Cytological, Phytohormone, and Transcriptome Analyses Provide Insights into Persimmon Fruit Shape Formation (Diospyros kaki Thunb.)
Abstract
:1. Introduction
2. Results
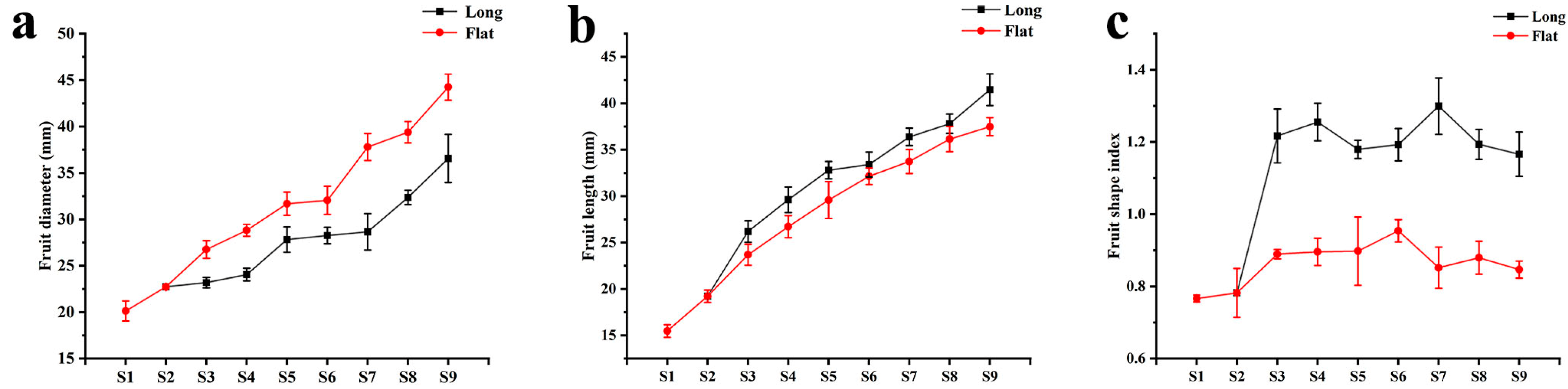
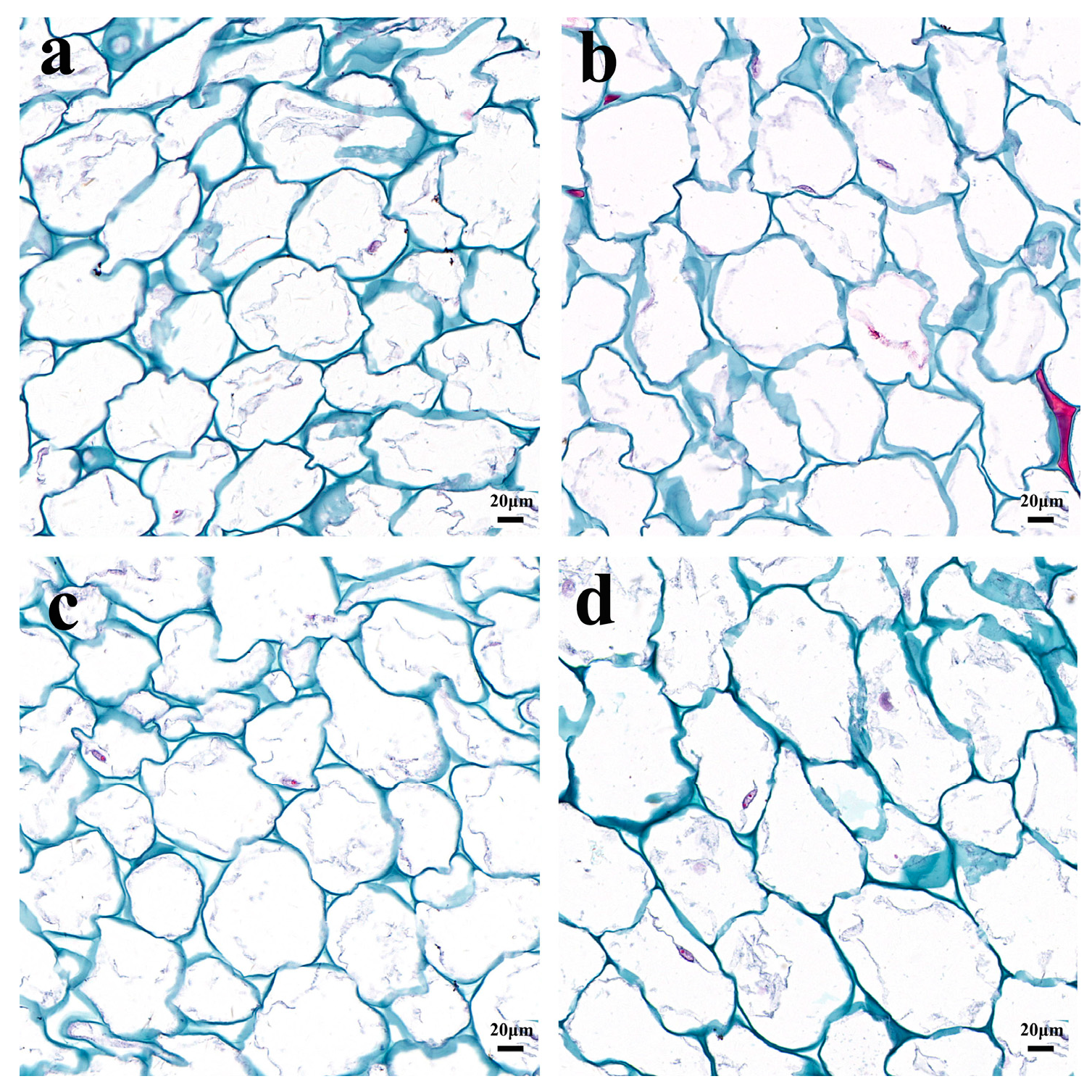
2.1. Morphological Comparison of Fruit
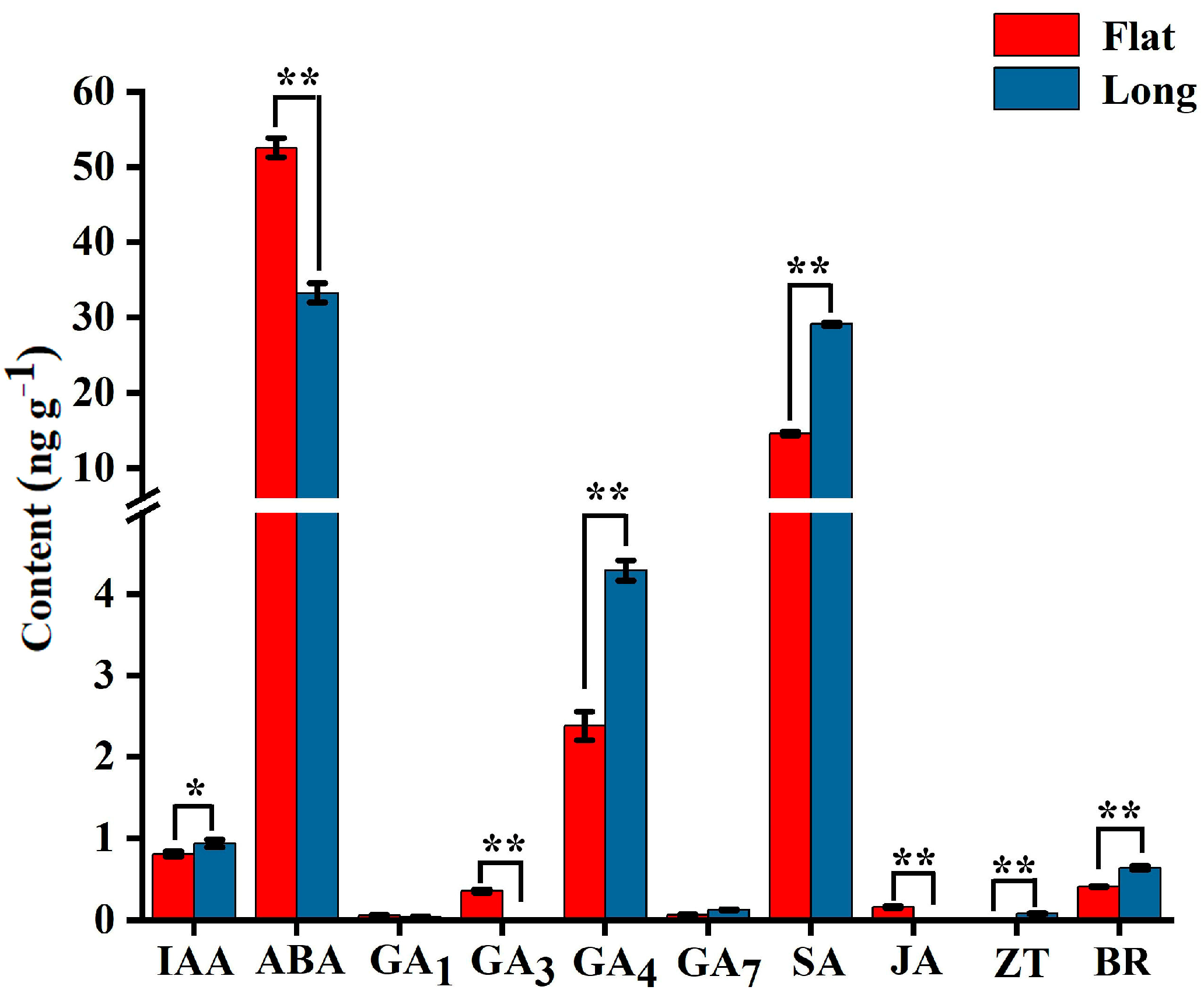
2.2. Phytohormone Content in Flat and Long Fruits
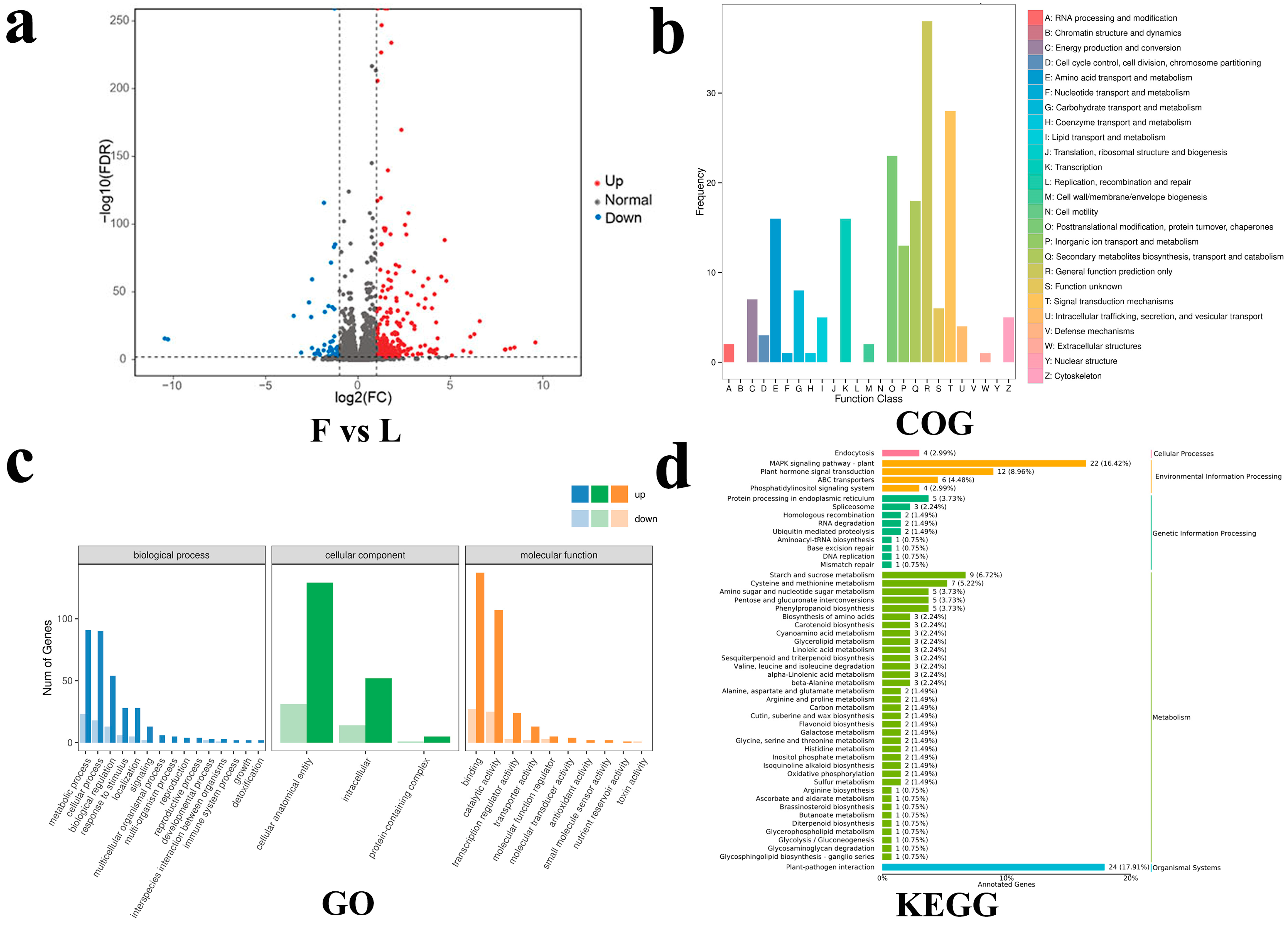
2.3. Transcriptome
2.4. DEGs Related to Phytohormones
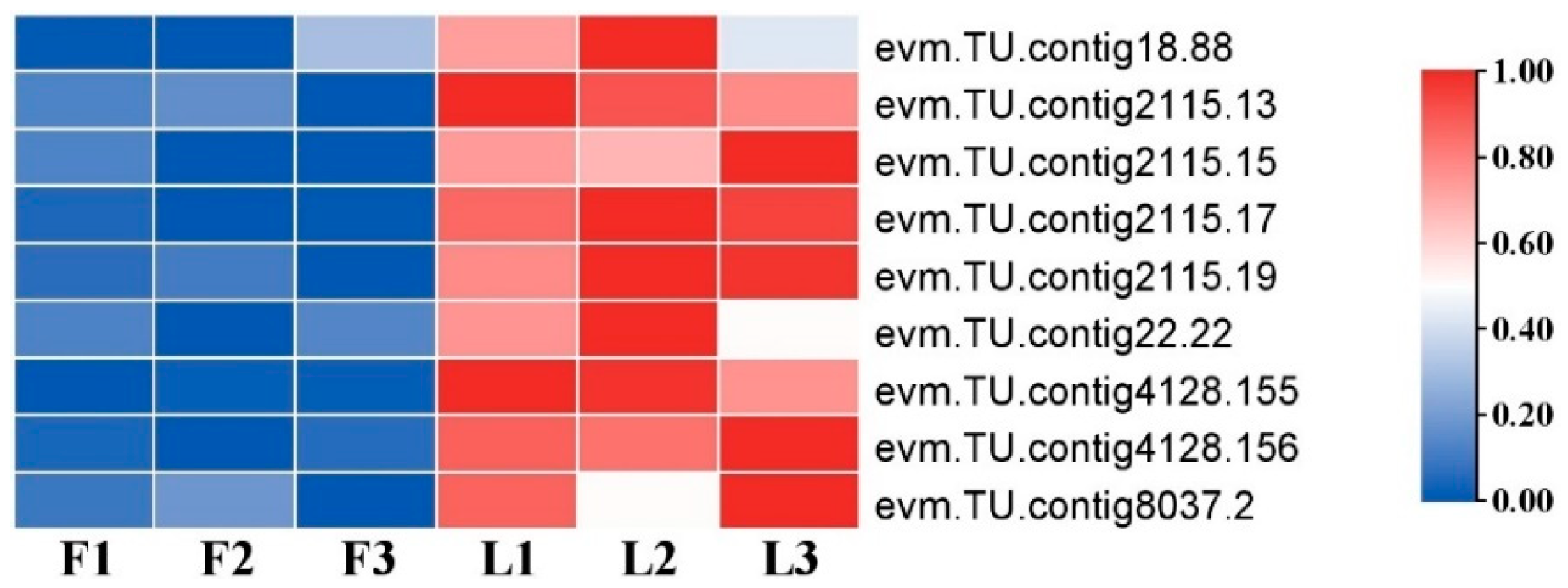
2.5. DEGs Related to Cell Division and Cell Expansion
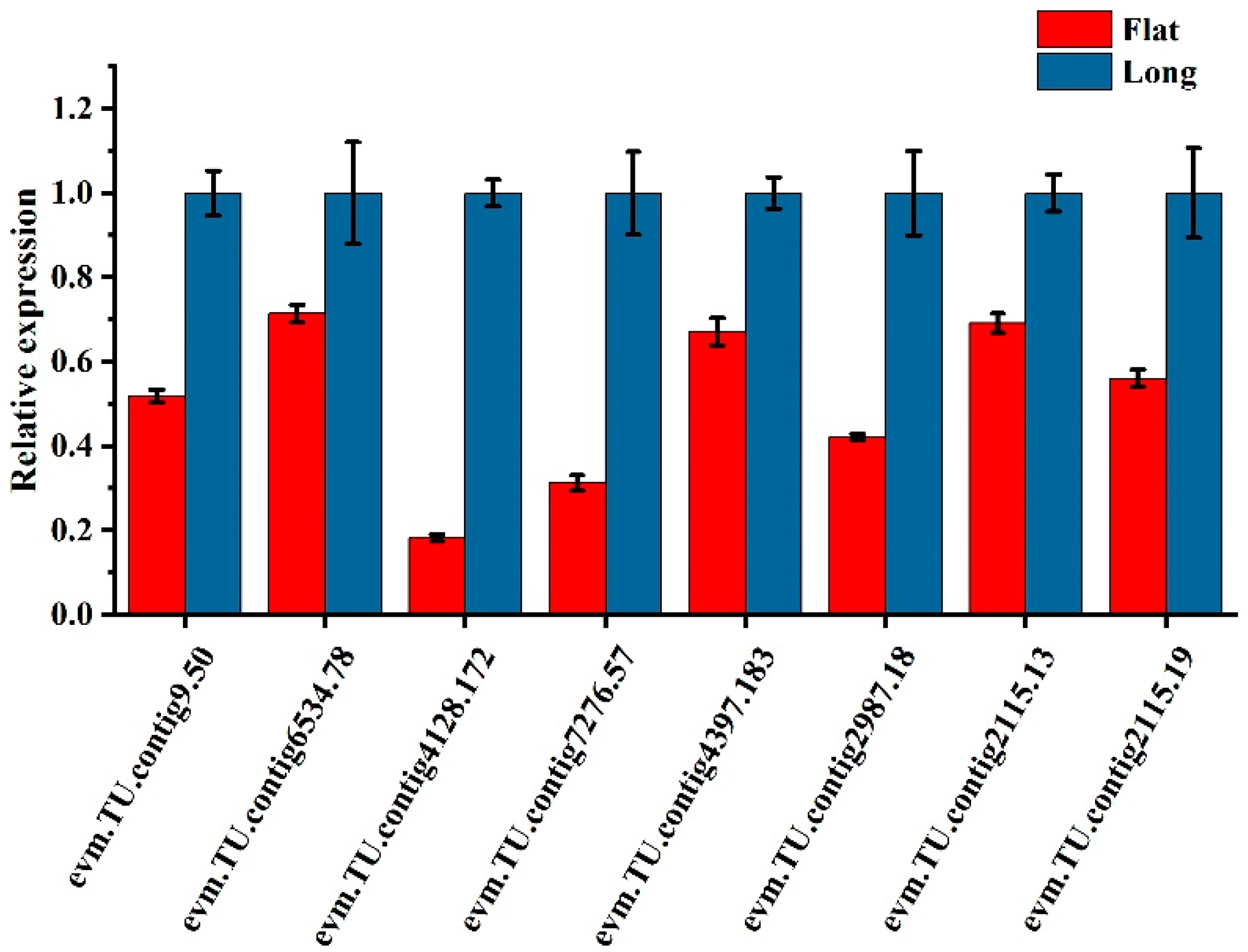
2.6. DEGs Validation by RT-qPCR
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Material
5.2. Paraffin Section
5.3. Phytohormone Assay
5.4. Transcriptome
5.5. Quantitative RT-PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, Z.; Wang, R. Persimmon in China: Domestication and traditional utilization of genetic resources. Adv. Hortic. Sci. 2008, 22, 239–243. [Google Scholar]
- Rodriguez, G.R.; Muños, S.; Anderson, C.; Sim, S.-C.; Michel, A.; Causse, M.; Gardener, B.B.M.; Francis, D.; van Der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodríguez, G.R.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Díaz, A. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, Y.; McGregor, C.; Liu, S.; Luan, F.; Gao, M.; Weng, Y. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 2020, 133, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Cirilli, M.; Baccichet, I.; Chiozzotto, R.; Silvestri, C.; Rossini, L.; Bassi, D. Genetic and phenotypic analyses reveal major quantitative loci associated to fruit size and shape traits in a non-flat peach collection (P. persica L. Batsch). Hortic. Res. 2021, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Clevenger, J.P.; Illa-Berenguer, E.; Meulia, T.; van der Knaap, E.; Sun, L. A comparison of sun, ovate, fs8. 1 and auxin application on tomato fruit shape and gene expression. Plant Cell Physiol. 2019, 60, 1067–1081. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, F.; Deng, Y.; Zhong, F.; Tian, P.; Lin, D.; Deng, J.; Zhang, Y.; Huang, T. Sly-miR159 regulates fruit morphology by modulating GA biosynthesis in tomato. Plant Biotechnol. J. 2022, 20, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, X.-J.; Tan, G.-F.; Zhou, W.-Q.; Wang, G.-L. Gibberellin and the plant growth retardant Paclobutrazol altered fruit shape and ripening in tomato. Protoplasma 2020, 257, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 2004, 16 (Suppl. 1), S181–S189. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef]
- Liu, J.; Van Eck, J.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302–13306. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Liu, X.; Sun, C.; Song, X.; Li, X.; Cui, H.; Guo, J.; Liu, L.; Ying, A.; Zhang, Z. CsTRM5 regulates fruit shape via mediating cell division direction and cell expansion in cucumber. Hortic. Res. 2023, 10, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cao, K.; Li, Y.; Yao, J.-L.; Deng, C.; Wang, Q.; Zhu, G.; Fang, W.; Chen, C.; Wang, X. Comparative transcriptome and microscopy analyses provide insights into flat shape formation in peach (Prunus persica). Front. Plant Sci. 2018, 8, 309093. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Akagi, T.; Tao, R. Quantitative characterization of fruit shape and its differentiation pattern in diverse persimmon (Diospyros kaki) cultivars. Sci. Hortic. 2018, 228, 41–48. [Google Scholar] [CrossRef]
- Li, H.; Sun, P.; Wang, Y.; Zhang, Z.; Yang, J.; Suo, Y.; Han, W.; Diao, S.; Li, F.; Fu, J. Allele-aware chromosome-level genome assembly of the autohexaploid Diospyros kaki Thunb. Sci. Data 2023, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Colle, M.; Weng, Y.; Kang, Y.; Ophir, R.; Sherman, A.; Grumet, R. Variation in cucumber (Cucumis sativus L.) fruit size and shape results from multiple components acting pre-anthesis and post-pollination. Planta 2017, 246, 641–658. [Google Scholar] [CrossRef]
- Fu, F.Q.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Asami, T.; Yu, J.Q. A role of brassinosteroids in early fruit development in cucumber. J. Exp. Bot. 2008, 59, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Higashi, K.; Hosoya, K.; Ezura, H. Histological analysis of fruit development between two melon (Cucumis melo L. reticulatus) genotypes setting a different size of fruit. J. Exp. Bot. 1999, 50, 1593–1597. [Google Scholar] [CrossRef]
- Olmstead, J.W.; Iezzoni, A.F.; Whiting, M.D. Genotypic differences in sweet cherry fruit size are primarily a function of cell number. J. Am. Soc. Hortic. Sci. 2007, 132, 697–703. [Google Scholar] [CrossRef]
- Scorzal, R.; May, L.G.; Purnell, B.; Upchurch, B. Differences in number and area of mesocarp cells between small-and large-fruited peach cultivars. J. Am. Soc. Hortic. Sci. 1991, 116, 861–864. [Google Scholar] [CrossRef]
- Yang, X.Y.; Wang, Y.; Jiang, W.J.; Liu, X.L.; Zhang, X.M.; Yu, H.J.; Huang, S.W.; Liu, G.Q. Characterization and expression profiling of cucumber kinesin genes during early fruit development: Revealing the roles of kinesins in exponential cell production and enlargement in cucumber fruit. J. Exp. Bot. 2013, 64, 4541–4557. [Google Scholar] [CrossRef]
- Zhang, C.; Tanabe, K.; Tani, H.; Nakajima, H.; Mori, M.; Sakuno, E. Biologically active gibberellins and abscisic acid in fruit of two late-maturing Japanese pear cultivars with contrasting fruit size. J. Am. Soc. Hortic. Sci. 2007, 132, 452–458. [Google Scholar] [CrossRef]
- Dash, M.; Malladi, A. The AINTEGUMENTA genes, MdANT1 and MdANT2, are associated with the regulation of cell production during fruit growth in apple (Malus × domestica Borkh.). BMC Plant Biol. 2012, 12, 98. [Google Scholar] [CrossRef]
- Huang, K.; Wang, D.; Duan, P.; Zhang, B.; Xu, R.; Li, N.; Li, Y. WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 2017, 91, 849–860. [Google Scholar] [CrossRef]
- De Jong, M.; Mariani, C.; Vriezen, W.H. The role of auxin and gibberellin in tomato fruit set. J. Exp. Bot. 2009, 60, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Hendelman, A.; Buxdorf, K.; Stav, R.; Kravchik, M.; Arazi, T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol. Biol. 2012, 78, 561–576. [Google Scholar] [CrossRef]
- Ren, Z.; Liu, R.; Gu, W.; Dong, X. The Solanum lycopersicum auxin response factor SlARF2 participates in regulating lateral root formation and flower organ senescence. Plant Sci. 2017, 256, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Yang, R.; Wang, Y.; Ma, Y.; Lin, Y.; Li, Z.; Song, Y.; Feng, X.; Li, L. Candidate gene mining of GA-mediated regulation of pear fruit shape. Hortic. Environ. Biotechnol. 2024. [Google Scholar] [CrossRef]
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, G.; Liu, D.; Niu, M.; Tong, H.; Chu, C. GSK2 stabilizes OFP3 to suppress brassinosteroid responses in rice. Plant J. 2020, 102, 1187–1201. [Google Scholar] [CrossRef]
- Gallego-Bartolomé, J.; Minguet, E.G.; Grau-Enguix, F.; Abbas, M.; Locascio, A.; Thomas, S.G.; Alabadí, D.; Blázquez, M.A. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 13446–13451. [Google Scholar] [CrossRef]
- Stewart Lilley, J.L.; Gan, Y.; Graham, I.A.; Nemhauser, J.L. The effects of DELLAs on growth change with developmental stage and brassinosteroid levels. Plant J. 2013, 76, 165–173. [Google Scholar] [CrossRef]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q.; Chu, C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef]
- Li, Q.-F.; Wang, C.; Jiang, L.; Li, S.; Sun, S.S.; He, J.-X. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 2012, 5, ra72. [Google Scholar] [CrossRef]
- Liao, X.; Li, M.; Liu, B.; Yan, M.; Yu, X.; Zi, H.; Liu, R.; Yamamuro, C. Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. USA 2018, 115, E11542–E11550. [Google Scholar] [CrossRef]
- True, J.H.; Shaw, S.L. Exogenous auxin induces transverse microtubule arrays through TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX receptors. Plant Physiol. 2020, 182, 892–907. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, B.; Wang, S.; Lin, T.; Yang, L.; Zhao, Z.; Zhang, Z.; Huang, S.; Yang, X. Genome-wide target mapping shows histone deacetylase complex1 regulates cell proliferation in cucumber fruit. Plant Physiol. 2020, 182, 167–184. [Google Scholar] [CrossRef]
- Nardozza, S.; Cooney, J.; Boldingh, H.L.; Hewitt, K.G.; Trower, T.; Jones, D.; Thrimawithana, A.H.; Allan, A.C.; Richardson, A.C. Phytohormone and transcriptomic analysis reveals endogenous cytokinins affect kiwifruit growth under restricted carbon supply. Metabolites 2020, 10, 23. [Google Scholar] [CrossRef]
- Debi, B.R.; Taketa, S.; Ichii, M. Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa). J. Plant Physiol. 2005, 162, 507–515. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, Y.; Tu, Y.; Wang, Y.; Cheng, W.; Yang, Y. Overexpression of an EIN3-binding F-box protein2-like gene caused elongated fruit shape and delayed fruit development and ripening in tomato. Plant Sci. 2018, 272, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004, 7, 40–49. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Floková, K.; Feussner, K.; Herrfurth, C.; Miersch, O.; Mik, V.; Tarkowská, D.; Strnad, M.; Feussner, I.; Wasternack, C.; Novák, O. A previously undescribed jasmonate compound in flowering Arabidopsis thaliana–The identification of cis-(+)-OPDA-Ile. Phytochemistry 2016, 122, 230–237. [Google Scholar] [CrossRef]
- Zhang, Y.; Park, C.; Bennett, C.; Thornton, M.; Kim, D. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021, 31, 1290–1295. [Google Scholar] [CrossRef]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved Transcriptome Assembly Using a Hybrid of Long and Short Reads with StringTie. PLoS Comput. Biol. 2022, 18, e1009730. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Du, G.; Wang, L.; Li, H.; Sun, P.; Fu, J.; Suo, Y.; Han, W.; Diao, S.; Mai, Y.; Li, F. Selection and validation of reference genes for quantitative gene expression analyses in persimmon (Diospyros kaki Thunb.) using real-time quantitative PCR. Biol. Futur. 2019, 70, 261–267. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Suo, Y.; Li, H.; Sun, P.; Han, W.; Fu, J. Cytological, Phytohormone, and Transcriptome Analyses Provide Insights into Persimmon Fruit Shape Formation (Diospyros kaki Thunb.). Int. J. Mol. Sci. 2024, 25, 4812. https://doi.org/10.3390/ijms25094812
Li H, Suo Y, Li H, Sun P, Han W, Fu J. Cytological, Phytohormone, and Transcriptome Analyses Provide Insights into Persimmon Fruit Shape Formation (Diospyros kaki Thunb.). International Journal of Molecular Sciences. 2024; 25(9):4812. https://doi.org/10.3390/ijms25094812
Chicago/Turabian StyleLi, Huawei, Yujing Suo, Hui Li, Peng Sun, Weijuan Han, and Jianmin Fu. 2024. "Cytological, Phytohormone, and Transcriptome Analyses Provide Insights into Persimmon Fruit Shape Formation (Diospyros kaki Thunb.)" International Journal of Molecular Sciences 25, no. 9: 4812. https://doi.org/10.3390/ijms25094812
APA StyleLi, H., Suo, Y., Li, H., Sun, P., Han, W., & Fu, J. (2024). Cytological, Phytohormone, and Transcriptome Analyses Provide Insights into Persimmon Fruit Shape Formation (Diospyros kaki Thunb.). International Journal of Molecular Sciences, 25(9), 4812. https://doi.org/10.3390/ijms25094812





