OsCAMTA3 Negatively Regulates Disease Resistance to Magnaporthe oryzae by Associating with OsCAMTAPL in Rice
Abstract
1. Introduction
2. Results
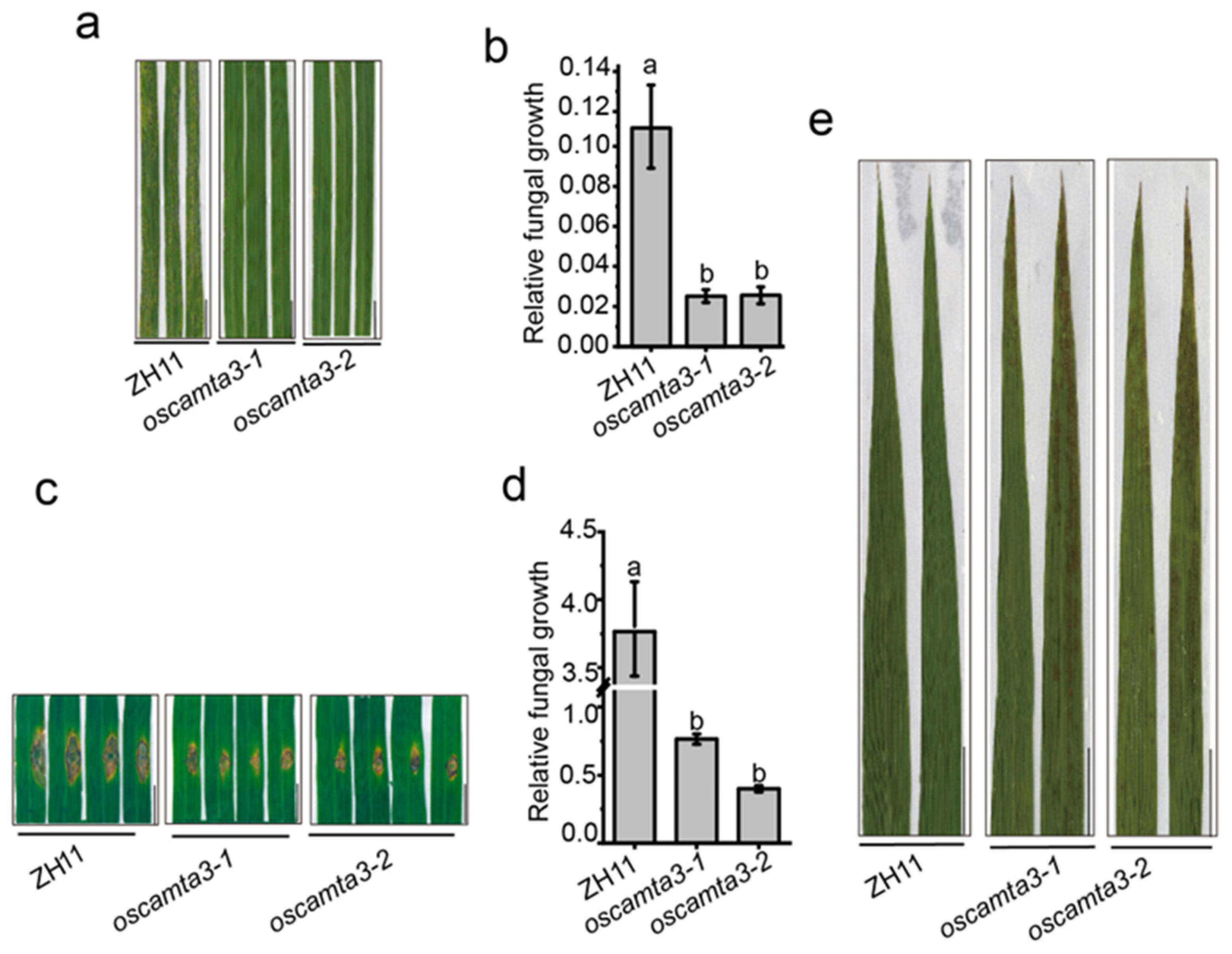
2.1. OsCAMTA3 Is a Negative Immune Regulator in Rice
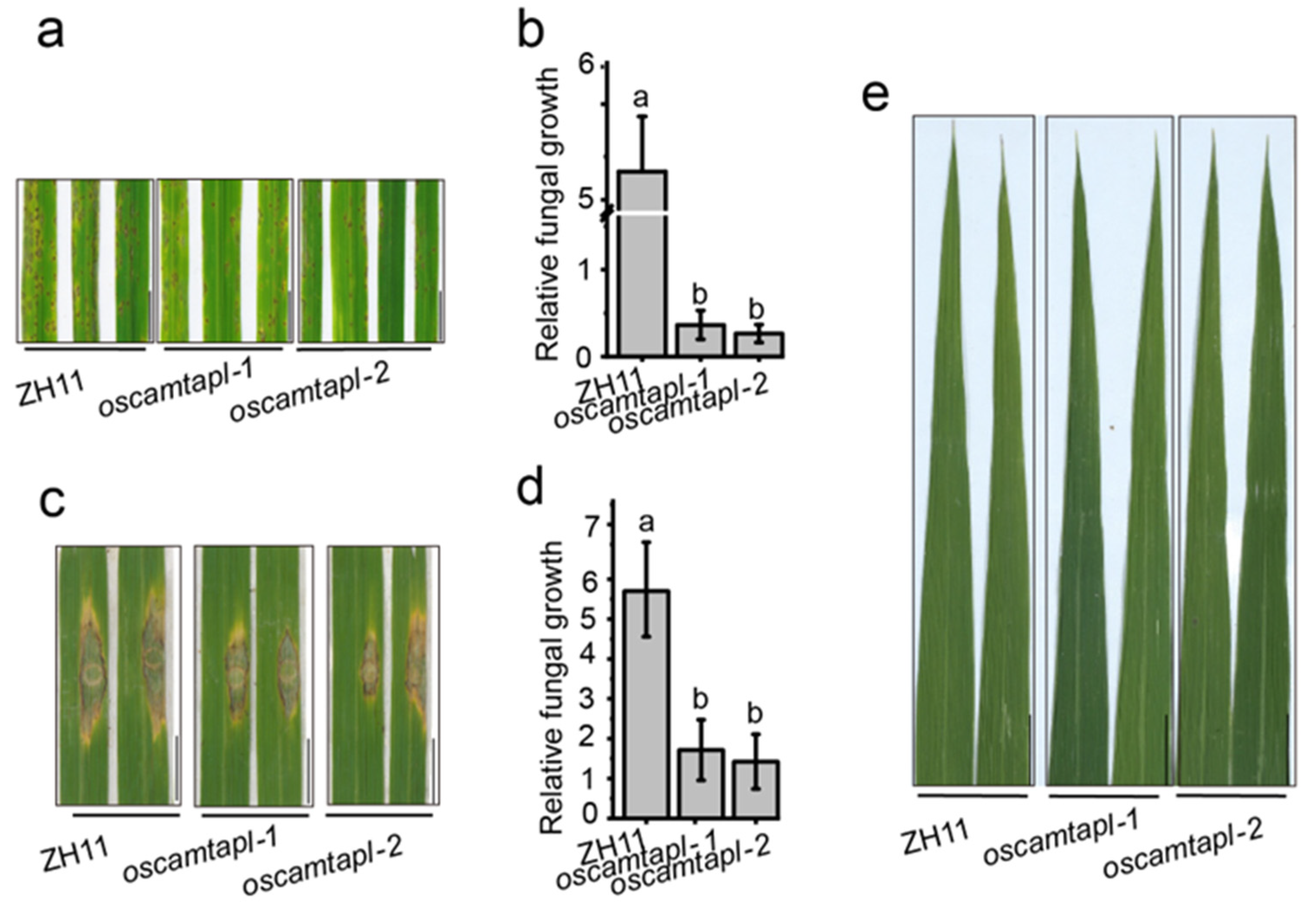
2.2. OsCAMTAPL Negatively Regulates Disease Resistance in Rice
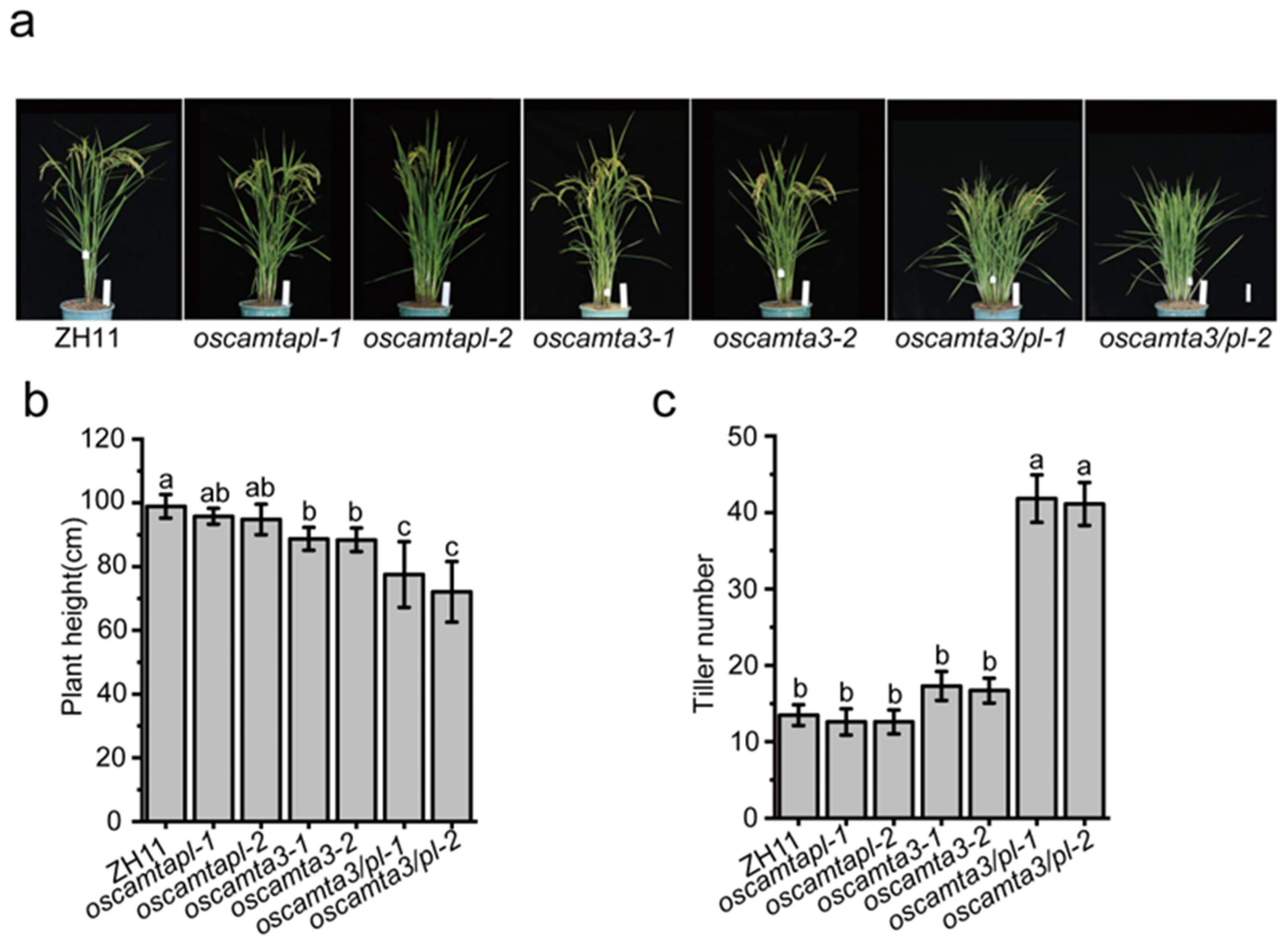
2.3. OsCAMTA3 and OsCAMTAPL Function Together to Negatively Regulate Disease Resistance in Rice
2.4. OsCAMTA3 Associates with OsCAMTAPL
2.5. OsCAMTA3 and OsCAMTAPL Contribute to Plant Architecture
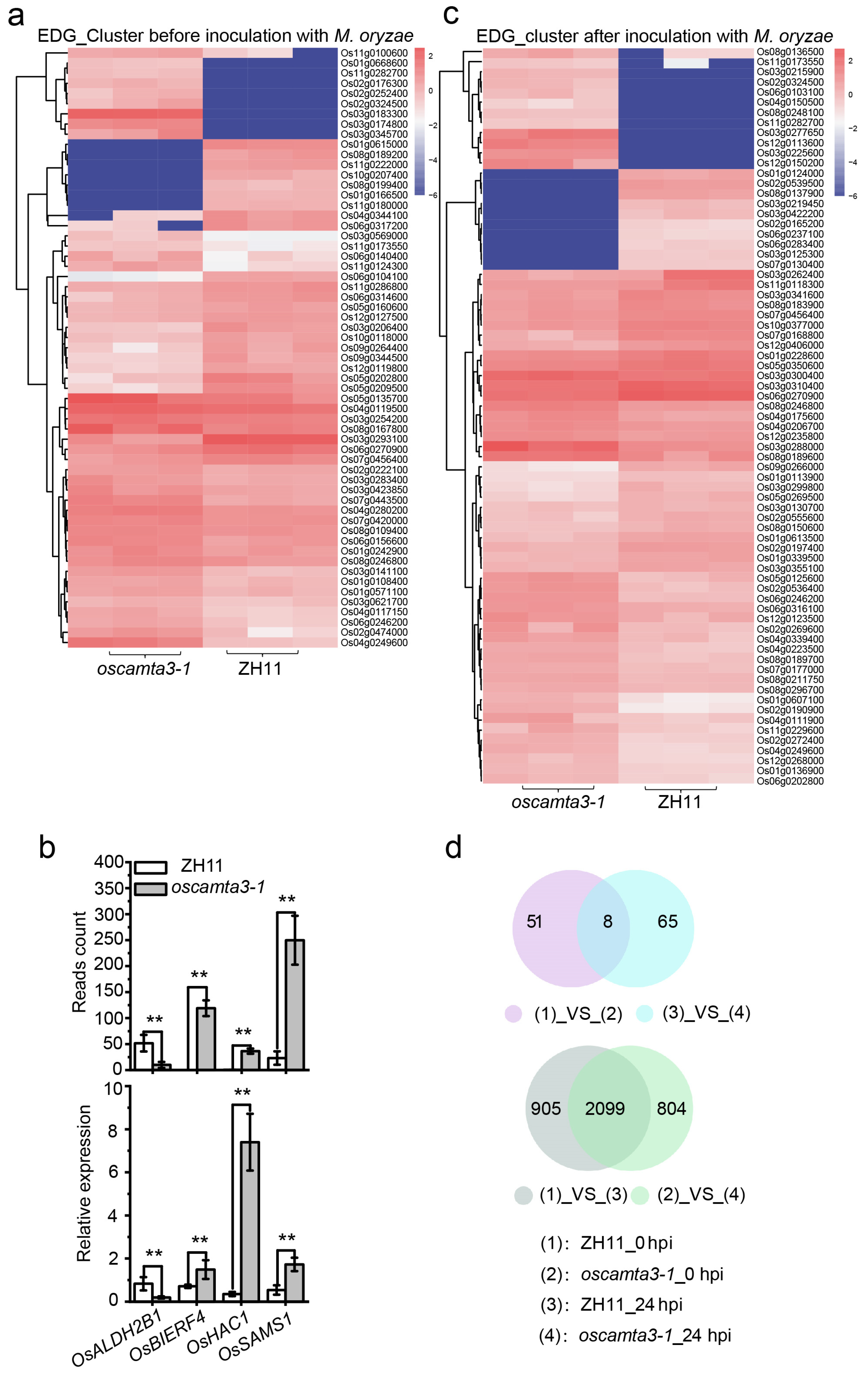
2.6. Many Genes Are Differentially Expressed between ZH11 and oscamta3-1
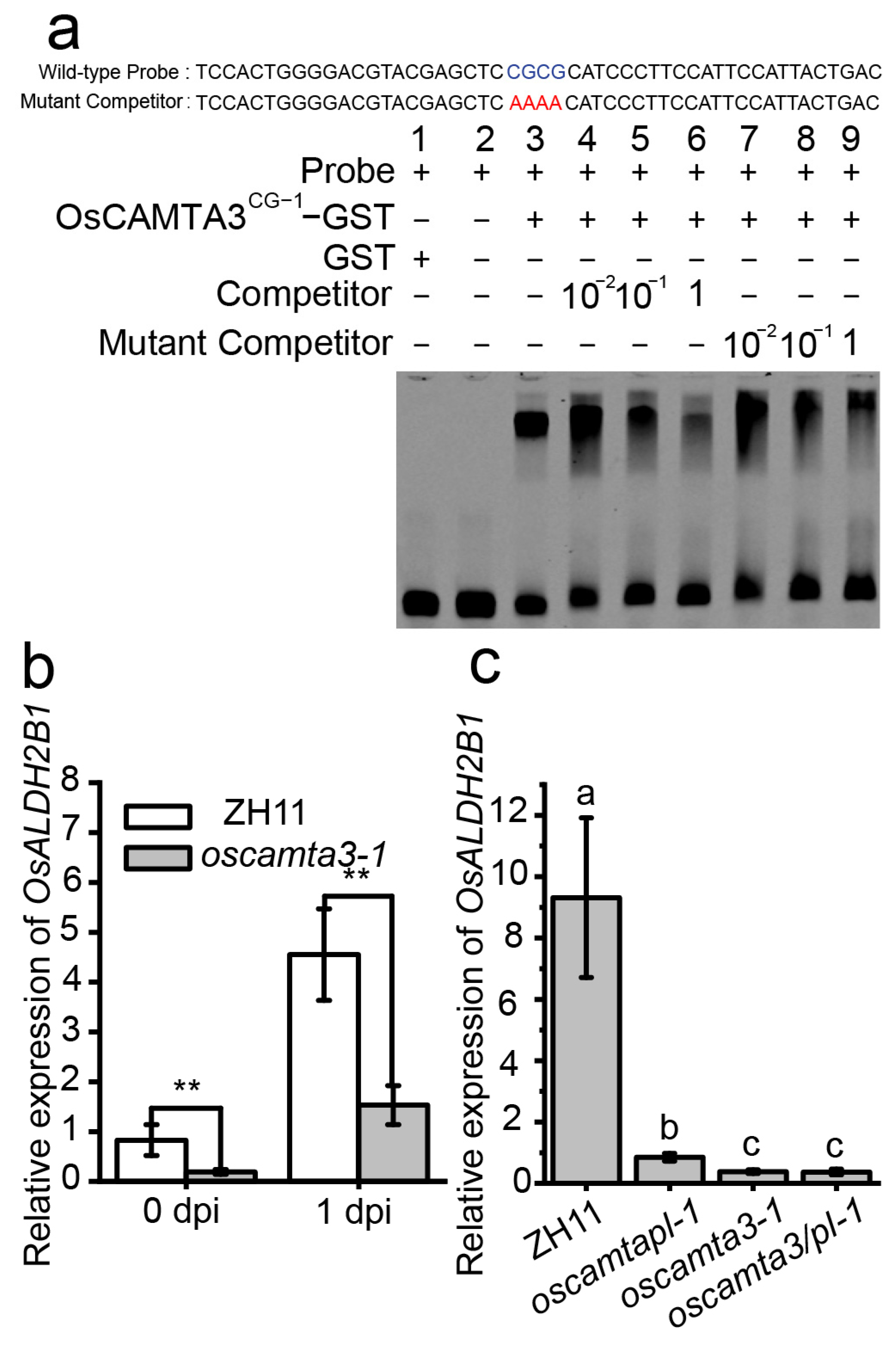
2.7. OsCAMTA3 Binds to the Promoter of OsALDH2B1 to Regulate Its Expression
3. Discussion
3.1. OsCAMTA3 and OsCAMTAPL Negatively Regulate Disease Resistance in Rice
3.2. Trade-Off Regulation of Rice Growth and Development by OsCAMTA3
3.3. Regulation of CAMTAs in Plants
3.4. The Potential Application of OsCAMTA3 and OsCAMTAPL in Breeding
4. Materials and Methods
4.1. Plant Materials and Growth Conditions [67]
4.2. Inoculation of Blast Fungus and Fungal Biomass Assays [67]
4.3. qRT-PCR Analysis [68]
4.4. Y2H Assay [69]
4.5. Firefly Split-LUC Assay [69]
4.6. Co-IP Assay [70]
4.7. RNA-Sequencing Analysis [71]
4.8. Recombinant Protein Expression [64]
4.9. Electrophoretic Mobility Shift Assay (EMSA) [62]
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ngou, B.P.M.; Jones, J.D.G.; Ding, P. Plant immune networks. Trends Plant Sci. 2022, 27, 255–273. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for nlr-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef]
- Wang, W.; Feng, B.; Zhou, J.M.; Tang, D. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Chen, K.; Xiao, Z.; Liang, X.; Lu, D. Phosphorylation status of cpk28 affects its ubiquitination and protein stability. New Phytol. 2023, 237, 1270–1284. [Google Scholar] [CrossRef]
- van Loon, L.C. The intelligent behavior of plants. Trends Plant Sci. 2016, 21, 286–294. [Google Scholar] [CrossRef] [PubMed]
- O’Day, D.H.; Huber, R.J. Calmodulin binding proteins and neuroinflammation in multiple neurodegenerative diseases. BMC Neurosci. 2022, 23, 10. [Google Scholar] [CrossRef]
- Wanford, J.J.; Odendall, C. Ca(2+)-calmodulin signalling at the host-pathogen interface. Curr. Opin. Microbiol. 2023, 72, 102267. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, C.; Robison, A.J. Calmodulin acetylation: A modification to remember. J. Biol. Chem. 2021, 297, 101273. [Google Scholar] [CrossRef] [PubMed]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/nadph oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Hake, K.; Wang, W.; Zhao, T.; Romeis, T.; Tang, D. Calcium-dependent protein kinase5 associates with the truncated nlr protein tir-nbs2 to contribute to exo70b1-mediated immunity. Plant Cell 2017, 29, 746–759. [Google Scholar] [CrossRef]
- Zhu, X.; Dunand, C.; Snedden, W.; Galaud, J.P. Cam and cml emergence in the green lineage. Trends Plant Sci 2015, 20, 483–489. [Google Scholar] [CrossRef]
- McCormack, E.; Tsai, Y.C.; Braam, J. Handling calcium signaling: Arabidopsis cams and cmls. Trends Plant Sci 2005, 10, 383–389. [Google Scholar] [CrossRef]
- Batistic, O.; Kudla, J. Plant calcineurin b-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 2009, 1793, 985–992. [Google Scholar] [CrossRef]
- Huang, W.; Wu, Z.; Tian, H.; Li, X.; Zhang, Y. Arabidopsis calmodulin-binding protein 60b plays dual roles in plant immunity. Plant Commun. 2021, 2, 100213. [Google Scholar] [CrossRef]
- Li, L.S.; Ying, J.; Li, E.; Ma, T.; Li, M.; Gong, L.M.; Wei, G.; Zhang, Y.; Li, S. Arabidopsis cbp60b is a central transcriptional activator of immunity. Plant Physiol. 2021, 186, 1645–1659. [Google Scholar] [CrossRef] [PubMed]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Jauregui, E.; Du, L.; Tanaka, K.; Poovaiah, B.W. Calcium signatures and signaling events orchestrate plant-microbe interactions. Curr. Opin. Plant Biol. 2017, 38, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.N.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.; Kim, Y.; Gilmour, S.J.; Thomashow, M.F. Temperature modulation of camta3 gene induction activity is mediated through the DNA binding domain. Plant J. Cell Mol. Biol. 2022, 112, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, S.; Gilmour, S.J.; Thomashow, M.F. Roles of camta transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of arabidopsis. Plant J. Cell Mol. Biol. 2013, 75, 364–376. [Google Scholar] [CrossRef]
- Bouché, N.; Scharlat, A.; Snedden, W.; Bouchez, D.; Fromm, H. A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 2002, 277, 21851–21861. [Google Scholar] [CrossRef] [PubMed]
- Büyük, İ.; İlhan, E.; Şener, D.; Özsoy, A.U.; Aras, S. Genome-wide identification of camta gene family members in phaseolus vulgaris l. And their expression profiling during salt stress. Mol. Biol. Rep. 2019, 46, 2721–2732. [Google Scholar] [CrossRef]
- Pandey, N.; Ranjan, A.; Pant, P.; Tripathi, R.K.; Ateek, F.; Pandey, H.P.; Patre, U.V.; Sawant, S.V. Camta 1 regulates drought responses in arabidopsis thaliana. BMC Genom. 2013, 14, 216. [Google Scholar] [CrossRef]
- Kidokoro, S.; Yoneda, K.; Takasaki, H.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 2017, 29, 760–774. [Google Scholar] [CrossRef]
- Nie, H.; Zhao, C.; Wu, G.; Wu, Y.; Chen, Y.; Tang, D. Sr1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating ndr1 and ein3. Plant Physiol. 2012, 158, 1847–1859. [Google Scholar] [CrossRef]
- Kim, Y.S.; An, C.; Park, S.; Gilmour, S.J.; Wang, L.; Renna, L.; Brandizzi, F.; Grumet, R.; Thomashow, M.F. Camta-mediated regulation of salicylic acid immunity pathway genes in arabidopsis exposed to low temperature and pathogen infection. Plant Cell 2017, 29, 2465–2477. [Google Scholar] [CrossRef]
- Liu, N.; Xu, Y.; Li, Q.; Cao, Y.; Yang, D.; Liu, S.; Wang, X.; Mi, Y.; Liu, Y.; Ding, C.; et al. A lncrna fine-tunes salicylic acid biosynthesis to balance plant immunity and growth. Cell Host Microbe 2022, 30, 1124–1138.e8. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Castroverde, C.D.M.; Huang, S.; Li, C.; Hilleary, R.; Seroka, A.; Sohrabi, R.; Medina-Yerena, D.; Huot, B.; Wang, J.; et al. Increasing the resilience of plant immunity to a warming climate. Nature 2022, 607, 339–344. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Locci, F.; Wanke, F.; Zhang, L.; Saile, S.C.; Joe, A.; Karelina, D.; Hua, C.; Fröhlich, K.; Wan, W.L.; et al. The eds1-pad4-adr1 node mediates arabidopsis pattern-triggered immunity. Nature 2021, 598, 495–499. [Google Scholar] [CrossRef]
- Sun, T.; Huang, J.; Xu, Y.; Verma, V.; Jing, B.; Sun, Y.; Ruiz Orduna, A.; Tian, H.; Huang, X.; Xia, S.; et al. Redundant camta transcription factors negatively regulate the biosynthesis of salicylic acid and n-hydroxypipecolic acid by modulating the expression of sard1 and cbp60g. Mol. Plant 2020, 13, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Gilmour, S.J.; Chao, L.; Park, S.; Thomashow, M.F. Arabidopsis camta transcription factors regulate pipecolic acid biosynthesis and priming of immunity genes. Mol. Plant 2020, 13, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Jing, B.; Xu, S.; Xu, M.; Li, Y.; Li, S.; Ding, J.; Zhang, Y. Brush and spray: A high-throughput systemic acquired resistance assay suitable for large-scale genetic screening. Plant Physiol 2011, 157, 973–980. [Google Scholar] [CrossRef]
- Kumar, S.; Zavaliev, R.; Wu, Q.; Zhou, Y.; Cheng, J.; Dillard, L.; Powers, J.; Withers, J.; Zhao, J.; Guan, Z.; et al. Structural basis of npr1 in activating plant immunity. Nature 2022, 605, 561–566. [Google Scholar] [CrossRef]
- Backer, R.; Naidoo, S.; van den Berg, N. The nonexpressor of pathogenesis-related genes 1 (npr1) and related family: Mechanistic insights in plant disease resistance. Front. Plant Sci. 2019, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Tanaka, K.; Poovaiah, B.W. Calmodulin-binding transcription activator atsr1/camta3 fine-tunes plant immune response by transcriptional regulation of the salicylate receptor npr1. Plant Cell Environ. 2021, 44, 3140–3154. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Poovaiah, B.W. Interplay between Ca2+/calmodulin-mediated signaling and atsr1/camta3 during increased temperature resulting in compromised immune response in plants. Int. J. Mol. Sci. 2022, 23, 2175. [Google Scholar] [CrossRef] [PubMed]
- Lolle, S.; Greeff, C.; Petersen, K.; Roux, M.; Jensen, M.K.; Bressendorff, S.; Rodriguez, E.; Sømark, K.; Mundy, J.; Petersen, M. Matching nlr immune receptors to autoimmunity in camta3 mutants using antimorphic nlr alleles. Cell Host Microbe 2017, 21, 518–529.e4. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microrna. eLife 2015, 4, e05733. [Google Scholar] [CrossRef] [PubMed]
- Eseola, A.B.; Ryder, L.S.; Osés-Ruiz, M.; Findlay, K.; Yan, X.; Cruz-Mireles, N.; Molinari, C.; Garduño-Rosales, M.; Talbot, N.J. Investigating the cell and developmental biology of plant infection by the rice blast fungus magnaporthe oryzae. Fungal Genet. Biol. 2021, 154, 103562. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Orth, K. Rise of a cereal killer: The biology of magnaporthe oryzae biotrophic growth. Trends Microbiol. 2018, 26, 582–597. [Google Scholar] [CrossRef]
- Sha, G.; Sun, P.; Kong, X.; Han, X.; Sun, Q.; Fouillen, L.; Zhao, J.; Li, Y.; Yang, L.; Wang, Y.; et al. Genome editing of a rice cdp-dag synthase confers multipathogen resistance. Nature 2023, 618, 1017–1023. [Google Scholar] [CrossRef]
- Xiao, G.; Wang, W.; Liu, M.; Li, Y.; Liu, J.; Franceschetti, M.; Yi, Z.; Zhu, X.; Zhang, Z.; Lu, G.; et al. The piks allele of the nlr immune receptor pik breaks the recognition of avrpik effectors of rice blast fungus. J. Integr. Plant Biol. 2022, 65, 810–824. [Google Scholar] [CrossRef]
- Park, C.H.; Shirsekar, G.; Bellizzi, M.; Chen, S.; Songkumarn, P.; Xie, X.; Shi, X.; Ning, Y.; Zhou, B.; Suttiviriya, P.; et al. The e3 ligase apip10 connects the effector avrpiz-t to the nlr receptor piz-t in rice. PLoS Pathog. 2016, 12, e1005529. [Google Scholar] [CrossRef]
- Li, J.; Wang, Q.; Li, C.; Bi, Y.; Fu, X.; Wang, R. Novel haplotypes and networks of avr-pik alleles in magnaporthe oryzae. BMC Plant Biol. 2019, 19, 204. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair rga4/rga5 recognizes the magnaporthe oryzae effectors avr-pia and avr1-co39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.S.; Koo, S.C.; Jin, B.J.; Baek, D.; Kim, M.C. Rice cam-binding transcription factor (oscbt) mediates defense signaling via transcriptional reprogramming. Plant Biotechnol. Rep. 2020, 14, 309–321. [Google Scholar] [CrossRef]
- Choi, M.S.; Kim, M.C.; Yoo, J.H.; Moon, B.C.; Koo, S.C.; Park, B.O.; Lee, J.H.; Koo, Y.D.; Han, H.J.; Lee, S.Y.; et al. Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 2005, 280, 40820–40831. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Yuan, M.; Liu, H.; Hui, S.; Wang, S. The versatile functions of osaldh2b1 provide a genic basis for growth–defense trade-offs in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 201918994. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Song, F.; Goodman, R.M.; Zheng, Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J. Plant Physiol. 2006, 163, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Wang, T.; Chen, Z.; Tang, Z.; Wu, Z.; Salt, D.E.; Chao, D.Y.; Zhao, F.J. Oshac1;1 and oshac1;2 function as arsenate reductases and regulate arsenic accumulation. Plant Physiol. 2016, 172, 1708–1719. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, Y.; Luo, W.; Li, W.; Chen, N.; Zhang, D.; Chong, K. The f-box protein osfbk12 targets ossams1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 2013, 163, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef]
- Yuan, P.; Yang, S.; Feng, L.; Chu, J.; Dong, H.; Sun, J.; Chen, H.; Li, Z.; Yamamoto, N.; Zheng, A.; et al. Red-light receptor phytochrome b inhibits bzr1-nac028-cad8b signaling to negatively regulate rice resistance to sheath blight. Plant Cell Environ. 2023, 46, 1249–1263. [Google Scholar] [CrossRef]
- Unver, T.; Turktas, M.; Budak, H. In planta evidence for the involvement of a ubiquitin conjugating enzyme (UBC E2 clade) in negative regulation of disease resistance. Plant Mol. Biol. Rep. 2013, 31, 323–334. [Google Scholar] [CrossRef]
- Kan, Y.; Mu, X.R.; Zhang, H.; Gao, J.; Shan, J.X.; Ye, W.W.; Lin, H.X. Tt2 controls rice thermotolerance through sct1-dependent alteration of wax biosynthesis. Nat. Plants 2022, 8, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, X.; He, Y.; Sang, T.; Wang, P.; Dai, S.; Zhang, S.; Meng, X. Differential phosphorylation of the transcription factor wrky33 by the protein kinases cpk5/cpk6 and mpk3/mpk6 cooperatively regulates camalexin biosynthesis in arabidopsis. Plant Cell 2020, 32, 2621–2638. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Jiang, X.; Zhong, G.; Wang, W.; Hake, K.; Matschi, S.; Lederer, S.; Hoehenwarter, W.; Sun, Q.; Lee, J.; et al. Camta3 repressor destabilization triggers tir-domain protein tn2-mediated autoimmunity in the arabidopsis exo70b1 mutant. Plant Cell 2024, 36, 2021–2040. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhao, C.; Zhao, M.; Li, Y.; Wen, G. Phylogeny and evolution of calcineurin b-like (cbl) gene family in grass and functional analyses of rice cbls. J. Plant Biol. 2020, 63, 117–130. [Google Scholar] [CrossRef]
- Gao, C.; Lu, S.; Zhou, R.; Wang, Z.; Li, Y.; Fang, H.; Wang, B.; Chen, M.; Cao, Y. The oscbl8-oscipk17 module regulates seedling growth and confers resistance to heat and drought in rice. Int. J. Mol. Sci. 2022, 23, 12451. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Diao, Z.; Yang, D.; Wang, X.; Zheng, X.; Xiang, X.; Xiao, Y.; Chen, Z.; Wang, W.; Wu, Y.; et al. The 14-3-3 protein gf14c positively regulates immunity by modulating the protein homoeostasis of the gras protein osscl7 in rice. Plant Cell Environ. 2022, 45, 1065–1081. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, G.; Shi, H.; Tang, D. Receptor-like kinase 902 associates with and phosphorylates brassinosteroid-signaling kinase1 to regulate plant immunity. Mol. Plant 2019, 12, 59–70. [Google Scholar] [CrossRef]
- Chen, R.; Sun, P.; Zhong, G.; Wang, W.; Tang, D. The receptor-like protein53 immune complex associates with llg1 to positively regulate plant immunity. J. Integr. Plant Biol. 2022, 64, 1833–1846. [Google Scholar] [CrossRef]
- Gao, C.; Sun, P.; Wang, W.; Tang, D. Arabidopsis e3 ligase keg associates with and ubiquitinates mkk4 and mkk5 to regulate plant immunity. J. Integr. Plant Biol. 2021, 63, 327–339. [Google Scholar] [CrossRef]
- Niu, Y.; Huang, X.; He, Z.; Zhang, Q.; Meng, H.; Shi, H.; Feng, B.; Zhou, Y.; Zhang, J.; Lu, G.; et al. Phosphorylation of ostga5 by casein kinase ii compromises its suppression of defense-related gene transcription in rice. Plant Cell 2022, 34, 3425–3442. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Li, S.; Wang, W.; Tang, D. OsCAMTA3 Negatively Regulates Disease Resistance to Magnaporthe oryzae by Associating with OsCAMTAPL in Rice. Int. J. Mol. Sci. 2024, 25, 5049. https://doi.org/10.3390/ijms25095049
Yu S, Li S, Wang W, Tang D. OsCAMTA3 Negatively Regulates Disease Resistance to Magnaporthe oryzae by Associating with OsCAMTAPL in Rice. International Journal of Molecular Sciences. 2024; 25(9):5049. https://doi.org/10.3390/ijms25095049
Chicago/Turabian StyleYu, Shibo, Shengping Li, Wei Wang, and Dingzhong Tang. 2024. "OsCAMTA3 Negatively Regulates Disease Resistance to Magnaporthe oryzae by Associating with OsCAMTAPL in Rice" International Journal of Molecular Sciences 25, no. 9: 5049. https://doi.org/10.3390/ijms25095049
APA StyleYu, S., Li, S., Wang, W., & Tang, D. (2024). OsCAMTA3 Negatively Regulates Disease Resistance to Magnaporthe oryzae by Associating with OsCAMTAPL in Rice. International Journal of Molecular Sciences, 25(9), 5049. https://doi.org/10.3390/ijms25095049





