C. elegans Cytoplasmic Isocitrate Dehydrogenase Neomorphic G98N and R133H Mutants Produce the Oncometabolite 2-Hydroxyglutarate
Abstract
1. Introduction
2. Results
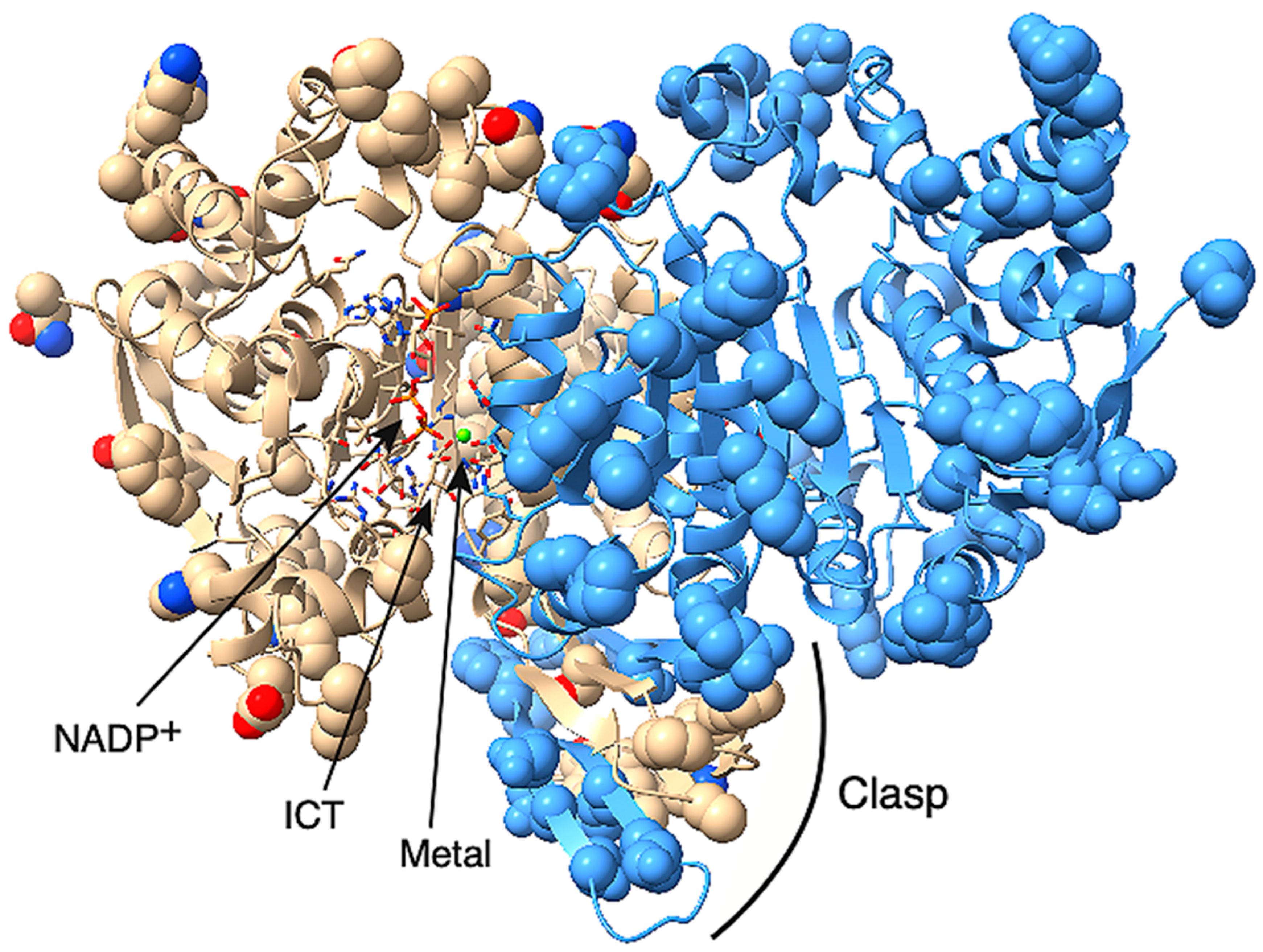
2.1. IDH-1 Structure Prediction
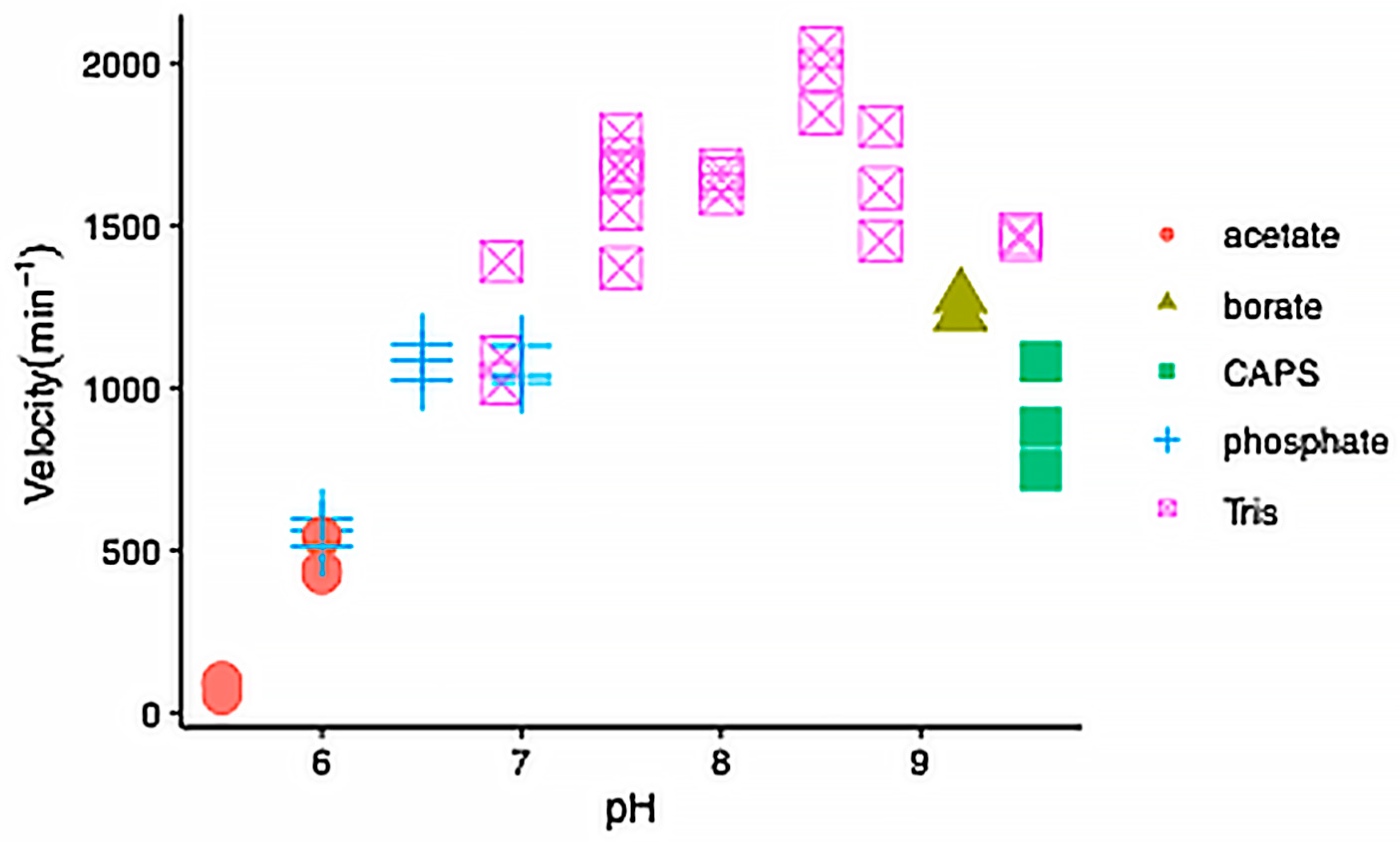
2.2. Enzyme Kinetics
2.3. Analysis of G98N and R133H
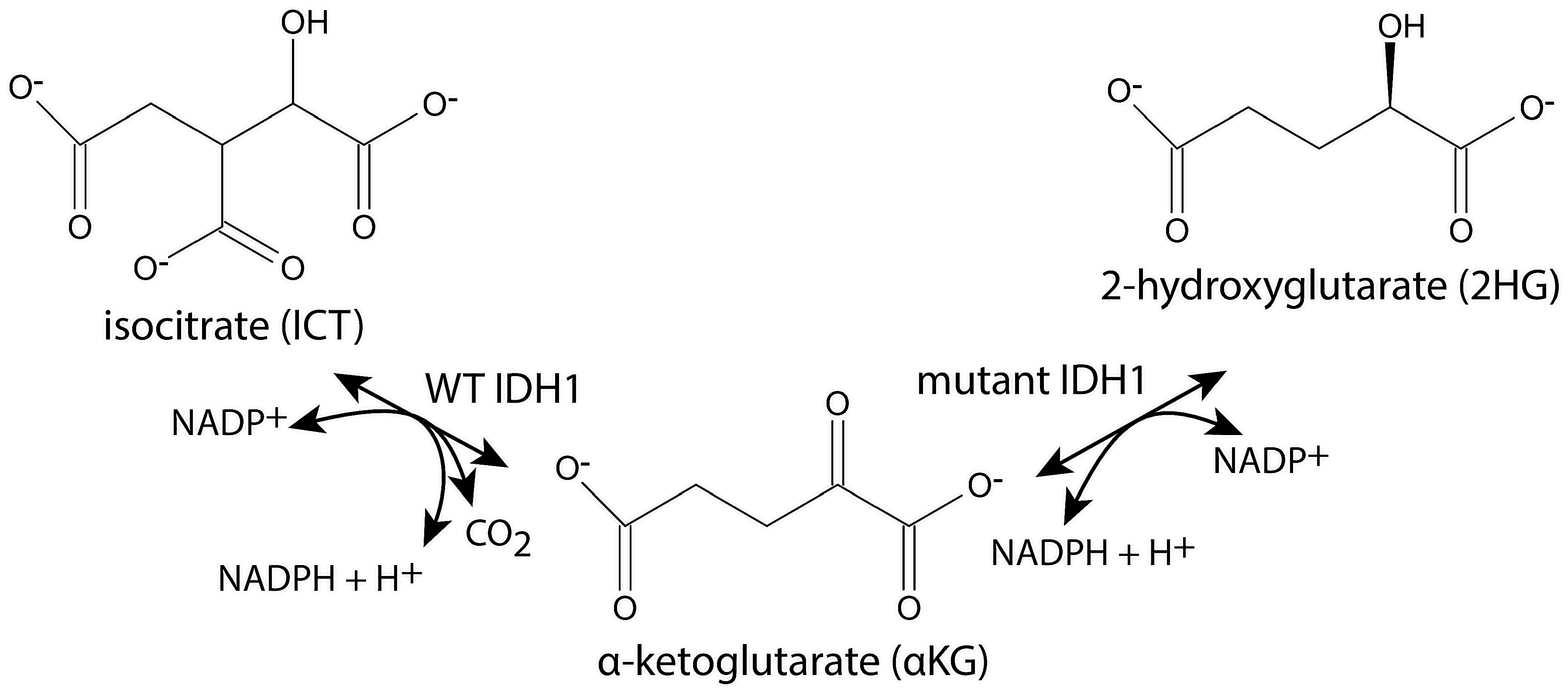
2.4. 2-Hydroxyglutarate Production
3. Discussion
3.1. Enzyme Kinetics Results
3.2. Implications for In Vivo Experiments
3.3. Metal Binding
3.4. Potential for C. elegans as a Cancer Model
4. Materials and Methods
4.1. Sequence Analysis
4.2. Plasmid Construction
4.3. Recombinant Enzyme Expression and Purification
4.4. Steady-State Enzyme Kinetics Measurements
4.5. Liquid Chromatography—Mass Spectrometry (LC-MS) Procedure
4.6. Protein Molecular Modeling and Structure Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| αKG | α-ketoglutarate |
| CBD | chitin binding domain |
| G97N | mutation in IDH1 |
| G98N | mutation in IDH-1 |
| ICT | isocitrate |
| IDH1 | human cytoplasmic isocitrate dehydrogenase |
| IDH-1 | C. elegans cytoplasmic isocitrate dehydrogenase |
| LC-MS | liquid chromatography—mass spectrometry |
| 6 mA | methylation at N6 of adenosine |
| NAD+ | oxidized nicotinamide adenine dinucleotide |
| NADP+ | oxidized nicotinamide adenine dinucleotide phosphate |
| NADPH | reduced nicotinamide adenine dinucleotide phosphate |
| R132H | mutation in IDH1 |
| R133H | mutation in IDH-1 |
| TET | ten-eleven translocation (TET) oxygenase |
| TCA | tricarboxylic acid |
References
- Badur, M.G.; Muthusamy, T.; Parker, S.J.; Ma, S.; McBrayer, S.K.; Cordes, T.; Magana, J.H.; Guan, K.-L.; Metallo, C.M. Oncogenic R132 IDH1 Mutations Limit NADPH for De Novo Lipogenesis through (D)2-Hydroxyglutarate Production in Fibrosarcoma Cells. Cell Rep. 2018, 25, 1018–1026.e4. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, L.; Liu, L.; Su, X.; Rabinowitz, J.D. Chemical Basis for Deuterium Labeling of Fat and NADPH. J. Am. Chem. Soc. 2017, 139, 14368–14371. [Google Scholar] [CrossRef]
- Koh, H.-J.; Lee, S.-M.; Son, B.-G.; Lee, S.-H.; Ryoo, Z.Y.; Chang, K.-T.; Park, J.-W.; Park, D.-C.; Song, B.J.; Veech, R.L.; et al. Cytosolic NADP+-Dependent Isocitrate Dehydrogenase Plays a Key Role in Lipid Metabolism. J. Biol. Chem. 2004, 279, 39968–39974. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine Reliance in Cell Metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef] [PubMed]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive Glutamine Metabolism by IDH1 Mediates Lipogenesis under Hypoxia. Nature 2011, 481, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Sjöblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Bleeker, F.E.; Lamba, S.; Leenstra, S.; Troost, D.; Hulsebos, T.; Vandertop, W.P.; Frattini, M.; Molinari, F.; Knowles, M.; Cerrato, A.; et al. IDH1 Mutations at Residue p.R132 (IDH1) Occur Frequently in High-Grade Gliomas but Not in Other Solid Tumors. Hum. Mutat. 2009, 30, 7–11. [Google Scholar] [CrossRef]
- Huang, J.; Tseng, L.-H.; Parini, V.; Lokhandwala, P.M.; Pallavajjala, A.; Rodriguez, E.; Xian, R.; Chen, L.; Gocke, C.D.; Eshleman, J.R.; et al. IDH1 and IDH2 Mutations in Colorectal Cancers. Am. J. Clin. Pathol. 2021, 156, 777–786. [Google Scholar] [CrossRef]
- Hvinden, I.C.; Cadoux-Hudson, T.; Schofield, C.J.; McCullagh, J.S.O. Metabolic Adaptations in Cancers Expressing Isocitrate Dehydrogenase Mutations. Cell Rep. Med. 2021, 2, 100469. [Google Scholar] [CrossRef]
- Kang, M.R.; Kim, M.S.; Oh, J.E.; Kim, Y.R.; Song, S.Y.; Seo, S.I.; Lee, J.Y.; Yoo, N.J.; Lee, S.H. Mutational Analysis of IDH1 Codon 132 in Glioblastomas and Other Common Cancers. Int. J. Cancer 2009, 125, 353–355. [Google Scholar] [CrossRef]
- Amary, M.F.; Bacsi, K.; Maggiani, F.; Damato, S.; Halai, D.; Berisha, F.; Pollock, R.; O’Donnell, P.; Grigoriadis, A.; Diss, T.; et al. IDH1 and IDH2 Mutations Are Frequent Events in Central Chondrosarcoma and Central and Periosteal Chondromas but Not in Other Mesenchymal Tumours. J. Pathol. 2011, 224, 334–343. [Google Scholar] [CrossRef]
- Cairns, R.A.; Iqbal, J.; Lemonnier, F.; Kucuk, C.; de Leval, L.; Jais, J.-P.; Parrens, M.; Martin, A.; Xerri, L.; Brousset, P.; et al. IDH2 Mutations Are Frequent in Angioimmunoblastic T-Cell Lymphoma. Blood 2012, 119, 1901–1903. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Maharry, K.; Wu, Y.-Z.; Radmacher, M.D.; Mrózek, K.; Margeson, D.; Holland, K.B.; Whitman, S.P.; Becker, H.; Schwind, S.; et al. IDH1 and IDH2 Gene Mutations Identify Novel Molecular Subsets within de Novo Cytogenetically Normal Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2010, 28, 2348–2355. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-Associated IDH1 Mutations Produce 2-Hydroxyglutarate. Nature 2009, 462, 739. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Cairns, R.A.; Minden, M.D.; Driggers, E.M.; Bittinger, M.A.; Jang, H.G.; Sasaki, M.; Jin, S.; Schenkein, D.P.; Su, S.M.; et al. Cancer-Associated Metabolite 2-Hydroxyglutarate Accumulates in Acute Myelogenous Leukemia with Isocitrate Dehydrogenase 1 and 2 Mutations. J. Exp. Med. 2010, 207, 339–344. [Google Scholar] [CrossRef]
- Gelman, S.J.; Naser, F.; Mahieu, N.G.; McKenzie, L.D.; Dunn, G.P.; Chheda, M.G.; Patti, G.J. Consumption of NADPH for 2-HG Synthesis Increases Pentose Phosphate Pathway Flux and Sensitizes Cells to Oxidative Stress. Cell Rep. 2018, 22, 512–522. [Google Scholar] [CrossRef]
- Shi, J.; Sun, B.; Shi, W.; Zuo, H.; Cui, D.; Ni, L.; Chen, J. Decreasing GSH and Increasing ROS in Chemosensitivity Gliomas with IDH1 Mutation. Tumour Biol. 2015, 36, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cadoux-Hudson, T.; Schofield, C.J. Isocitrate Dehydrogenase Variants in Cancer-Cellular Consequences and Therapeutic Opportunities. Curr. Opin. Chem. Biol. 2020, 57, 122–134. [Google Scholar] [CrossRef]
- Chou, F.-J.; Liu, Y.; Lang, F.; Yang, C. D-2-Hydroxyglutarate in Glioma Biology. Cells 2021, 10, 2345. [Google Scholar] [CrossRef]
- Gunn, K.; Losman, J.-A. Isocitrate Dehydrogenase Mutations in Cancer: Mechanisms of Transformation and Metabolic Liability. Cold Spring Harb. Perspect. Med. 2024, 14, a041537. [Google Scholar] [CrossRef]
- Hao, J.; Huang, Z.; Zhang, S.; Song, K.; Wang, J.; Gao, C.; Fang, Z.; Zhang, N. Deciphering the Multifaceted Roles and Clinical Implications of 2-Hydroxyglutarate in Cancer. Pharmacol. Res. 2024, 209, 107437. [Google Scholar] [CrossRef] [PubMed]
- Grassian, A.R.; Parker, S.J.; Davidson, S.M.; Divakaruni, A.S.; Green, C.R.; Zhang, X.; Slocum, K.L.; Pu, M.; Lin, F.; Vickers, C.; et al. IDH1 Mutations Alter Citric Acid Cycle Metabolism and Increase Dependence on Oxidative Mitochondrial Metabolism. Cancer Res. 2014, 74, 3317–3331. [Google Scholar] [CrossRef]
- Salamanca-Cardona, L.; Shah, H.; Poot, A.J.; Correa, F.M.; Gialleonardo, V.D.; Lui, H.; Miloushev, V.Z.; Granlund, K.L.; Tee, S.S.; Cross, J.R.; et al. In Vivo Imaging of Glutamine Metabolism to the Oncometabolite 2-Hydroxyglutarate in IDH1/2 Mutant Tumors. Cell Metab. 2017, 26, 830–841.e3. [Google Scholar] [CrossRef]
- Seltzer, M.J.; Bennett, B.D.; Joshi, A.D.; Gao, P.; Thomas, A.G.; Ferraris, D.V.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Rabinowitz, J.D.; et al. Inhibition of Glutaminase Preferentially Slows Growth of Glioma Cells with Mutant IDH1. Cancer Res. 2010, 70, 8981–8987. [Google Scholar] [CrossRef]
- Ohka, F.; Ito, M.; Ranjit, M.; Senga, T.; Motomura, A.; Motomura, K.; Saito, K.; Kato, K.; Kato, Y.; Wakabayashi, T.; et al. Quantitative Metabolome Analysis Profiles Activation of Glutaminolysis in Glioma with IDH1 Mutation. Tumour Biol. 2014, 35, 5911–5920. [Google Scholar] [CrossRef] [PubMed]
- Berger, R.S.; Ellmann, L.; Reinders, J.; Kreutz, M.; Stempfl, T.; Oefner, P.J.; Dettmer, K. Degradation of D-2-Hydroxyglutarate in the Presence of Isocitrate Dehydrogenase Mutations. Sci. Rep. 2019, 9, 7436. [Google Scholar] [CrossRef] [PubMed]
- Vinekar, R.; Verma, C.; Ghosh, I. Functional Relevance of Dynamic Properties of Dimeric NADP-Dependent Isocitrate Dehydrogenases. BMC Bioinform. 2012, 13 (Suppl. S17), S2. [Google Scholar] [CrossRef]
- Rendina, A.R.; Pietrak, B.; Smallwood, A.; Zhao, H.; Qi, H.; Quinn, C.; Adams, N.D.; Concha, N.; Duraiswami, C.; Thrall, S.H.; et al. Mutant IDH1 Enhances the Production of 2-Hydroxyglutarate Due to Its Kinetic Mechanism. Biochemistry 2013, 52, 4563–4577. [Google Scholar] [CrossRef]
- Liu, S.; Abboud, M.I.; John, T.; Mikhailov, V.; Hvinden, I.; Walsby-Tickle, J.; Liu, X.; Pettinati, I.; Cadoux-Hudson, T.; McCullagh, J.S.O.; et al. Roles of Metal Ions in the Selective Inhibition of Oncogenic Variants of Isocitrate Dehydrogenase 1. Commun. Biol. 2021, 4, 1243. [Google Scholar] [CrossRef]
- Song, P.; Wei, H.; Cao, Z.; Wang, P.; Zhu, G. Single Arginine Mutation in Two Yeast Isocitrate Dehydrogenases: Biochemical Characterization and Functional Implication. PLoS ONE 2014, 9, e115025. [Google Scholar] [CrossRef]
- Song, P.; Li, S.; Wu, Y.; Lv, C.; Wang, P.; Zhu, G. Point Mutation (R153H or R153C) in Escherichia Coli Isocitrate Dehydrogenase: Biochemical Characterization and Functional Implication. J. Basic Microbiol. 2017, 57, 41–49. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Ho, H.-L.; Lin, S.-C.; Hsu, C.-Y.; Ho, D.M.-T. Loss of BCAT1 Expression Is a Sensitive Marker for IDH-Mutant Diffuse Glioma. Neurosurgery 2019, 85, 335. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-M.; Colaiácovo, M.P. CRISPR-Cas9-Guided Genome Engineering in Caenorhabditis elegans. Curr. Protoc. Mol. Biol. 2019, 129, e106. [Google Scholar] [CrossRef]
- Nance, J.; Frøkjær-Jensen, C. The Caenorhabditis elegans Transgenic Toolbox. Genetics 2019, 212, 959–990. [Google Scholar] [CrossRef] [PubMed]
- Shaye, D.D.; Greenwald, I. OrthoList: A Compendium of C. elegans Genes with Human Orthologs. PLoS ONE 2011, 6, e20085. [Google Scholar] [CrossRef]
- Ponomarova, O.; Starbard, A.N.; Belfi, A.; Anderson, A.V.; Sundaram, M.V.; Walhout, A.J. Idh-1 Neomorphic Mutation Confers Sensitivity to Vitamin B12 in Caenorhabditis elegans. Life Sci. Alliance 2024, 7, e202402924. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhao, J.; Xu, Z.; Peng, B.; Huang, Q.; Arnold, E.; Ding, J. Structures of Human Cytosolic NADP-Dependent Isocitrate Dehydrogenase Reveal a Novel Self-Regulatory Mechanism of Activity. J. Biol. Chem. 2004, 279, 33946–33957. [Google Scholar] [CrossRef]
- Huang, Y.C.; Grodsky, N.B.; Kim, T.-K.; Colman, R.F. Ligands of the Mn2+ Bound to Porcine Mitochondrial NADP-Dependent Isocitrate Dehydrogenase, as Assessed by Mutagenesis. Biochemistry 2004, 43, 2821–2828. [Google Scholar] [CrossRef]
- Grodsky, N.B.; Soundar, S.; Colman, R.F. Evaluation by Site-Directed Mutagenesis of Aspartic Acid Residues in the Metal Site of Pig Heart NADP-Dependent Isocitrate Dehydrogenase. Biochemistry 2000, 39, 2193–2200. [Google Scholar] [CrossRef]
- Ceccarelli, C.; Grodsky, N.B.; Ariyaratne, N.; Colman, R.F.; Bahnson, B.J. Crystal Structure of Porcine Mitochondrial NADP+-Dependent Isocitrate Dehydrogenase Complexed with Mn2+ and Isocitrate. J. Biol. Chem. 2002, 277, 43454–43462. [Google Scholar] [CrossRef]
- Soundar, S.; Danek, B.L.; Colman, R.F. Identification by Mutagenesis of Arginines in the Substrate Binding Site of the Porcine NADP-Dependent Isocitrate Dehydrogenase. J. Biol. Chem. 2000, 275, 5606–5612. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-K.; Colman, R.F. Ser95, Asn97, and Thr78 Are Important for the Catalytic Function of Porcine NADP-Dependent Isocitrate Dehydrogenase. Protein Sci. 2005, 14, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, S.; Miller, S.P.; Carrondo, M.A.; Dean, A.M.; Matias, P.M. Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase. Biochemistry 2012, 51, 7098–7115. [Google Scholar] [CrossRef]
- Aktas, D.F.; Cook, P.F. A Lysine-Tyrosine Pair Carries out Acid-Base Chemistry in the Metal Ion-Dependent Pyridine Dinucleotide-Linked Beta-Hydroxyacid Oxidative Decarboxylases. Biochemistry 2009, 48, 3565–3577. [Google Scholar] [CrossRef] [PubMed]
- Pietrak, B.; Zhao, H.; Qi, H.; Quinn, C.; Gao, E.; Boyer, J.G.; Concha, N.; Brown, K.; Duraiswami, C.; Wooster, R.; et al. A Tale of Two Subunits: How the Neomorphic R132H IDH1 Mutation Enhances Production of αHG. Biochemistry 2011, 50, 4804–4812. [Google Scholar] [CrossRef]
- Vander Meersche, Y.; Cretin, G.; de Brevern, A.G.; Gelly, J.-C.; Galochkina, T. MEDUSA: Prediction of Protein Flexibility from Sequence. J. Mol. Biol. 2021, 433, 166882. [Google Scholar] [CrossRef]
- Luna, L.A.; Lesecq, Z.; White, K.A.; Hoang, A.; Scott, D.A.; Zagnitko, O.; Bobkov, A.A.; Barber, D.L.; Schiffer, J.M.; Isom, D.G.; et al. An Acidic Residue Buried in the Dimer Interface of Isocitrate Dehydrogenase 1 (IDH1) Helps Regulate Catalysis and pH Sensitivity. Biochem. J. 2020, 477, 2999–3018. [Google Scholar] [CrossRef]
- Leonardi, R.; Subramanian, C.; Jackowski, S.; Rock, C.O. Cancer-Associated Isocitrate Dehydrogenase Mutations Inactivate NADPH-Dependent Reductive Carboxylation. J. Biol. Chem. 2012, 287, 14615–14620. [Google Scholar] [CrossRef]
- Stern, A.; Fokra, M.; Sarvin, B.; Alrahem, A.A.; Lee, W.D.; Aizenshtein, E.; Sarvin, N.; Shlomi, T. Inferring Mitochondrial and Cytosolic Metabolism by Coupling Isotope Tracing and Deconvolution. Nat. Commun. 2023, 14, 7525. [Google Scholar] [CrossRef]
- Jin, G.; Reitman, Z.J.; Duncan, C.G.; Spasojevic, I.; Gooden, D.M.; Rasheed, B.A.; Yang, R.; Lopez, G.Y.; He, Y.; McLendon, R.E.; et al. Disruption of Wild Type IDH1 Suppresses D-2-Hydroxyglutarate Production in IDH1-Mutated Gliomas. Cancer Res. 2013, 73, 496–501. [Google Scholar] [CrossRef]
- Ward, P.S.; Lu, C.; Cross, J.R.; Abdel-Wahab, O.; Levine, R.L.; Schwartz, G.K.; Thompson, C.B. The Potential for Isocitrate Dehydrogenase Mutations to Produce 2-Hydroxyglutarate Depends on Allele Specificity and Subcellular Compartmentalization. J. Biol. Chem. 2013, 288, 3804–3815. [Google Scholar] [CrossRef] [PubMed]
- Sesanto, R.; Kuehn, J.F.; Barber, D.L.; White, K.A. Low pH Facilitates Heterodimerization of Mutant Isocitrate Dehydrogenase IDH1-R132H and Promotes Production of 2-Hydroxyglutarate. Biochemistry 2021, 60, 1983–1994. [Google Scholar] [CrossRef]
- Pusch, S.; Schweizer, L.; Beck, A.-C.; Lehmler, J.-M.; Weissert, S.; Balss, J.; Miller, A.K.; von Deimling, A. D-2-Hydroxyglutarate Producing Neo-Enzymatic Activity Inversely Correlates with Frequency of the Type of Isocitrate Dehydrogenase 1 Mutations Found in Glioma. Acta Neuropathol. Commun. 2014, 2, 19. [Google Scholar] [CrossRef]
- Brown, J.B.; Celniker, S.E. Lessons from modENCODE. Annu. Rev. Genom. Hum. Genet. 2015, 16, 31–53. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Garcia, B.A. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb. Perspect. Biol. 2015, 7, a025064. [Google Scholar] [CrossRef]
- Simpson, V.J.; Johnson, T.E.; Hammen, R.F.C. elegans DNA Does Not Contain 5-Methylcytosine at Any Time during Development or Aging. Nucleic Acids Res. 1986, 14, 6711–6717. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.-H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Shang, G.; Yang, M.; Li, M.; Ma, L.; Liu, Y.; Ma, J.; Chen, Y.; Wang, X.; Fan, S.; Xie, M.; et al. Structural Basis of Nucleic Acid Recognition and 6mA Demethylation by Caenorhabditis elegans NMAD-1A. Int. J. Mol. Sci. 2024, 25, 686. [Google Scholar] [CrossRef]
- Colman, R.F. Role of Metal Ions in Reactions Catalyzed by Pig Heart Triphosphopyridine Nucleotide-Dependent Isocitrate Dehydrogenase. II. Effect on Catalytic Properties and Reactivity of Amino Acid Residues. J. Biol. Chem. 1972, 247, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Grubbs, R.D. Intracellular Magnesium and Magnesium Buffering. Biometals 2002, 15, 251–259. [Google Scholar] [CrossRef]
- Benedetto, A.; Au, C.; Avila, D.S.; Milatovic, D.; Aschner, M. Extracellular Dopamine Potentiates Mn-Induced Oxidative Stress, Lifespan Reduction, and Dopaminergic Neurodegeneration in a BLI-3–Dependent Manner in Caenorhabditis elegans. PLoS Genet. 2010, 6, e1001084. [Google Scholar] [CrossRef]
- Bornhorst, J.; Chakraborty, S.; Meyer, S.; Lohren, H.; Brinkhaus, S.G.; Knight, A.L.; Caldwell, K.A.; Caldwell, G.A.; Karst, U.; Schwerdtle, T.; et al. The Effects of Pdr1, Djr1.1 and Pink1 Loss in Manganese-Induced Toxicity and the Role of α-Synuclein in C. elegans. Metallomics 2014, 6, 476–490. [Google Scholar] [CrossRef]
- Martins, A.C.; Gubert, P.; Li, J.; Ke, T.; Nicolai, M.M.; Moura, A.V.; Bornhorst, J.; Bowman, A.B.; Aschner, M. Caenorhabditis elegans as a Model to Study Manganese-Induced Neurotoxicity. Biomolecules 2022, 12, 1396. [Google Scholar] [CrossRef]
- Au, C.; Benedetto, A.; Anderson, J.; Labrousse, A.; Erikson, K.; Ewbank, J.J.; Aschner, M. SMF-1, SMF-2 and SMF-3 DMT1 Orthologues Regulate and Are Regulated Differentially by Manganese Levels in C. elegans. PLoS ONE 2009, 4, e7792. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, A.-M.; Besir, H. Staining of Proteins in Gels with Coomassie G-250 without Organic Solvent and Acetic Acid. J. Vis. Exp. 2009, e1350. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling Protein Quaternary Structure of Homo- and Hetero-Oligomers beyond Binary Interactions by Homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef]
- Studer, G.; Tauriello, G.; Bienert, S.; Biasini, M.; Johner, N.; Schwede, T. ProMod3-A Versatile Homology Modelling Toolbox. PLoS Comput. Biol. 2021, 17, e1008667. [Google Scholar] [CrossRef]
- Jakob, C.G.; Upadhyay, A.K.; Donner, P.L.; Nicholl, E.; Addo, S.N.; Qiu, W.; Ling, C.; Gopalakrishnan, S.M.; Torrent, M.; Cepa, S.P.; et al. Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315. J. Med. Chem. 2018, 61, 6647–6657. [Google Scholar] [CrossRef]
- Cho, H.J.; Cho, H.Y.; Park, J.-W.; Kwon, O.-S.; Lee, H.-S.; Huh, T.L.; Kang, B.S. NADP+-Dependent Cytosolic Isocitrate Dehydrogenase Provides NADPH in the Presence of Cadmium Due to the Moderate Chelating Effect of Glutathione. J. Biol. Inorg. Chem. 2018, 23, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—Distance Constraints Applied on Model Quality Estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
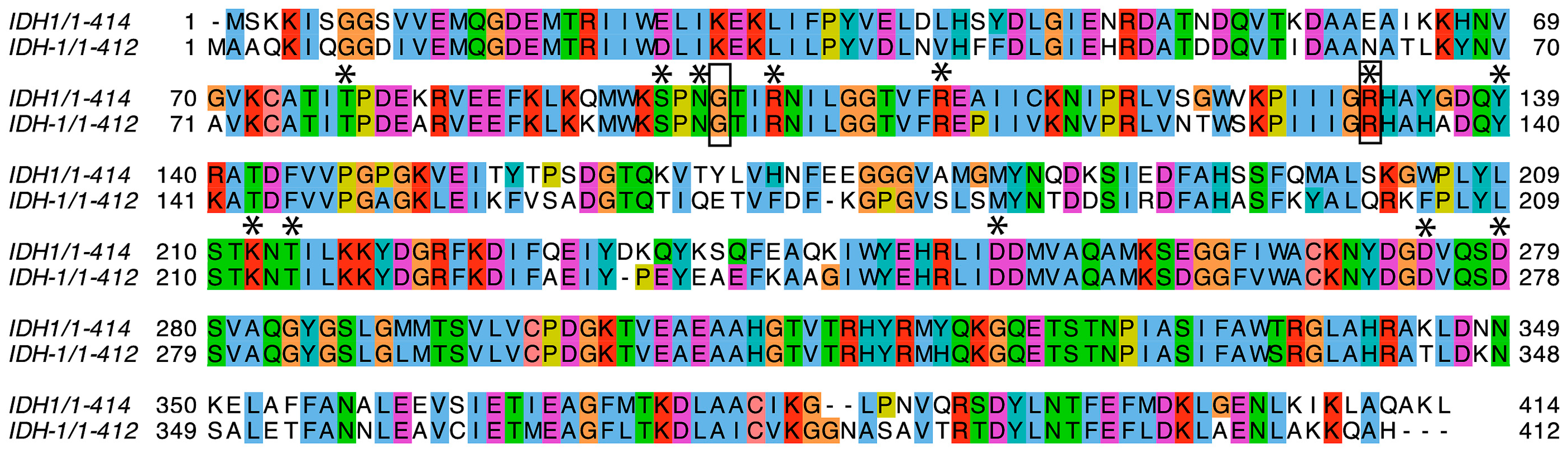

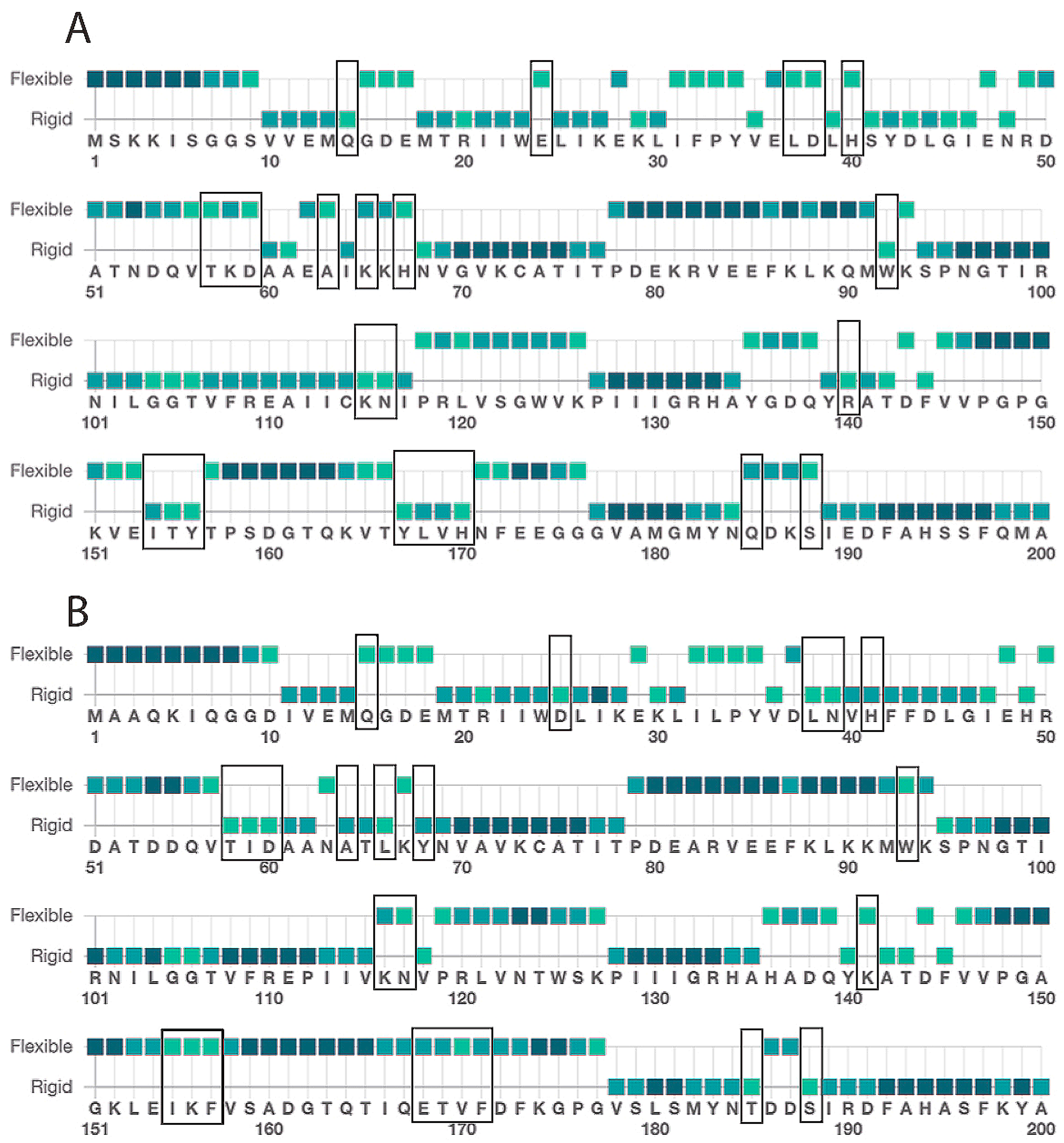

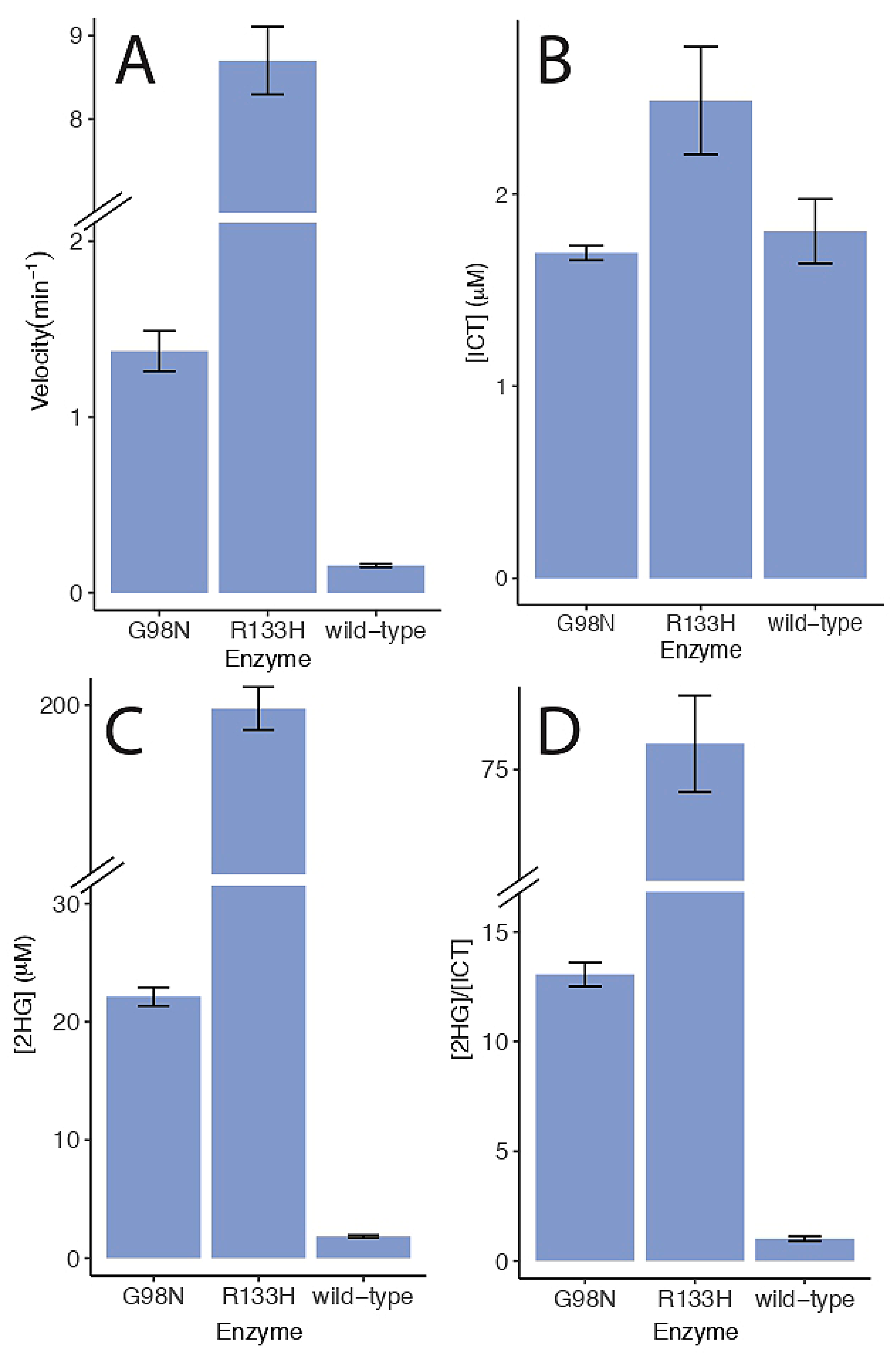
| Enzyme | Varied Substrate | kcat, min−1 | KM, μM | kcat/KM, μM−1 min−1 |
|---|---|---|---|---|
| Wild-type IDH-1 | ICT | 2510 ± 70 | 6.9 ± 0.8 | 360 ± 40 |
| Wild-type IDH-1 | NADP+ | 1260 ± 70 | 3.3 ± 0.5 | 380 ± 60 |
| Wild-type IDH-1 * | Mn2+ | 1590 ± 90 | 9 ± 2 | 176 ± 30 |
| G98N | ICT | 190 ± 6 | 13 ± 1 | 15 ± 1 |
| G98N | NADP+ | 104 ± 3 | 2.8 ± 0.3 | 37 ± 4 |
| R133H | ICT | 11.7 ± 0.6 | 2160 ± 250 | 0.0054 ± 0.0007 |
| R133H | αKG | 18.1 ± 0.7 | 14 ± 2 | 1.3 ± 0.2 |
| R133H | NADPH | 16.7 ± 0.7 | 1.7 ± 0.2 | 10 ± 1 |
| Enzyme | Varied Substrate | Metal | kcat, min−1 | KM, μM | kcat/KM, μM−1 min−1 | Citation |
|---|---|---|---|---|---|---|
| Wild-type IDH1 | ICT | Mg2+ | 517 ± 9 | 5.0 ± 0.3 | 103 ± 6 | [30] |
| Wild-type IDH1 | ICT | Mg2+ | 749 | 7 | 107 | [47] |
| Wild-type IDH1 | ICT | Mg2+ | 3000 ± 240 | 38 ± 6 | 78 ± 18 | [31] |
| Wild-type IDH1 | ICT | Mn2+ | 4860 ± 660 | 35 ± 4 | 138 ± 36 | [31] |
| Wild-type IDH1 | NADP+ | Mg2+ | 3000 ± 240 | 27 ± 2 | 114 ± 18 | [31] |
| Wild-type IDH1 | NADP+ | Mg2+ | 517 ± 9 | 6.2 ± 0.3 | 83 ± 4 | [30] |
| Wild-type IDH1 | Mn2+ | Mn2+ | 4860 ± 660 | 2.0 ± 0.3 | 2460 ± 720 | [31] |
| Wild-type IDH1 | aKG | Mg2+ | 0.07 | 43 | 0.0015 | [47] |
| Wild-type IDH1 | aKG | Mg2+ | 134 ± 5 | 138 ± 38 | 0.97 ± 0.27 | [30] |
| Wild-type IDH1 | aKG | Mg2+ | 126 ± 6 | 1100 ± 300 | 0.11 ± 0.04 | [31] |
| G97N | ICT | Mg2+ | 6.1 ± 0.3 | 0.46 ± 0.07 | 13 ± 2 | [30] |
| G97N | NADP+ | Mg2+ | 6.1 ± 0.3 | 0.06 ± 0.03 | 10.2 ± 0.5 | [30] |
| G97N | aKG | Mg2+ | 2.52 ± 0.05 | 295 ± 13 | 0.0090 ± 0.0004 | [30] |
| R132H | ICT | Mg2+ | 7.7 ± 0.4 | 6600 ± 860 | 0.0012 ± 0.0002 | [30] |
| R132H | ICT | Mg2+ | 6.0 ± 0.6 | 257 ± 50 | 0.023 ± 0.007 | [31] |
| R132H | ICT | Mn2+ | 47 ± 5 | 219 ± 35 | 0.22 ± 0.05 | [31] |
| R132H | αKG | Mg2+ | 42.8 ± 0.7 | 1080 ± 67 | 0.040 ± 0.003 | [30] |
| R132H | αKG | Mg2+ | 37 ± 4 | 652 ± 116 | 0.06 ± 0.02 | [31] |
| R132H | αKG | Mn2+ | 62 ± 3 | 175 ± 26 | 0.35 ± 0.07 | [31] |
| R132H | NADPH | Mg2+ | 37 ± 4 | 15 ± 2 | 2.5 ± 0.6 | [31] |
| R132H | NADPH | Mg2+ | 45 ± 0.9 | 0.43 ± 0.04 | 105 ± 10 | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouchard, M.; McAllister, A.; Bourlett, N.S.; Hoyt, C.; Calcul, L.; Walstrom, K.M. C. elegans Cytoplasmic Isocitrate Dehydrogenase Neomorphic G98N and R133H Mutants Produce the Oncometabolite 2-Hydroxyglutarate. Int. J. Mol. Sci. 2025, 26, 8238. https://doi.org/10.3390/ijms26178238
Bouchard M, McAllister A, Bourlett NS, Hoyt C, Calcul L, Walstrom KM. C. elegans Cytoplasmic Isocitrate Dehydrogenase Neomorphic G98N and R133H Mutants Produce the Oncometabolite 2-Hydroxyglutarate. International Journal of Molecular Sciences. 2025; 26(17):8238. https://doi.org/10.3390/ijms26178238
Chicago/Turabian StyleBouchard, Melissa, Anne McAllister, Noah S. Bourlett, Chelsea Hoyt, Laurent Calcul, and Katherine M. Walstrom. 2025. "C. elegans Cytoplasmic Isocitrate Dehydrogenase Neomorphic G98N and R133H Mutants Produce the Oncometabolite 2-Hydroxyglutarate" International Journal of Molecular Sciences 26, no. 17: 8238. https://doi.org/10.3390/ijms26178238
APA StyleBouchard, M., McAllister, A., Bourlett, N. S., Hoyt, C., Calcul, L., & Walstrom, K. M. (2025). C. elegans Cytoplasmic Isocitrate Dehydrogenase Neomorphic G98N and R133H Mutants Produce the Oncometabolite 2-Hydroxyglutarate. International Journal of Molecular Sciences, 26(17), 8238. https://doi.org/10.3390/ijms26178238







