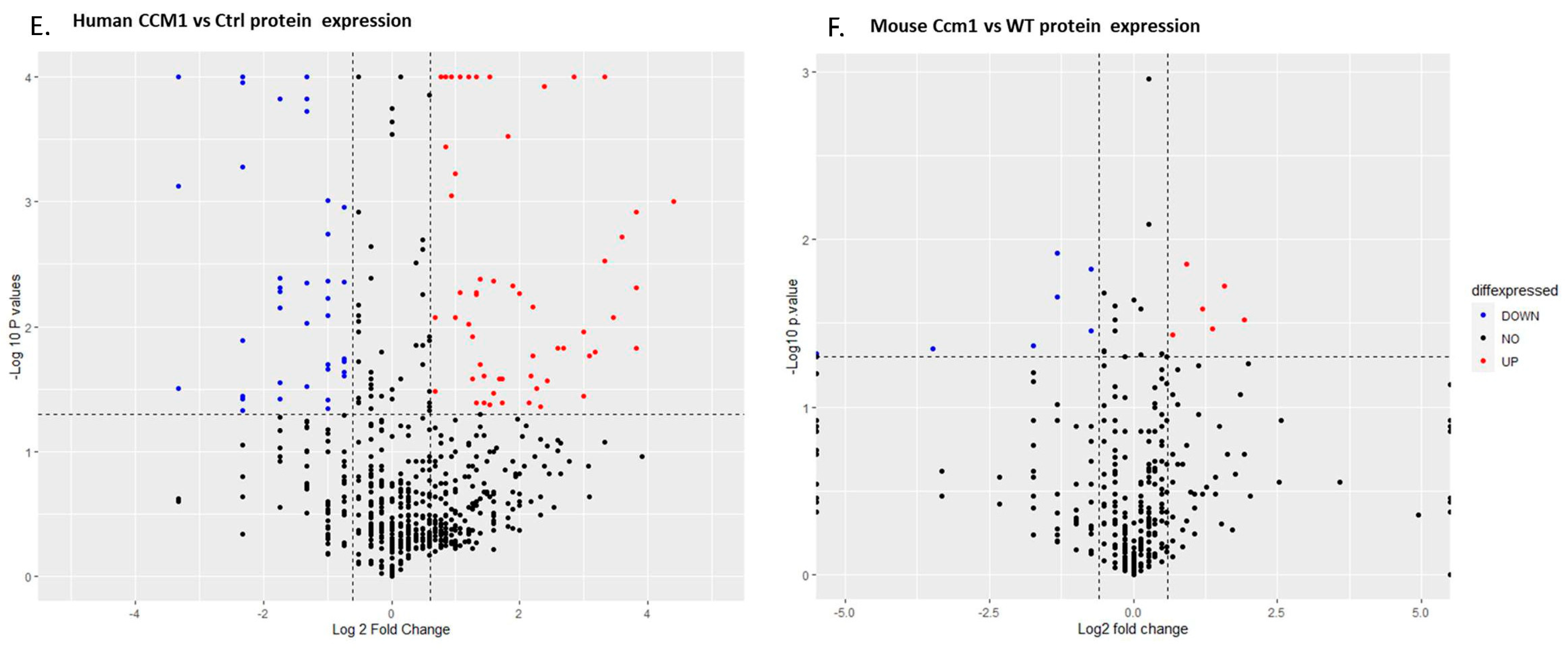
2.1. Differentially Expressed Serum Proteins Were Identified through Proteomics in Two Species
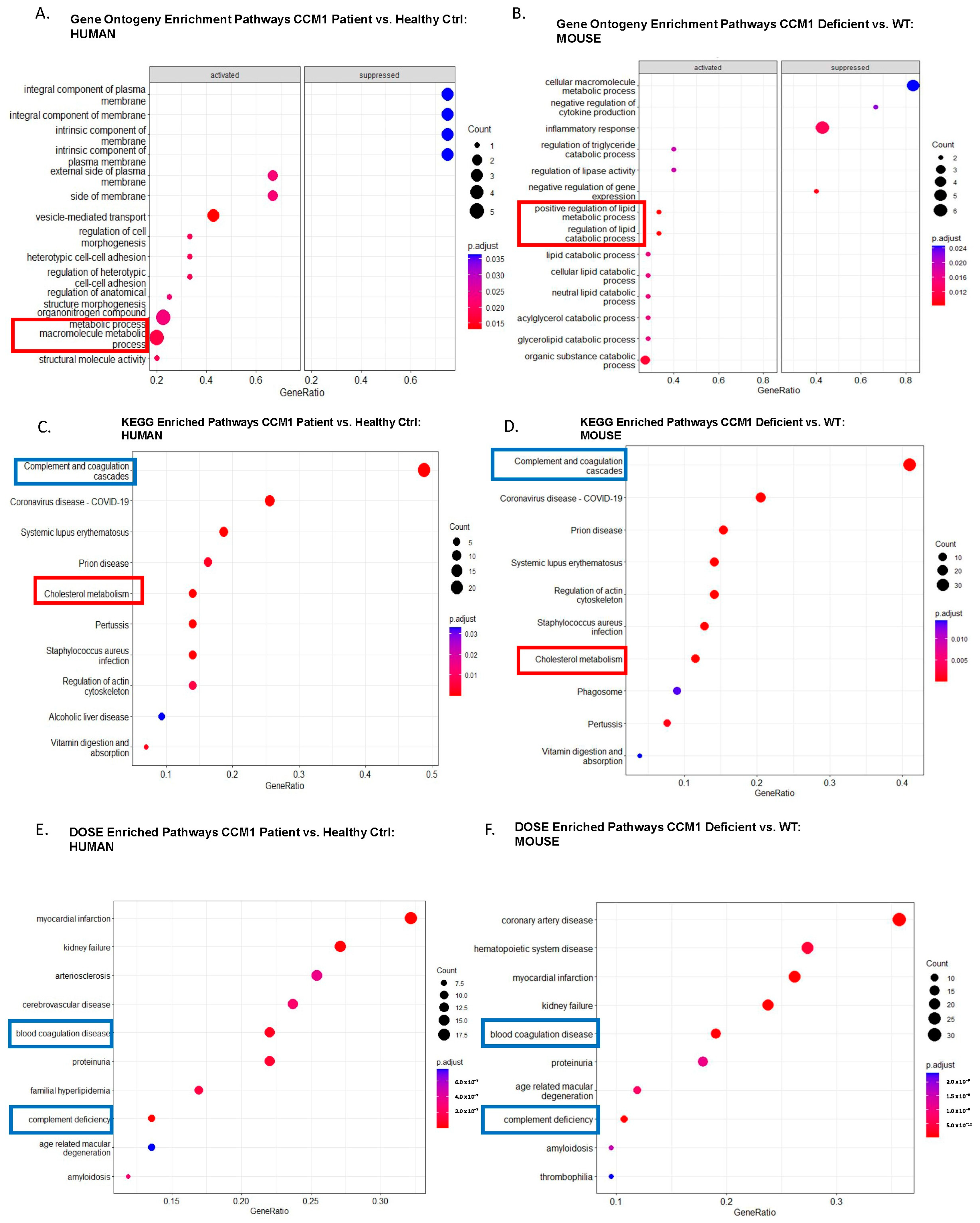
2.2. CCM1-Associated Signaling Pathways Were Identified through Pathway Analyses among DEPs in Two Species
2.3. Common Shared CCM1-Associated Signaling Pathways Were Identified in Two Species
2.4. Candidate Serum Circulating Biomarkers Were Identified through Comparative Analysis among Common Shared CCM1-Associated Signaling Pathways in Two Species
3. Discussion
4. Materials and Methods
4.1. fCCM Patient Cohort Recruitment Procedure
4.2. Data Collection
4.3. Biomarker Data Collection from Proteomics
4.4. Biomarker Identification through Pathway Analyses with Bioinformatics Tools
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Batra, S.; Lin, D.; Recinos, P.F.; Zhang, J.; Rigamonti, D. Cavernous malformations: Natural history, diagnosis and treatment. Nat. Rev. Neurol. 2009, 5, 659–670. [Google Scholar] [CrossRef]
- Padarti, A.; Zhang, J. Recent advances in cerebral cavernous malformation research. Vessel Plus 2018, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Rigamonti, D.; Drayer, B.P.; Johnson, P.C.; Hadley, M.N.; Zabramski, J.; Spetzler, R.F. The MRI appearance of cavernous malformations (angiomas). J. Neurosurg. 1987, 67, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Zlotoff, B.J.; Bang, R.H.; Padilla, R.S.; Morrison, L. Cutaneous angiokeratoma and venous malformations in a Hispanic-American patient with cerebral cavernous malformations. Br. J. Dermatol. 2007, 157, 210–212. [Google Scholar] [CrossRef] [PubMed]
- Manole, A.K.; Forrester, V.J.; Zlotoff, B.J.; Hart, B.L.; Morrison, L.A. Cutaneous findings of familial cerebral cavernous malformation syndrome due to the common Hispanic mutation. Am. J. Med. Genet. A 2020, 182, 1066–1072. [Google Scholar] [CrossRef] [PubMed]
- Choquet, H.; Nelson, J.; Pawlikowska, L.; Akers, A.; Baca, B.; Khan, Y.; Hart, B.; Morrison, L.; Kim, H. Clinical Factors Associated with Lesion Count in Familial Cerebral Cavernous Malformation Type 1 Patients with the Common Hispanic Mutation. Stroke 2013, 44, A188. [Google Scholar] [CrossRef]
- Abou-Fadel, J.; Jiang, X.; Padarti, A.; Goswami, D.G.; Smith, M.; Grajeda, B.; Bhalli, M.; Le, A.; Walker, W.E.; Zhang, J. mPR-Specific Actions Influence Maintenance of the Blood-Brain Barrier (BBB). Int. J. Mol. Sci. 2022, 23, 9684. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Molecular biology of cerebral cavernous malformation. In Cavernous Malformations of the Nervous System; Rigamonti, D., Ed.; Cambridge University Press: Cambridge, UK, 2011; pp. 31–40. [Google Scholar] [CrossRef]
- Zhang, J. Learn from the past, review the present, and look towards the future. Vessel Plus 2022, 6, 20. [Google Scholar] [CrossRef]
- Zhang, J.; Abou-Fadel, J. Calm the raging hormone—A new therapeutic strategy involving progesterone-signaling for hemorrhagic CCMs. Vessel Plus 2021, 5, 23. [Google Scholar] [CrossRef]
- Zhang, J.; Abou-Fadel, J.; Renteria, M.; Belkin, O.; Chen, B.; Zhu, Y.; Dammann, P.; Rigamonti, D. Cerebral Cavernous Malformations (CCMs) do not fall in the Spectrum of PIK3CA-Related Overgrowth (PROS). J. Neurol. Neurosurg. Psychiatry 2022. [Google Scholar] [CrossRef]
- Lopez-Serrano, R.; Martinez, N.E.; Kusak, M.E.; Quiros, A.; Martinez, R. Significant Hemorrhage Rate Reduction after Gamma Knife Radiosurgery in Symptomatic Cavernous Malformations: Long-Term Outcome in 95 Case Series and Literature Review. Stereotact. Funct. Neurosurg. 2017, 95, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Wang, W.H.; Yang, H.C.; Lin, C.J.; Wu, H.M.; Lin, Y.Y.; Hu, Y.S.; Chen, C.J.; Chen, Y.W.; Chou, C.C.; et al. Gamma Knife radiosurgery for cerebral cavernous malformation. Sci. Rep. 2019, 9, 19743. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.; Poorthuis, M.H.F.; Grover, P.; Kitchen, N.; Al-Shahi Salman, R. Surgery for cerebral cavernous malformations: A systematic review and meta-analysis. Neurosurg. Rev. 2022, 45, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yue, K.; Sun, J.; Cao, Y.; Zhao, B.; Zhang, H.; Dai, S.; Zhang, L.; Luo, P.; Jiang, X. Microsurgery vs. Gamma Knife Radiosurgery for the Treatment of Brainstem Cavernous Malformations: A Systematic Review and Meta-Analysis. Front. Neurol. 2021, 12, 600461. [Google Scholar] [CrossRef] [PubMed]
- Augello, C.J.; Noll, J.M.; Distel, T.J.; Wainright, J.D.; Stout, C.E.; Ford, B.D. Identification of novel blood biomarker panels to detect ischemic stroke in patients and their responsiveness to therapeutic intervention. Brain Res. 2018, 1698, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Fainardi, E.; Bortolotti, D.; Castellazzi, M.; Casetta, I.; Bellini, T.; Rizzo, R. Detection of serum soluble HLA-G levels in patients with acute ischemic stroke: A pilot study. Hum. Immunol. 2020, 81, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.Y.; Lou, B.; Zhou, J.J.; Zhong, R.; Wang, R.J.; Zang, X.P.; Shen, H.M.; Li, Y.H. Blood biomarkers in ischemic stroke: Role of biomarkers in differentiation of clinical phenotype. Eur. J. Inflamm. 2018, 16, 2058739218780058. [Google Scholar] [CrossRef]
- Makris, K.; Haliassos, A.; Chondrogianni, M.; Tsivgoulis, G. Blood biomarkers in ischemic stroke: Potential role and challenges in clinical practice and research. Crit. Rev. Clin. Lab. Sci. 2018, 55, 294–328. [Google Scholar] [CrossRef] [PubMed]
- Ramesha, S.; Lui, M.; Richardson, B.; Frankel, M.; Rangaraju, S. Human plasma proteomics for biomarker discovery for ischemic stroke and TIA. Neurology 2020, 94, 5081. [Google Scholar] [CrossRef]
- Girard, R.; Zeineddine, H.A.; Fam, M.D.; Mayampurath, A.; Cao, Y.; Shi, C.; Shenkar, R.; Polster, S.P.; Jesselson, M.; Duggan, R.; et al. Plasma Biomarkers of Inflammation Reflect Seizures and Hemorrhagic Activity of Cerebral Cavernous Malformations. Transl. Stroke Res. 2018, 9, 34–43. [Google Scholar] [CrossRef]
- Maiuri, F.; Mariniello, G.; Corvino, S.; Somma, T.; Guadagno, E.; Teodonno, G.; Del Basso De Caro, M.; Cappabianca, P. Cavernous Malformations to Be Investigated for Familiarity: The Role of Ki67 MIB1. World Neurosurg. 2021, 155, e75–e82. [Google Scholar] [CrossRef]
- Sone, J.Y.; Li, Y.; Hobson, N.; Romanos, S.G.; Srinath, A.; Lyne, S.B.; Shkoukani, A.; Carrion-Penagos, J.; Stadnik, A.; Piedad, K.; et al. Perfusion and permeability as diagnostic biomarkers of cavernous angioma with symptomatic hemorrhage. J. Cereb. Blood Flow. Metab. 2021, 41, 2944–2956. [Google Scholar] [CrossRef] [PubMed]
- Lyne, S.B.; Girard, R.; Koskimaki, J.; Zeineddine, H.A.; Zhang, D.; Cao, Y.; Li, Y.; Stadnik, A.; Moore, T.; Lightle, R.; et al. Biomarkers of cavernous angioma with symptomatic hemorrhage. JCI Insight 2019, 4, e128577. [Google Scholar] [CrossRef]
- Chen, N.; Zhang, H.; Zang, E.; Liu, Z.X.; Lan, Y.F.; Hao, W.L.; He, S.; Fan, X.; Sun, G.L.; Wang, Y.L. Adaptation insights from comparative transcriptome analysis of two Opisthopappus species in the Taihang mountains. BMC Genom. 2022, 23, 466. [Google Scholar] [CrossRef]
- Bluthgen, N.; Brand, K.; Cajavec, B.; Swat, M.; Herzel, H.; Beule, D. Biological profiling of gene groups utilizing Gene Ontology. Genome Inform. 2005, 16, 106–115. [Google Scholar]
- Wang, X.; Li, N.; Guo, H.; Yin, X.; Zheng, Y. Correlation Analysis of Gene and Radiomic Features in Colorectal Cancer Liver Metastases. Comput. Math. Methods Med. 2022, 2022, 8559011. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Yan, G.R.; He, Q.Y. DOSE: An R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015, 31, 608–609. [Google Scholar] [CrossRef]
- Li, W.; Guo, J.; Chen, J.; Yao, H.; Mao, R.; Li, C.; Zhang, G.; Chen, Z.; Xu, X.; Wang, C. Identification of Immune Infiltration and the Potential Biomarkers in Diabetic Peripheral Neuropathy through Bioinformatics and Machine Learning Methods. Biomolecules 2022, 13, 39. [Google Scholar] [CrossRef]
- Antognelli, C.; Trapani, E.; Delle Monache, S.; Perrelli, A.; Daga, M.; Pizzimenti, S.; Barrera, G.; Cassoni, P.; Angelucci, A.; Trabalzini, L.; et al. KRIT1 loss-of-function induces a chronic Nrf2-mediated adaptive homeostasis that sensitizes cells to oxidative stress: Implication for Cerebral Cavernous Malformation disease. Free. Radic. Biol. Med. 2018, 115, 202–218. [Google Scholar] [CrossRef]
- Antognelli, C.; Trapani, E.; Delle Monache, S.; Perrelli, A.; Fornelli, C.; Retta, F.; Cassoni, P.; Talesa, V.N.; Retta, S.F. Data in support of sustained upregulation of adaptive redox homeostasis mechanisms caused by KRIT1 loss-of-function. Data Brief. 2018, 16, 929–938. [Google Scholar] [CrossRef]
- Goitre, L.; De Luca, E.; Braggion, S.; Trapani, E.; Guglielmotto, M.; Biasi, F.; Forni, M.; Moglia, A.; Trabalzini, L.; Retta, S.F. KRIT1 loss of function causes a ROS-dependent upregulation of c-Jun. Free. Radic. Biol. Med. 2014, 68, 134–147. [Google Scholar] [CrossRef]
- Goitre, L.; DiStefano, P.V.; Moglia, A.; Nobiletti, N.; Baldini, E.; Trabalzini, L.; Keubel, J.; Trapani, E.; Shuvaev, V.V.; Muzykantov, V.R.; et al. Up-regulation of NADPH oxidase-mediated redox signaling contributes to the loss of barrier function in KRIT1 deficient endothelium. Sci. Rep. 2017, 7, 8296. [Google Scholar] [CrossRef]
- Simao, F.; Feener, E.P. The Effects of the Contact Activation System on Hemorrhage. Front. Med. 2017, 4, 121. [Google Scholar] [CrossRef]
- Loczi, L.; Orban-Kalmandi, R.; Arokszallasi, T.; Fekete, I.; Fekete, K.; Heja, M.; Toth, J.; Csiba, L.; Bagoly, Z. Thrombin generation as a predictor of outcomes in patients with non-traumatic intracerebral hemorrhage. Front. Neurol. 2022, 13, 912664. [Google Scholar] [CrossRef]
- Choudhri, T.F.; Hoh, B.L.; Prestigiacomo, C.J.; Huang, J.; Kim, L.J.; Schmidt, A.M.; Kisiel, W.; Connolly, E.S., Jr.; Pinsky, D.J. Targeted inhibition of intrinsic coagulation limits cerebral injury in stroke without increasing intracerebral hemorrhage. J. Exp. Med. 1999, 190, 91–99. [Google Scholar] [CrossRef]
- Burchell, S.R.; Tang, J.; Zhang, J.H. Hematoma Expansion Following Intracerebral Hemorrhage: Mechanisms Targeting the Coagulation Cascade and Platelet Activation. Curr. Drug Targets 2017, 18, 1329–1344. [Google Scholar] [CrossRef]
- Li, Z.; Khan, S.; Liu, Y.; Wei, R.; Yong, V.W.; Xue, M. Therapeutic strategies for intracerebral hemorrhage. Front. Neurol. 2022, 13, 1032343. [Google Scholar] [CrossRef]
- Jansen van Vuuren, J.; Pillay, S.; Naidoo, A. Circulating Biomarkers in Long-Term Stroke Prognosis: A Scoping Review Focusing on the South African Setting. Cureus 2022, 14, e23971. [Google Scholar] [CrossRef]
- Zheng, Y.; Fan, L.; Xia, S.; Yang, Q.; Zhang, Z.; Chen, H.; Zeng, H.; Fu, X.; Peng, Y.; Xu, C.; et al. Role of complement C1q/C3-CR3 signaling in brain injury after experimental intracerebral hemorrhage and the effect of minocycline treatment. Front. Immunol. 2022, 13, 919444. [Google Scholar] [CrossRef]
- Hua, Y.; Xi, G.; Keep, R.F.; Hoff, J.T. Complement activation in the brain after experimental intracerebral hemorrhage. J. Neurosurg. 2000, 92, 1016–1022. [Google Scholar] [CrossRef]
- Holste, K.; Xia, F.; Garton, H.J.L.; Wan, S.; Hua, Y.; Keep, R.F.; Xi, G. The role of complement in brain injury following intracerebral hemorrhage: A review. Exp. Neurol. 2021, 340, 113654. [Google Scholar] [CrossRef]
- Clarke, A.R.; Christophe, B.R.; Khahera, A.; Sim, J.L.; Connolly, E.S., Jr. Therapeutic Modulation of the Complement Cascade in Stroke. Front. Immunol. 2019, 10, 1723. [Google Scholar] [CrossRef]
- Schmaier, A.H. Plasma Prekallikrein: Its Role in Hereditary Angioedema and Health and Disease. Front. Med. 2018, 5, 3. [Google Scholar] [CrossRef]
- Wang, J.K.; Li, Y.; Zhao, X.L.; Liu, Y.B.; Tan, J.; Xing, Y.Y.; Adi, D.; Wang, Y.T.; Fu, Z.Y.; Ma, Y.T.; et al. Ablation of Plasma Prekallikrein Decreases Low-Density Lipoprotein Cholesterol by Stabilizing Low-Density Lipoprotein Receptor and Protects Against Atherosclerosis. Circulation 2022, 145, 675–687. [Google Scholar] [CrossRef]
- Sun, Y.; Langer, H.F. Platelets, Thromboinflammation and Neurovascular Disease. Front. Immunol. 2022, 13, 843404. [Google Scholar] [CrossRef]
- Shariat-Madar, Z.; Mahdi, F.; Schmaier, A.H. Assembly and activation of the plasma kallikrein/kinin system: A new interpretation. Int. Immunopharmacol. 2002, 2, 1841–1849. [Google Scholar] [CrossRef]
- Barco, S.; Sollfrank, S.; Trinchero, A.; Adenaeuer, A.; Abolghasemi, H.; Conti, L.; Hauser, F.; Kremer Hovinga, J.A.; Lackner, K.J.; Loewecke, F.; et al. Severe plasma prekallikrein deficiency: Clinical characteristics, novel KLKB1 mutations, and estimated prevalence. J. Thromb. Haemost. 2020, 18, 1598–1617. [Google Scholar] [CrossRef]
- Jiao, W.; Ji, J.F.; Xu, W.; Bu, W.; Zheng, Y.; Ma, A.; Zhao, B.; Fan, Q. Distinct downstream signaling and the roles of VEGF and PlGF in high glucose-mediated injuries of human retinal endothelial cells in culture. Sci. Rep. 2019, 9, 15339. [Google Scholar] [CrossRef]
- Abdel-Magid, A.F. Plasma Kallikrein Inhibitors for the Treatment of Retinal Vascular Permeability Associated with Diabetic Retinopathy and Diabetic Macular Edema. ACS Med. Chem. Lett. 2017, 8, 776–777. [Google Scholar] [CrossRef]
- Bossi, F.; Peerschke, E.I.; Ghebrehiwet, B.; Tedesco, F. Cross-talk between the complement and the kinin system in vascular permeability. Immunol. Lett. 2011, 140, 7–13. [Google Scholar] [CrossRef]
- Bossi, F.; Fischetti, F.; Regoli, D.; Durigutto, P.; Frossi, B.; Gobeil, F., Jr.; Ghebrehiwet, B.; Peerschke, E.I.; Cicardi, M.; Tedesco, F. Novel pathogenic mechanism and therapeutic approaches to angioedema associated with C1 inhibitor deficiency. J. Allergy Clin. Immunol. 2009, 124, 1303–1310.e1304. [Google Scholar] [CrossRef]
- Guan, J.; Clermont, A.C.; Pham, L.D.; Ustunkaya, T.; Revenko, A.S.; MacLeod, A.R.; Feener, E.P.; Simao, F. Plasma Kallikrein Contributes to Intracerebral Hemorrhage and Hypertension in Stroke-Prone Spontaneously Hypertensive Rats. Transl. Stroke Res. 2022, 13, 287–299. [Google Scholar] [CrossRef]
- Simao, F.; Ustunkaya, T.; Clermont, A.C.; Feener, E.P. Plasma kallikrein mediates brain hemorrhage and edema caused by tissue plasminogen activator therapy in mice after stroke. Blood 2017, 129, 2280–2290. [Google Scholar] [CrossRef]
- Sanchez-Navarro, A.; Gonzalez-Soria, I.; Caldino-Bohn, R.; Bobadilla, N.A. An integrative view of serpins in health and disease: The contribution of SerpinA3. Am. J. Physiol. Cell Physiol. 2021, 320, C106–C118. [Google Scholar] [CrossRef]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef]
- Richardson, J.; Viswanathan, K.; Lucas, A. Serpins, the vasculature, and viral therapeutics. Front. Biosci. 2006, 11, 1042–1056. [Google Scholar] [CrossRef][Green Version]
- He, X.; Cheng, R.; Benyajati, S.; Ma, J.X. PEDF and its roles in physiological and pathological conditions: Implication in diabetic and hypoxia-induced angiogenic diseases. Clin. Sci. 2015, 128, 805–823. [Google Scholar] [CrossRef]
- Grover, S.P.; Mackman, N. Anticoagulant SERPINs: Endogenous Regulators of Hemostasis and Thrombosis. Front. Cardiovasc. Med. 2022, 9, 878199. [Google Scholar] [CrossRef]
- Bianchini, E.P.; Auditeau, C.; Razanakolona, M.; Vasse, M.; Borgel, D. Serpins in Hemostasis as Therapeutic Targets for Bleeding or Thrombotic Disorders. Front. Cardiovasc. Med. 2020, 7, 622778. [Google Scholar] [CrossRef]
- Iwaki, T.; Tanaka, A.; Miyawaki, Y.; Suzuki, A.; Kobayashi, T.; Takamatsu, J.; Matsushita, T.; Umemura, K.; Urano, T.; Kojima, T.; et al. Life-threatening hemorrhage and prolonged wound healing are remarkable phenotypes manifested by complete plasminogen activator inhibitor-1 deficiency in humans. J. Thromb. Haemost. 2011, 9, 1200–1206. [Google Scholar] [CrossRef]
- Barnstable, C.J.; Tombran-Tink, J. Neuroprotective and antiangiogenic actions of PEDF in the eye: Molecular targets and therapeutic potential. Prog. Retin. Eye Res. 2004, 23, 561–577. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, X.; Li, X. Peptidoglycan-based immunomodulation. Appl. Microbiol. Biotechnol. 2022, 106, 981–993. [Google Scholar] [CrossRef]
- Dziarski, R.; Gupta, D. Peptidoglycan recognition in innate immunity. J. Endotoxin Res. 2005, 11, 304–310. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Hozo, I.; Rangarajan, S.; Mariuzza, R.A.; Dziarski, R.; Gupta, D. Genetic Association of Peptidoglycan Recognition Protein Variants with Inflammatory Bowel Disease. PLoS ONE 2013, 8, e67393. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Jigami, T.; Furukawa, S.; Sagara, M.; Echizen, K.; Shibata, Y.; Sato, R.; Akiyama, T. The adenomatous polyposis coli-associated guanine nucleotide exchange factor Asef is involved in angiogenesis. J. Biol. Chem. 2010, 285, 1199–1207. [Google Scholar] [CrossRef]
- Hankey, W.; Frankel, W.L.; Groden, J. Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: Implications for therapeutic targeting. Cancer Metastasis Rev. 2018, 37, 159–172. [Google Scholar] [CrossRef]
- Harris, E.S.; Nelson, W.J. Adenomatous polyposis coli regulates endothelial cell migration independent of roles in beta-catenin signaling and cell-cell adhesion. Mol. Biol. Cell 2010, 21, 2611–2623. [Google Scholar] [CrossRef][Green Version]
- Nar, G.; Sanlialp, S.C.; Nar, R. Retinol binding protein 4 levels relate to the presence and severity of coronary artery disease. J. Med. Biochem. 2021, 40, 384–389. [Google Scholar] [CrossRef]
- Steinhoff, J.S.; Lass, A.; Schupp, M. Biological Functions of RBP4 and Its Relevance for Human Diseases. Front. Physiol. 2021, 12, 659977. [Google Scholar] [CrossRef]
- Ji, Y.; Song, J.; Su, T.; Gu, X. Adipokine Retinol Binding Protein 4 and Cardiovascular Diseases. Front. Physiol. 2022, 13, 856298. [Google Scholar] [CrossRef] [PubMed]
- Codoner-Franch, P.; Carrasco-Luna, J.; Allepuz, P.; Codoner-Alejos, A.; Guillem, V. Association of RBP4 genetic variants with childhood obesity and cardiovascular risk factors. Pediatr. Diabetes 2016, 17, 576–583. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peyre, M.; Miyagishima, D.; Bielle, F.; Chapon, F.; Sierant, M.; Venot, Q.; Lerond, J.; Marijon, P.; Abi-Jaoude, S.; Le Van, T.; et al. Somatic PIK3CA Mutations in Sporadic Cerebral Cavernous Malformations. N. Engl. J. Med. 2021, 385, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Dammann, P.; Wrede, K.; Zhu, Y.; Matsushige, T.; Maderwald, S.; Umutlu, L.; Quick, H.H.; Hehr, U.; Rath, M.; Ladd, M.E.; et al. Correlation of the venous angioarchitecture of multiple cerebral cavernous malformations with familial or sporadic disease: A susceptibility-weighted imaging study with 7-Tesla MRI. J. Neurosurg. 2017, 126, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Yang, Y.; Song, D.; Huo, R.; Li, H.; Chen, Y.; Nam, Y.; Zhou, Q.; Jiao, Y.; Fu, W.; et al. Somatic MAP3K3 mutation defines a subclass of cerebral cavernous malformation. Am. J. Hum. Genet. 2021, 108, 942–950. [Google Scholar] [CrossRef]
- Petersen, T.A.; Morrison, L.A.; Schrader, R.M.; Hart, B.L. Familial versus sporadic cavernous malformations: Differences in developmental venous anomaly association and lesion phenotype. AJNR Am. J. Neuroradiol. 2010, 31, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Serdar, C.C.; Cihan, M.; Yucel, D.; Serdar, M.A. Sample size, power and effect size revisited: Simplified and practical approaches in pre-clinical, clinical and laboratory studies. Biochem. Med. 2021, 31, 010502. [Google Scholar] [CrossRef] [PubMed]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gotzsche, P.C.; Vandenbroucke, J.P.; Initiative, S. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. Lancet 2007, 370, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Abou-Fadel, J.; Jiang, X.; Padarti, A.; Goswami, D.; Marchuk, D.; Walker, W.; Grajeda, B.; Zhang, J. CCM signaling complex (CSC) is a master regulator governing homeostasis of progesterone and its mediated signaling cascades. bioRxiv 2020. [Google Scholar] [CrossRef]
- Abou-Fadel, J.; Jiang, X.; Padarti, A.; Grajeda, B.; Zhang, J. CCM signaling complex (CSC) coupling both classic and non-classic progesterone receptor signaling. bioRxiv 2020. [Google Scholar] [CrossRef]
- Abou-Fadel, J.; Qu, Y.; Gonzalez, E.; Smith, M.; Zhang, J. Emerging roles of CCM genes during tumorigenesis with potential application as novel biomarkers across major types of cancers. Oncol. Rep. 2020, 43, 1945–1963. [Google Scholar] [CrossRef] [PubMed]
- Abou-Fadel, J.; Smith, M.; Falahati, K.; Zhang, J. Comparative omics of CCM signaling complex (CSC). Chin. Neurosurg. J. 2020, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, J.L. Using R and Bioconductor in Clinical Genomics and Transcriptomics. J. Mol. Diagn. 2020, 22, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Carey, V.J.; Morgan, M.; Falcon, S.; Lazarus, R.; Gentleman, R. GGtools: Analysis of genetics of gene expression in bioconductor. Bioinformatics 2007, 23, 522–523. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.C.; Colaprico, A.; Olsen, C.; D’Angelo, F.; Bontempi, G.; Ceccarelli, M.; Noushmehr, H. TCGA Workflow: Analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Research 2016, 5, 1542. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology, C. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.; Sedeno-Cortes, A.E.; Pavlidis, P. Monitoring changes in the Gene Ontology and their impact on genomic data analysis. Gigascience 2018, 7, giy103. [Google Scholar] [CrossRef]
- Gene Ontology, C. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Zhang, J.D.; Wiemann, S. KEGGgraph: A graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics 2009, 25, 1470–1471. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Sartor, M.A.; Jagadish, H.V. Appearance frequency modulated gene set enrichment testing. BMC Bioinform. 2011, 12, 81. [Google Scholar] [CrossRef]
- Glass, K.; Quackenbush, J.; Spentzos, D.; Haibe-Kains, B.; Yuan, G.C. A network model for angiogenesis in ovarian cancer. BMC Bioinform. 2015, 16, 115. [Google Scholar] [CrossRef]
- Brun, Y.F.; Varma, R.; Hector, S.M.; Pendyala, L.; Tummala, R.; Greco, W.R. Simultaneous modeling of concentration-effect and time-course patterns in gene expression data from microarrays. Cancer Genom. Proteom. 2008, 5, 43–53. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Cham, Switzerland, 2016. [Google Scholar]

| (A) | ||
| Ontology | Description | Genes Involved |
| BP | negative regulation of multicellular organismal process | FN1 APOD PGLYRP2 RBP4 KNG1 APOA2 APCS VTN APOE KLKB1 SERPINF2 APOC3 |
| BP | vesicle-mediated transport | APOA2 VTN APOE PCLO APOC3 |
| BP | regulation of vesicle-mediated transport | APOA2 VTN APOE PCLO APOC3 |
| MF | phospholipid binding | APOE PCLO APOC3 |
| BP | regulation of wound healing | KNG1 APCS VTN APOE KLKB1 SERPINF2 |
| BP | negative regulation of cellular process | APOD PGLYRP2 RBP4 KNG1 C5 APOA2 APCS APOE APOC3 |
| (B) | ||
| Ontology | Description | Genes Involved |
| BP | Cytokine Production | Thbs1 Serpinf1 B2m |
| BP | Regulation of Cytokine Production | Thbs1 Serpinf1 B2m |
| BP | Behavior | Thbs1 Serpinf1 B2m |
| BP | Negative Regulation of Gene Expression | Thbs1 Pglyrp2 Serpinf1 |
| BP | Immune Response | Pglyrp2 Igkv1-135 Kng2 Igkv4-70 APCS Ighv6-6 B2m |
| CC | Extracellular region | C8b Pf4 Klkb1 Gpx3 Qsox1 F12 RBP4 Gsn Cd5l F5 Lcat Agt Thbs1 Pglyrp2 Igkv1-135 Serpinf1 Kng2 F13a1 Igkv4-70 APCS Ighv6-6 B2m |
| (A) | ||
| DO:ID | Description | Gene Symbol |
| DOID:6262 | Complement Deficiency | APOA1, C3, C4B, C5, C6, C7, C8A, C8B, CFH, CFHR1 |
| DOID:1247 | Blood Coagulation Disease | C3, C4B, C5, CFH, CPB2, F11, F13B, FGA, PROS1, SELL, SERPINA10, SERPIND1, SERPINF2, VWF |
| DOID:5844 | Myocardial Infarction | APOA1, APOA4, APOE, C3, C4B, C7, C8A, C8B, CFH, CPB2, F11, F13B, FGA, GSN, ITIH4, LPA, SERPIND1, VWF |
| DOID:10871 | Age Related Macular Degeneration | APOE, C3, C5, CFD, CFHR1, SELL, SERPINF1, TF |
| DO: 576 | Proteinuria | APOA1, APOA4, APOE, C3, C4B, C5,C6, CFH, CPB2, HPX, LPA, VTN |
| DO:1074 | Kidney Failure | AMBP, APOA1, APOA4, APOE, C3, C6, CFH, CPB2, FGA, PROS1, S100A8, SELL, SERPINF1, TFRC, VWF |
| (B) | ||
| DO: ID | Description | Gene Symbol |
| DOID:3393 | Coronary Artery Disease | APOA1, APOA2, APOA4, APOC3, APOC4, APOE, APOM, C3, C7, C8A, C8B, CFH, CPB2, F11, F13B, F5, FGA, GSN, ITIH3, ITH4, LCAT, LPA, LUM, PON3, SELL, SERPIND1, SERPINF2, SERPING1, VTN, VWF |
| DOID:626 | Complement Deficiency | APOA1, C3,C5, C6, C7, C8A, C8B, CFH, CFHR1 |
| DOID:1074 | Kidney Failure | AMBP, APOA1, APOA4, APOC3, APOE, B2M, C3, C6, CFH, CPB2, F5, FGA, LCAT, PROS1, PTGDS, S100A8, SELL, SERPINF1, TFRC, VWF |
| DOID:5844 | Myocardial Infarction | APOA1, APOA4, APOC3, APOE, C3, C7, C8A, C8B, CFH, CPB2, F11, F13B, F5, FGA, GSN, ITIH3, ITIH4, LCAT, LPA, SERPIND1, SERPING1, VWF |
| DOID:1247 | Blood Coagulation Disease | C3,C5, CFH, CP, CPB2, F11, F13B, F5, FGA, KLKB1, PROS1, SERPINA10, SERPIND1, SERPINF2, VWF |
| DOID:74 | Hematopoietic System Disease | APOE, C3,C5,CA1,CP, CPB2, F11, F13B, F5, FGA, KLKB1, MASP2, SELL, SERPINA10, SERPIND1, SERPINF1, SERPINF2, TF, TFRC, VWF |
| (A) | ||
| ID | Description | Gene Symbol |
| hsa04610 | Complement and coagulation cascades | A2M, C1QA, C1QC, C3, C4BP, C6, C8A, F11, F13B, FGA, MASP2, SERPIND1, SERPINF2, VTN VWF |
| hsa05171 | Coronavirus disease—COVID-19 | C1QA, C1QC, C3, C6, C8A, F13B, FGA, MASP2, VWF |
| hsa05322 | Systemic lupus erythematosus | C1QA, C1QC, C3, C6, C8A, H2AJ |
| hsa05150 | Staphylococcus aureus infection | C1QA, C1QC, C3, KRT9, MASP2 |
| hsa05133 | Pertussis | C1QA, C1QC, C3, C4BPB |
| hsa04613 | Neutrophil extracellular trap formation | C3, FGA, H2AJ, VWF |
| hsa04512 | ECM-receptor interaction | CD44, VTN, VWF |
| hsa05142 | Chagas disease | C1QA, C1QC, C3 |
| (B) | ||
| ID | Description | Gene Symbol |
| hsa04610 | Complement and coagulation cascades | C1QA, C1QB, C1QC, C3, C4BPB, C5, C6, C7, C8A, C8B, CFD, CFH, CFHR1, CPB2, F11, F13B, F5, FGA, KLKB1, MASP2, PROS1, SERPIND1, SERPINF2, SERPING1, VTN, VWF |
| hsa05171 | Coronavirus disease—COVID-19 | C1QA, C1QB, C1QX, C3, C5, C6, C7, C8A, C8B, CFD, F13B, FGA, IKBKG, MASP2,VWF |
| hsa05150 | Staphylococcus aureus infection | C1QA, C1QB, C1QC, C3, C5, CFD,CFH, KRT9, MASP2 |
| hsa04979 | Cholesterol metabolism | APOA1, APOA2, APOA4, APOC3, APOE, LCAT, LPA |
| hsa05322 | Systemic lupus erythematosus | C1QA, C1QB, C1QC, C3, C5, C6, C7, C8A, C8B, H2AJ |
| hsa05133 | Pertussis | C1QA, C1QB, C1QC, C3, C4BPB, C5, SERPING1 |
| Gene Symbol | Gene Name | Pathway |
|---|---|---|
| APCS | Amyloid P Component | Complement and coagulation cascades |
| SERPINF1 | Serpin Family F Member 1 | Complement and coagulation cascades |
| SERPINF2 | Serpin Family F Member 2 | Complement and coagulation cascades |
| THBS1 | Thrombospondin 1 | Complement and coagulation cascades |
| VTN | Vitronectin | Complement and coagulation cascades |
| FN1 | Fibronectin 1 | Complement and coagulation cascades |
| F12 | Coagulation Factor XII | Complement and coagulation cascades |
| GSN | Gelsolin | Complement and coagulation cascades |
| KLKB1 | Kallikrein B1 | Complement and coagulation cascades |
| KNG1 | Kininogen 1 | Complement and coagulation cascades |
| KNG2 | Kininogen 2 | Complement and coagulation cascades |
| LCAT | Lecithin-Cholesterol Acyltransferase | Cholesterol metabolism |
| APOA2 | Apolipoprotein A2 | Lipoprotein metabolism |
| APOC3 | Apolipoprotein C3 | Lipoprotein metabolism |
| APOD | Apolipoprotein D | Lipoprotein metabolism |
| APOE | Apolipoprotein E | Lipoprotein metabolism |
| PCLO | Piccolo Presynaptic Cytomatrix Protein | Cytoskeletal matrix |
| QSOX1 | Quiescin Sulfhydryl Oxidase 1 | Extracellular matrix |
| B2M | β2-Microglobulin | Immunopathways |
| RBP4 | Retinol Binding Protein 4 | Membrane transporter |
| PGLYRP2 | Peptidoglycan Recognition Protein 2 | N-acetylmuramoyl-l-alanine amidase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Croft, J.; Grajeda, B.; Aguirre, L.A.; Abou-Fadel, J.S.; Ellis, C.C.; Estevao, I.; Almeida, I.C.; Zhang, J. Circulating Blood Prognostic Biomarker Signatures for Hemorrhagic Cerebral Cavernous Malformations (CCMs). Int. J. Mol. Sci. 2024, 25, 4740. https://doi.org/10.3390/ijms25094740
Croft J, Grajeda B, Aguirre LA, Abou-Fadel JS, Ellis CC, Estevao I, Almeida IC, Zhang J. Circulating Blood Prognostic Biomarker Signatures for Hemorrhagic Cerebral Cavernous Malformations (CCMs). International Journal of Molecular Sciences. 2024; 25(9):4740. https://doi.org/10.3390/ijms25094740
Chicago/Turabian StyleCroft, Jacob, Brian Grajeda, Luis A. Aguirre, Johnathan S. Abou-Fadel, Cameron C. Ellis, Igor Estevao, Igor C. Almeida, and Jun Zhang. 2024. "Circulating Blood Prognostic Biomarker Signatures for Hemorrhagic Cerebral Cavernous Malformations (CCMs)" International Journal of Molecular Sciences 25, no. 9: 4740. https://doi.org/10.3390/ijms25094740
APA StyleCroft, J., Grajeda, B., Aguirre, L. A., Abou-Fadel, J. S., Ellis, C. C., Estevao, I., Almeida, I. C., & Zhang, J. (2024). Circulating Blood Prognostic Biomarker Signatures for Hemorrhagic Cerebral Cavernous Malformations (CCMs). International Journal of Molecular Sciences, 25(9), 4740. https://doi.org/10.3390/ijms25094740









