DNA Methylation in Rice: Mechanisms, Regulatory Roles, and Beyond
Abstract
1. Introduction
2. Molecular Mechanism of DNA Methylation
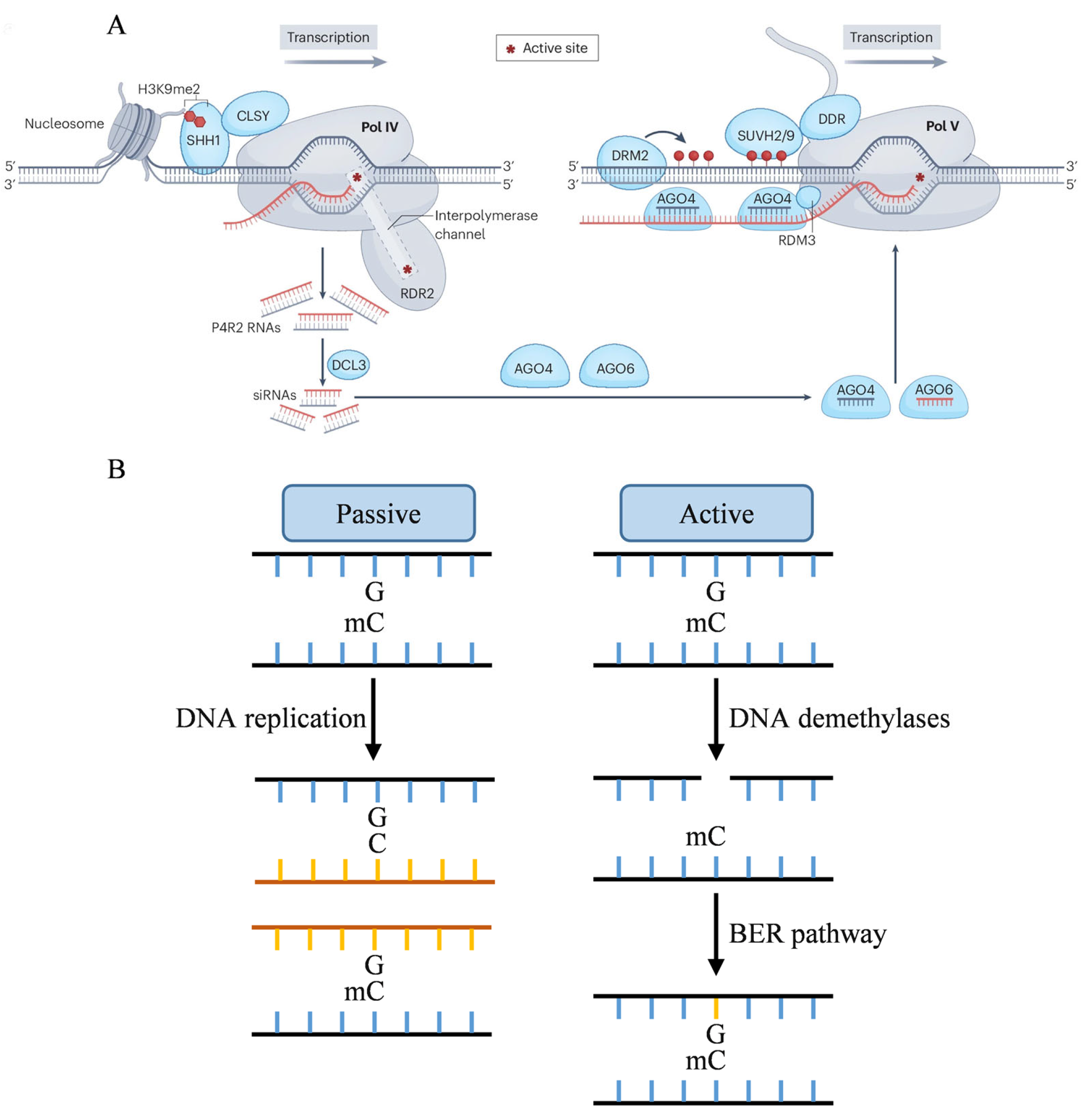
2.1. De Novo Methylation
2.2. Maintenance Methylation
2.3. DNA Demethylation
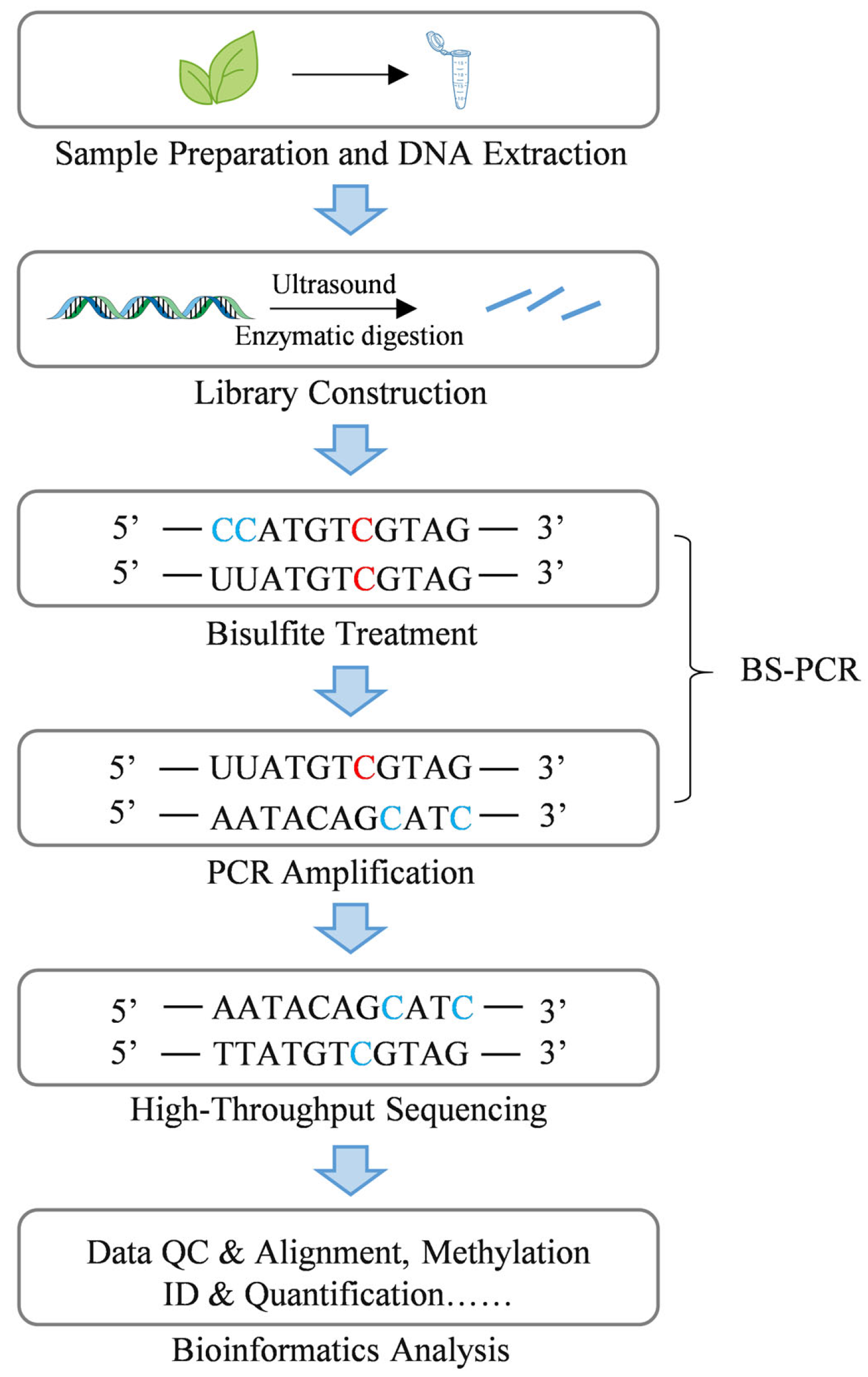
3. DNA Methylation Detection Methods
4. Role of DNA Methylation in Rice Growth and Development
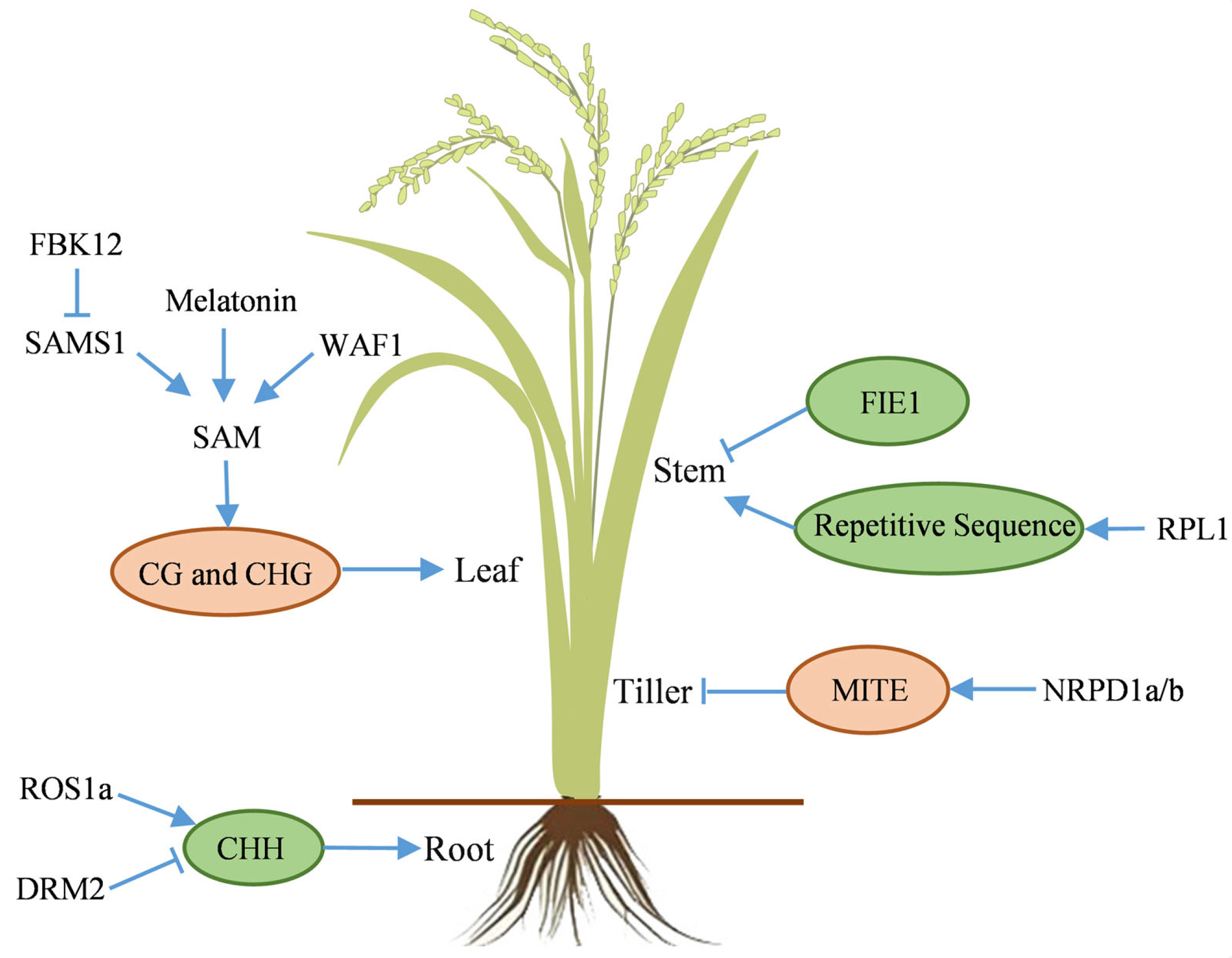
4.1. Vegetative Growth Stage
4.2. Reproductive Growth Stage
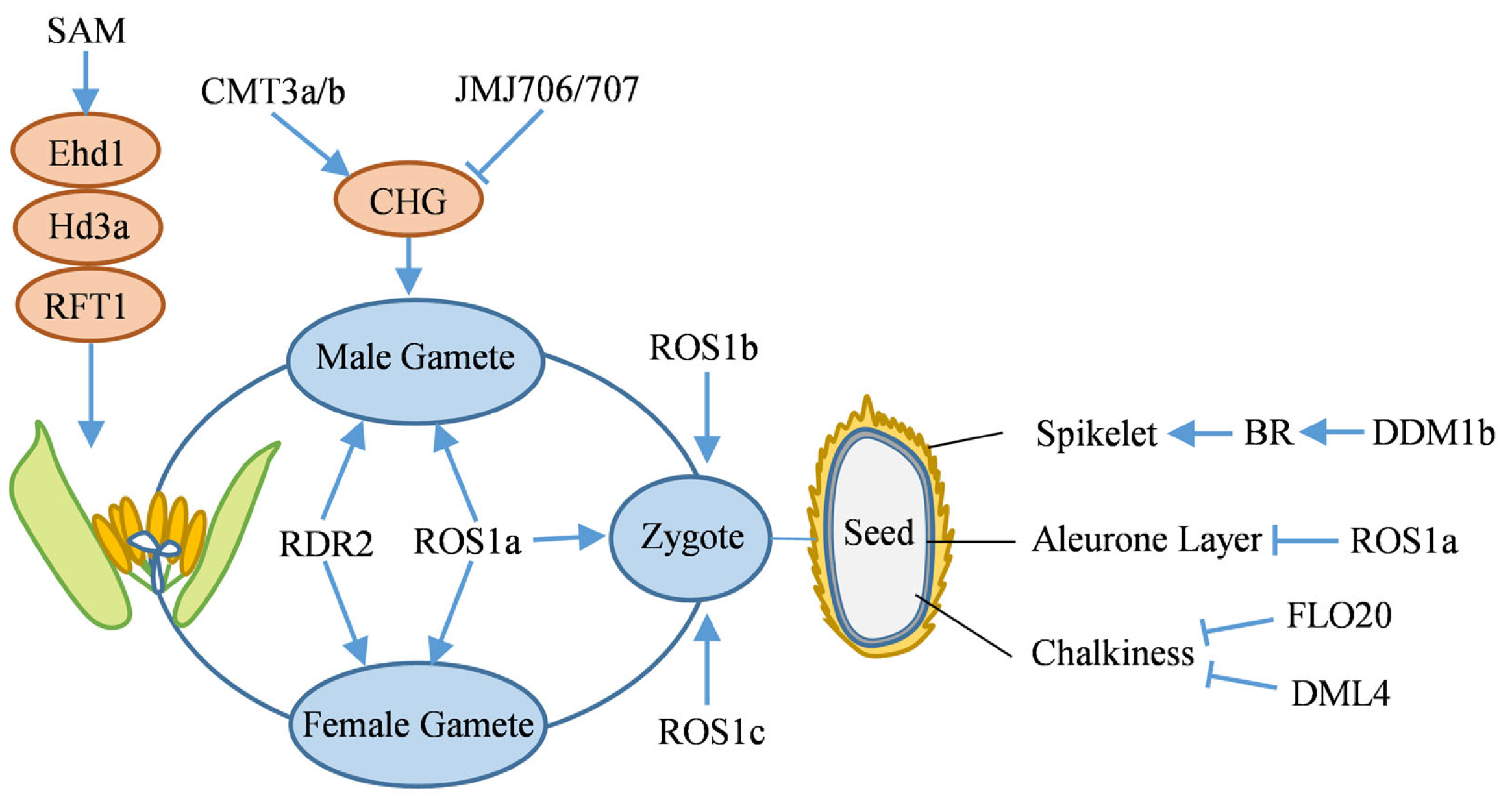
4.2.1. Heading and Flowering
4.2.2. Gamete and Zygote Development
4.2.3. Seed Development
5. Role of DNA Methylation in Rice Stress Response
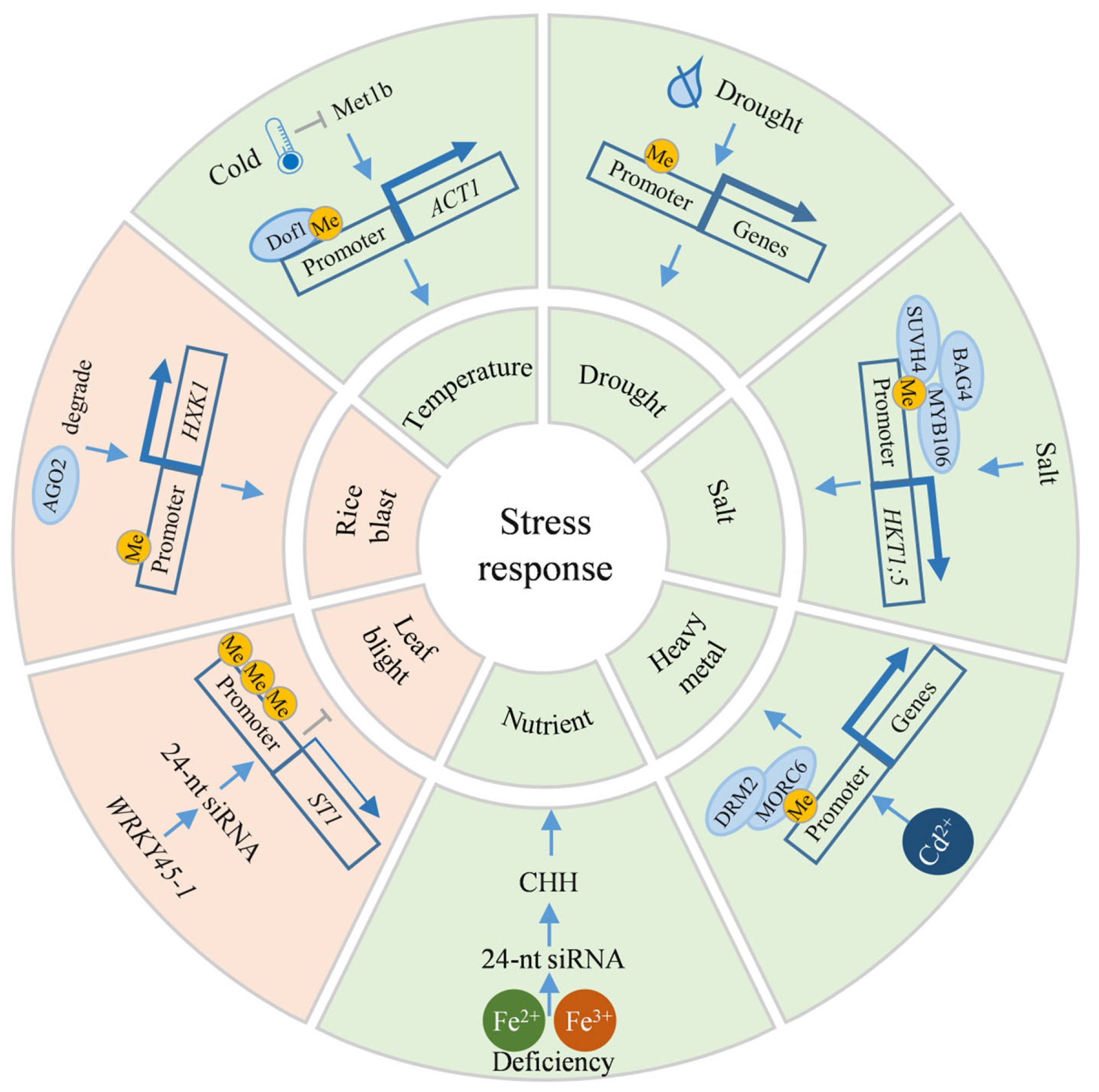
5.1. Abiotic Stress
5.2. Biotic Stress
6. Role of DNA Methylation in Other Biological Process
7. Summary and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Jablonka, E.; Lamb, M.J. Précis of evolution in four dimensions. Behav. Brain Sci. 2007, 30, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Niu, Q.; Xu, Y.; Huang, H.; Li, L.; Tang, D.; Wu, S.; Liu, P.; Liu, R.; Ma, Y.; Zhang, B.; et al. Two transcription factors play critical roles in mediating epigenetic regulation of fruit ripening in tomato. Proc. Natl. Acad. Sci. 2025, 122, e2422798122. [Google Scholar] [CrossRef]
- Miao, L.; Xu, W.; Liu, Y.; Huang, X.; Chen, Z.; Wang, H.; Wang, Z.; Chen, Y.; Song, Q.; Zhang, J.; et al. Reshaped DNA methylation cooperating with homoeolog-divergent expression promotes improved root traits in synthesized tetraploid wheat. New Phytol. 2024, 242, 507–523. [Google Scholar] [CrossRef]
- Secco, D.; Wang, C.; Shou, H.; Schultz, M.D.; Chiarenza, S.; Nussaume, L.; Ecker, J.R.; Whelan, J.; Lister, R. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 2015, 4, e09343. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Gendrel, A.V.; Black, M.; Vaughn, M.W.; Dedhia, N.; Richard McCombie, W.; Lavine, K.; Mittal, V.; May, B.; Kasschau, K.D.; et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 2004, 430, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Chodavarapu, R.K.; Feng, S.; Bernatavichute, Y.V.; Chen, P.-Y.; Stroud, H.; Yu, Y.; Hetzel, J.A.; Kuo, F.; Kim, J.; Cokus, S.J.; et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010, 466, 388–392. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman Derr, D.; Eshed Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef]
- Lyons, D.B.; Zilberman, D. DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes. eLife 2017, 6, e30674. [Google Scholar] [CrossRef]
- He, L.; Huang, H.; Bradai, M.; Zhao, C.; You, Y.; Ma, J.; Zhao, L.; Lozano-Durán, R.; Zhu, J.K. DNA methylation-free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nat. Commun. 2022, 13, 1335. [Google Scholar] [CrossRef]
- Yang, L.; Lang, C.; Wu, Y.; Meng, D.; Yang, T.; Li, D.; Jin, T.; Zhou, X. ROS1-mediated decrease in DNA methylation and increase in expression of defense genes and stress response genes in Arabidopsis thaliana due to abiotic stresses. BMC Plant Biol. 2022, 22, 104. [Google Scholar] [CrossRef]
- Mohidem, N.A.; Hashim, N.; Shamsudin, R.; Che Man, H. Rice for food security: Revisiting its rroduction, diversity, rice milling process and nutrient content. Agriculture 2022, 12, 741. [Google Scholar] [CrossRef]
- Chen, R.; Deng, Y.; Ding, Y.; Guo, J.; Qiu, J.; Wang, B.; Wang, C.; Xie, Y.; Zhang, Z.; Chen, J.; et al. Rice functional genomics: Decades’ efforts and roads ahead. Sci. China Life Sci. 2022, 65, 33–92. [Google Scholar] [CrossRef] [PubMed]
- Tirnaz, S.; Batley, J. DNA methylation: Toward crop disease resistance improvement. Trends Plant Sci. 2019, 24, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Song, X.; Wei, L.; Liu, C.; Cao, X. Epigenetic regulation and epigenomic landscape in rice. Natl. Sci. Rev. 2016, 3, 309–327. [Google Scholar] [CrossRef]
- Xue, Y.; Cao, X.; Chen, X.; Deng, X.; Deng, X.W.; Ding, Y.; Dong, A.; Duan, C.G.; Fang, X.; Gong, L.; et al. Epigenetics in the modern era of crop improvements. Sci. China Life Sci. 2025, 68, 1570–1609. [Google Scholar] [CrossRef]
- Liu, S.; Sretenovic, S.; Fan, T.; Cheng, Y.; Li, G.; Qi, A.; Tang, X.; Xu, Y.; Guo, W.; Zhong, Z.; et al. Hypercompact CRISPR–Cas12j2 (CasΦ) enables genome editing, gene activation, and epigenome editing in plants. Plant Commun. 2022, 3, 100453. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Alariqi, M.; Li, B.; Hussain, A.; Zhou, H.; Wang, Q.; Wang, F.; Wang, G.; Zhu, X.; Hui, F.; et al. CRISPR/dCas13(Rx) derived RNA N6-methyladenosine (m6A) dynamic modification in plant. Adv. Sci. 2024, 11, 2401118. [Google Scholar] [CrossRef]
- Song, X.; Tang, S.; Liu, H.; Meng, Y.; Luo, H.; Wang, B.; Hou, X.L.; Yan, B.; Yang, C.; Guo, Z.; et al. Inheritance of acquired adaptive cold tolerance in rice through DNA methylation. Cell 2025, 188, 4213–4224.E12. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Shirane, K.; Toh, H.; Kobayashi, H.; Miura, F.; Chiba, H.; Ito, T.; Kono, T.; Sasaki, H. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013, 9, e1003439. [Google Scholar] [CrossRef]
- Xie, W.; Barr, C.L.; Kim, A.; Yue, F.; Lee, A.Y.; Eubanks, J.; Dempster, E.L.; Ren, B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 2012, 148, 816–831. [Google Scholar] [CrossRef]
- Niederhuth, C.E.; Schmitz, R.J. Covering your bases: Inheritance of DNA methylation in plant genomes. Mol. Plant 2014, 7, 472–480. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef]
- Vidalis, A.; Živković, D.; Wardenaar, R.; Roquis, D.; Tellier, A.; Johannes, F. Methylome evolution in plants. Genome Biol. 2016, 17, 264. [Google Scholar] [CrossRef]
- Li, X.; Zhu, J.; Hu, F.; Ge, S.; Ye, M.; Xiang, H.; Zhang, G.; Zheng, X.; Zhang, H.; Zhang, S.; et al. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genom. 2012, 13, 300. [Google Scholar] [CrossRef]
- Lloyd, J.P.B.; Lister, R. Epigenome plasticity in plants. Nat. Rev. Genet. 2022, 23, 55–68. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, Z.Y.; Zeng, L.; Tanaka, K.; Zhang, C.J.; Ma, J.; Bai, G.; Wang, P.; Zhang, S.W.; Liu, Z.W.; et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA 2013, 110, 8290–8295. [Google Scholar] [CrossRef]
- Law, J.A.; Du, J.; Hale, C.J.; Feng, S.; Krajewski, K.; Palanca, A.M.S.; Strahl, B.D.; Patel, D.J.; Jacobsen, S.E. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 2013, 498, 385–389. [Google Scholar] [CrossRef]
- Wang, Y.; Le, B.H.; Wang, J.; You, C.; Zhao, Y.; Galli, M.; Xu, Y.; Gallavotti, A.; Eulgem, T.; Mo, B.; et al. ZMP recruits and excludes Pol IV–mediated DNA methylation in a site-specific manner. Sci. Adv. 2022, 8, eadc9454. [Google Scholar] [CrossRef] [PubMed]
- Herr, A.J.; Jensen, M.B.; Dalmay, T.; Baulcombe, D.C. RNA polymerase IV directs silencing of endogenous DNA. Science 2005, 308, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Kanno, T.; Huettel, B.; Mette, M.F.; Aufsatz, W.; Jaligot, E.; Daxinger, L.; Kreil, D.P.; Matzke, M.; Matzke, A.J.M. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005, 37, 761–765. [Google Scholar] [CrossRef]
- Onodera, Y.; Haag, J.R.; Ream, T.; Nunes, P.C.; Pontes, O.; Pikaard, C.S. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 2005, 120, 613–622. [Google Scholar] [CrossRef]
- Huang, K.; Wu, X.X.; Fang, C.L.; Xu, Z.G.; Zhang, H.W.; Gao, J.; Zhou, C.M.; You, L.L.; Gu, Z.X.; Mu, W.H.; et al. Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA. Science 2021, 374, 1579–1586. [Google Scholar] [CrossRef]
- Zhai, J.; Bischof, S.; Wang, H.; Feng, S.; Lee, T.; Teng, C.; Chen, X.; Park, S.Y.; Liu, L.; Gallego-Bartolome, J.; et al. One precursor one siRNA model for Pol IV-dependent siRNAs biogenesis. Cell 2015, 163, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Blevins, T.; Podicheti, R.; Mishra, V.; Marasco, M.; Wang, J.; Rusch, D.; Tang, H.; Pikaard, C.S. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 2015, 4, e09591. [Google Scholar] [CrossRef]
- Li, S.; Vandivier, L.E.; Tu, B.; Gao, L.; Won, S.Y.; Li, S.; Zheng, B.; Gregory, B.D.; Chen, X. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 2015, 25, 235–245. [Google Scholar] [CrossRef]
- Wang, Q.; Xue, Y.; Zhang, L.; Zhong, Z.; Feng, S.; Wang, C.; Xiao, L.; Yang, Z.; Harris, C.J.; Wu, Z.; et al. Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation. Science 2021, 374, 1152–1157. [Google Scholar] [CrossRef]
- Loffer, A.; Singh, J.; Fukudome, A.; Mishra, V.; Wang, F.; Pikaard, C.S. A DCL3 dicing code within Pol IV-RDR2 transcripts diversifies the siRNA pool guiding RNA-directed DNA methylation. eLife 2022, 11, e73260. [Google Scholar] [CrossRef] [PubMed]
- Rajeswaran, R.; Pooggin, M.M. RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 2012, 40, 594–599. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Peragine, A.; Park, M.Y.; Poethig, R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005, 19, 2164–2175. [Google Scholar] [CrossRef]
- Xie, Z.; Allen, E.; Wilken, A.; Carrington, J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 12984–12989. [Google Scholar] [CrossRef] [PubMed]
- Elmayan, T.; Adenot, X.; Gissot, L.; Lauressergues, D.; Gy, I.; Vaucheret, H. A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J. 2009, 276, 835–844. [Google Scholar] [CrossRef]
- Yang, Z.; Ebright, Y.W.; Yu, B.; Chen, X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006, 34, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Sigman, M.J.; Panda, K.; Kirchner, R.; McLain, L.L.; Payne, H.; Peasari, J.R.; Husbands, A.Y.; Slotkin, R.K.; McCue, A.D. An siRNA-guided ARGONAUTE protein directs RNA polymerase V to initiate DNA methylation. Nat. Plants 2021, 7, 1461–1474. [Google Scholar] [CrossRef]
- Zhong, X.; Du, J.; Hale, C.J.; Gallego-Bartolome, J.; Feng, S.; Vashisht, A.A.; Chory, J.; Wohlschlegel, J.A.; Patel, D.J.; Jacobsen, S.E. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 2014, 157, 1050–1060. [Google Scholar] [CrossRef]
- Wierzbicki, A.T.; Haag, J.R.; Pikaard, C.S. Noncoding transcription by RNA Polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008, 135, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.K. Epigenetic gene regulation in plants and its potential applications in crop improvement. Nat. Rev. Mol. Cell Biol. 2025, 26, 51–67. [Google Scholar] [CrossRef]
- Naumann, U.; Daxinger, L.; Kanno, T.; Eun, C.; Long, Q.; Lorkovic, Z.J.; Matzke, M.; Matzke, A.J.M. Genetic evidence that DNA methyltransferase DRM2 has a direct catalytic role in RNA-directed DNA methylation in Arabidopsis thaliana. Genetics 2011, 187, 977–979. [Google Scholar] [CrossRef]
- Cao, X.; Jacobsen, S.E. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002, 12, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Lindroth, A.M.; Cao, X.; Jackson, J.P.; Zilberman, D.; McCallum, C.M.; Henikoff, S.; Jacobsen, S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001, 292, 2077–2080. [Google Scholar] [CrossRef]
- Du, J.; Zhong, X.; Bernatavichute, Y.V.; Stroud, H.; Feng, S.; Caro, E.; Vashisht, A.A.; Terragni, J.; Chin, H.G.; Tu, A.; et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 2012, 151, 167–180. [Google Scholar] [CrossRef]
- Moritoh, S.; Eun, C.H.; Ono, A.; Asao, H.; Okano, Y.; Yamaguchi, K.; Shimatani, Z.; Koizumi, A.; Terada, R. Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 2012, 71, 85–98. [Google Scholar] [CrossRef]
- Cheng, C.; Tarutani, Y.; Miyao, A.; Ito, T.; Yamazaki, M.; Sakai, H.; Fukai, E.; Hirochika, H. Loss of function mutations in the rice chromomethylase OsCMT3a cause a burst of transposition. Plant J. 2015, 83, 1069–1081. [Google Scholar] [CrossRef]
- Li, X.; Zhu, B.; Lu, Y.; Zhao, F.; Liu, Q.; Wang, J.; Ye, M.; Chen, S.; Nie, J.; Xiong, L.; et al. DNA methylation remodeling and the functional implication during male gametogenesis in rice. Genome Biol. 2024, 25, 84, https://doi.org/10.1186/s13059-024-03222-w. Correction in Genome Biol. 2024, 25, 196. [Google Scholar] [CrossRef]
- Yamauchi, T.; Moritoh, S.; Johzuka-Hisatomi, Y.; Ono, A.; Terada, R.; Nakamura, I.; Iida, S. Alternative splicing of the rice OsMET1 genes encoding maintenance DNA methyltransferase. J. Plant Physiol. 2008, 165, 1774–1782. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhang, W.; Mohammadi, M.A.; He, Z.; She, Z.; Yan, M.; Shi, C.; Lin, L.; Wang, A.; Liu, J.; et al. OsDDM1b controls grain size by influencing cell cycling and regulating homeostasis and signaling of brassinosteroid in rice. Front. Plant Sci. 2022, 873993. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, X.; Yao, X.; Yu, R.; Larkin, P.J.; Liu, C.M. Mutations in the DNA demethylase OsROS1 result in a thickened aleurone and improved nutritional value in rice grains. Proc. Natl. Acad. Sci. USA 2018, 115, 11327–11332. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Li, X.; Liu, Q.; Zhao, Y.; Jiang, W.; Wu, A.; Zhou, D.X. DNA demethylases remodel DNA methylation in rice gametes and zygote and are required for reproduction. Mol. Plant 2021, 14, 1569–1583. [Google Scholar] [CrossRef]
- Kim, M.Y.; Ono, A.; Scholten, S.; Kinoshita, T.; Zilberman, D.; Okamoto, T.; Fischer, R.L. DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc. Natl. Acad. Sci. USA 2019, 116, 9652–9657. [Google Scholar] [CrossRef]
- Li, C.; Kong, J.R.; Yu, J.; He, Y.Q.; Yang, Z.K.; Zhuang, J.J.; Ruan, C.C.; Yan, Y.; Xu, J.H. DNA demethylase gene OsDML4 controls salt tolerance by regulating the ROS homeostasis and the JA signaling in rice. Environ. Exp. Bot. 2023, 209, 105276. [Google Scholar] [CrossRef]
- Yan, Y.; Li, C.; Liu, Z.; Zhuang, J.J.; Kong, J.R.; Yang, Z.K.; Yu, J.; Shah Alam, M.; Ruan, C.C.; Zhang, H.M.; et al. A new demethylase gene, OsDML4, is involved in high temperature-increased grain chalkiness in rice. J. Exp. Bot. 2022, 73, 7273–7284. [Google Scholar] [CrossRef]
- Tan, F.; Zhou, C.; Zhou, Q.; Zhou, S.; Yang, W.; Zhao, Y.; Li, G.; Zhou, D.X. Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol. 2016, 171, 2041–2054. [Google Scholar] [CrossRef]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef]
- Haag, J.R.; Pikaard, C.S. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011, 12, 483–492. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhao, C.; Zhang, Q.; Zinta, G.; Wang, D.; Lozano-Durán, R.; Zhu, J.K. Pathway conversion enables a double-lock mechanism to maintain DNA methylation and genome stability. Proc. Natl. Acad. Sci. USA 2021, 118, e2107320118. [Google Scholar] [CrossRef]
- Yadav, N.S.; Khadka, J.; Domb, K.; Zemach, A.; Grafi, G. CMT3 and SUVH4/KYP silence the exonic Evelknievel retroelement to allow for reconstitution of CMT1 mRNA. Epigenetics Chromatin 2018, 11, 69. [Google Scholar] [CrossRef]
- Sasaki, E.; Kawakatsu, T.; Ecker, J.R.; Nordborg, M. Common alleles of CMT2 and NRPE1 are major determinants of CHH methylation variation in Arabidopsis thaliana. PLoS Genet. 2019, 15, e1008492. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.H.; Richards, E.J.; Chung, K.M.; Woo, H.R. Arabidopsis VIM proteins regulate epigenetic silencing by modulating DNA methylation and histone modification in cooperation with MET1. Mol. Plant 2014, 7, 1470–1485. [Google Scholar] [CrossRef]
- Higo, H.; Tahir, M.; Takashima, K.; Miura, A.; Watanabe, K.; Tagiri, A.; Ugaki, M.; Ishikawa, R.; Eiguchi, M.; Kurata, N.; et al. DDM1 (Decrease in DNA Methylation) genes in rice (Oryza sativa). Mol. Genet. Genom. 2012, 287, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Groth, M.; Moissiard, G.; Wirtz, M.; Wang, H.; Garcia-Salinas, C.; Ramos-Parra, P.A.; Bischof, S.; Feng, S.; Cokus, S.J.; John, A.; et al. MTHFD1 controls DNA methylation in Arabidopsis. Nat. Commun. 2016, 7, 11640. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.R.; Zhang, F.F.; Ma, Z.Y.; Huang, H.W.; Jiang, L.; Cai, T.; Zhu, J.K.; Zhang, C.; He, X.J. Folate polyglutamylation is involved in chromatin silencing by maintaining global DNA methylation and histone H3K9 dimethylation in Arabidopsis. Plant Cell 2013, 25, 2545–2559. [Google Scholar] [CrossRef] [PubMed]
- Bellacosa, A.; Drohat, A.C. Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites. DNA Repair 2015, 32, 33–42. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- He, Y.F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Maiti, A.; Drohat, A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine. J. Biol. Chem. 2011, 286, 35334–35338. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.R.; Krawczyk, C.; Robertson, A.B.; Kuśnierczyk, A.; Vågbø, C.B.; Schuermann, D.; Klungland, A.; Schär, P. Biochemical reconstitution of TET1–TDG–BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nat. Commun. 2016, 7, 10806. [Google Scholar] [CrossRef]
- Liu, S.; Dunwell, T.L.; Pfeifer, G.P.; Dunwell, J.M.; Ullah, I.; Wang, Y. Detection of oxidation products of 5-methyl-2′-deoxycytidine in Arabidopsis DNA. PLoS ONE 2013, 8, e84620. [Google Scholar] [CrossRef]
- Tang, Y.; Xiong, J.; Jiang, H.P.; Zheng, S.J.; Feng, Y.Q.; Yuan, B.F. Determination of oxidation products of 5-methylcytosine in plants by chemical derivatization coupled with liquid chromatography/tandem mass spectrometry analysis. Anal. Chem. 2014, 86, 7764–7772. [Google Scholar] [CrossRef]
- Kalinka, A.; Starczak, M.; Gackowski, D.; Stępień, E.; Achrem, M. Global DNA 5-hydroxymethylcytosine level and its chromosomal distribution in four rye species. J. Exp. Bot. 2023, 74, 3488–3502. [Google Scholar] [CrossRef]
- Hollwey, E.; Watson, M.; Meyer, P. Expression of the c-terminal domain of mammalian TET3 DNA dioxygenase in Arabidopsis thaliana induces heritable methylation changes at rDNA loci. Adv. Biosci. Biotechnol. 2016, 7, 243–250. [Google Scholar] [CrossRef][Green Version]
- Liu, Q.; Xue, Q.; Xu, J. Phylogenetic analysis of DNA demethylase genes in angiosperm. Hereditas 2014, 36, 276–285. [Google Scholar][Green Version]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef]
- Booth, M.J.; Branco, M.R.; Ficz, G.; Oxley, D.; Krueger, F.; Reik, W.; Balasubramanian, S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012, 336, 934–937. [Google Scholar] [CrossRef]
- Stroud, H.; Feng, S.; Morey Kinney, S.; Pradhan, S.; Jacobsen, S.E. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011, 12, R54. [Google Scholar] [CrossRef] [PubMed]
- Urich, M.A.; Nery, J.R.; Lister, R.; Schmitz, R.J.; Ecker, J.R. MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat. Protoc. 2015, 10, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Guan, X.; Hu, Y.; Zhang, Z.; Yang, H.; Shi, X.; Han, J.; Mei, H.; Wang, L.; Shao, L.; et al. Population-wide DNA methylation polymorphisms at single-nucleotide resolution in 207 cotton accessions reveal epigenomic contributions to complex traits. Cell Res. 2024, 34, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.; Saini, R.; Upadhyay, R.; Tiwari, K.N. Revealing the Epigenetic Mechanisms Underlying the Stress Response in Medicinal Plants. In Stress-Responsive Factors and Molecular Farming in Medicinal Plants; Singh, D., Mishra, A.K., Srivastava, A.K., Eds.; Springer Nature: Singapore, 2023; pp. 107–122. ISBN 978-981-9944-80-4. [Google Scholar]
- Simko, I. Differentially methylated genomic regions of lettuce seeds relate to divergence across morphologically distinct horticultural types. AoB Plants 2023, 15, plad060. [Google Scholar] [CrossRef]
- Tost, J. Current and Emerging Technologies for the Analysis of the Genome-Wide and Locus-Specific DNA Methylation Patterns. In DNA Methyltransferases-Role and Function; Jeltsch, A., Jurkowska, R.Z., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 343–430. ISBN 978-3-319-43624-1. [Google Scholar]
- Full Article: Serial Pyrosequencing for Quantitative DNA Methylation Analysis. Available online: https://www.tandfonline.com/doi/10.2144/000112190?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed (accessed on 16 August 2025).
- Deng, J.; Shoemaker, R.; Xie, B.; Gore, A.; LeProust, E.M.; Antosiewicz-Bourget, J.; Egli, D.; Maherali, N.; Park, I.H.; Yu, J.; et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009, 27, 353–360. [Google Scholar] [CrossRef]
- Xiong, Z.; Laird, P.W. COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997, 25, 2532–2534. [Google Scholar] [CrossRef]
- Yu, M.; Hon, G.C.; Szulwach, K.E.; Song, C.X.; Zhang, L.; Kim, A.; Li, X.; Dai, Q.; Park, B.; Min, J.H.; et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012, 149, 1368–1380. [Google Scholar] [CrossRef]
- Herman, J.G.; Graff, J.R.; Myöhänen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef]
- Melnikov, A.A.; Gartenhaus, R.B.; Levenson, A.S.; Motchoulskaia, N.A.; Levenson, V.V. MSRE-PCR for analysis of gene-specific DNA methylation. Nucleic Acids Res. 2005, 33, e93. [Google Scholar] [CrossRef]
- Shareef, S.J.; Bevill, S.M.; Raman, A.T.; Aryee, M.J.; van Galen, P.; Hovestadt, V.; Bernstein, B.E. Extended representation bisulfite sequencing of gene regulatory elements in multiplexed samples and single cells. Nat. Biotechnol. 2021, 39, 1086–1094. [Google Scholar] [CrossRef]
- Kawakatsu, T. Whole-Genome Bisulfite Sequencing and Epigenetic Variation in Cereal Methylomes. In Cereal Genomics: Methods and Protocols; Vaschetto, L.M., Ed.; Springer: New York, NY, USA, 2020; pp. 119–128. ISBN 978-1-4939-9865-4. [Google Scholar]
- Lentini, A.; Nestor, C.E. Mapping DNA Methylation in Mammals: The State of the Art. In DNA Modifications: Methods and Protocols; Ruzov, A., Gering, M., Eds.; Springer: New York, NY, USA, 2021; pp. 37–50. ISBN 978-1-07-160876-0. [Google Scholar]
- Gitan, R.S.; Shi, H.; Chen, C.M.; Yan, P.S.; Huang, T.H.M. Methylation-specific oligonucleotide microarray: A new potential for high-throughput methylation analysis. Genome Res. 2002, 12, 158–164. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weber, M.; Davies, J.J.; Wittig, D.; Oakeley, E.J.; Haase, M.; Lam, W.L.; Schübeler, D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005, 37, 853–862. [Google Scholar] [CrossRef]
- Tan, L.; Xiong, L.; Xu, W.; Wu, F.; Huang, N.; Xu, Y.; Kong, L.; Zheng, L.; Schwartz, L.; Shi, Y.; et al. Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res. 2013, 41, e84. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, M.S.; Shankar, R.; Garg, R.; Jain, M. Bisulphite sequencing reveals dynamic DNA methylation under desiccation and salinity stresses in rice cultivars. Genomics 2020, 112, 3537–3548. [Google Scholar] [CrossRef]
- Cui, N.; Chen, X.; Shi, Y.; Chi, M.; Hu, J.; Lai, K.; Wang, Z.; Wang, H. Changes in the epigenome and transcriptome of rice in response to Magnaporthe oryzae infection. Crop J. 2021, 9, 843–853. [Google Scholar] [CrossRef]
- Cao, S.; Chen, K.; Lu, K.; Chen, S.; Zhang, X.; Shen, C.; Zhu, S.; Niu, Y.; Fan, L.; Chen, Z.J.; et al. Asymmetric variation in DNA methylation during domestication and de-domestication of rice. Plant Cell 2023, 35, 3429–3443. [Google Scholar] [CrossRef]
- Sasaki, T. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [Google Scholar] [CrossRef]
- Hamilton, J.P.; Li, C.; Buell, C.R. The rice genome annotation project: An updated database for mining the rice genome. Nucleic Acids Res. 2025, 53, D1614–D1622. [Google Scholar] [CrossRef]
- Hu, J.; Chen, X.; Zhang, H.; Ding, Y. Genome-wide analysis of DNA methylation in photoperiod- and thermo-sensitive male sterile rice Peiai 64S. BMC Genom. 2015, 16, 102. [Google Scholar] [CrossRef]
- Zhang, L.; Cheng, Z.; Qin, R.; Qiu, Y.; Wang, J.L.; Cui, X.; Gu, L.; Zhang, X.; Guo, X.; Wang, D.; et al. Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 2012, 24, 4407–4421. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, P.; Chaudhuri, S. Analysis of DNA Methylation Profile in Plants by Chop-PCR. In Plant Innate Immunity: Methods and Protocols; Gassmann, W., Ed.; Springer: New York, NY, USA, 2019; pp. 79–90. ISBN 978-1-4939-9458-8. [Google Scholar]
- Li, J.; Yang, D.L.; Huang, H.; Zhang, G.; He, L.; Pang, J.; Lozano-Durán, R.; Lang, Z.; Zhu, J.K. Epigenetic memory marks determine epiallele stability at loci targeted by de novo DNA methylation. Nat. Plants 2020, 6, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Knowledge of Rice Growth Stages Important. Available online: https://www.lsuagcenter.com/portals/our_offices/research_stations/rice/features/publications/knowledge-of-rice-growth-stages-important (accessed on 18 June 2025).
- Jiang, W.; Zhou, Z.; Li, X.; Zhao, Y.; Zhou, S. DNA methylation dynamics play crucial roles in shaping the distinct transcriptomic profiles for different root-type initiation in rice. Genome Biol. 2025, 26, 99. [Google Scholar] [CrossRef]
- Zhang, C.C.; Yuan, W.Y.; Zhang, Q.F. RPL1, a gene involved in epigenetic processes regulates phenotypic plasticity in rice. Mol. Plant 2012, 5, 482–493. [Google Scholar] [CrossRef][Green Version]
- Lu, Y.; Gharib, A.; Chen, R.; Wang, H.; Tao, T.; Zuo, Z.; Bu, Q.; Su, Y.; Li, Y.; Luo, Y.; et al. Rice melatonin deficiency causes premature leaf senescence via DNA methylation regulation. Crop J. 2024, 12, 721–731. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, Y.; Luo, W.; Li, W.; Chen, N.; Zhang, D.; Chong, K. The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 2013, 163, 1673–1685. [Google Scholar] [CrossRef]
- Abe, M.; Yoshikawa, T.; Nosaka, M.; Sakakibara, H.; Sato, Y.; Nagato, Y.; Itoh, J. WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining microRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol. 2010, 154, 1335–1346. [Google Scholar] [CrossRef]
- Xu, L.; Yuan, K.; Yuan, M.; Meng, X.; Chen, M.; Wu, J.; Li, J.; Qi, Y. Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol. Plant 2020, 13, 851–863. [Google Scholar] [CrossRef]
- Li, W.; Han, Y.; Tao, F.; Chong, K. Knockdown of SAMS genes encoding S-adenosyl-l-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice. J. Plant Physiol. 2011, 168, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.C.; Shi, D.Q.; Chen, Y.H. Female gametophyte development in flowering plants. Annu. Rev. Plant Biol. 2010, 61, 89–108. [Google Scholar] [CrossRef]
- Hafidh, S.; Honys, D. Reproduction multitasking: The male gametophyte. Annu. Rev. Plant Biol. 2021, 72, 581–614. [Google Scholar] [CrossRef]
- Ono, A.; Yamaguchi, K.; Fukada-Tanaka, S.; Terada, R.; Mitsui, T.; Iida, S. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012, 71, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, K.; Zeng, L.; Xu, D.; Zhu, T.; Yin, Y.; Zhan, H.; Wu, Y.; Yang, D.-L. Reinforcement of CHH methylation through RNA-directed DNA methylation ensures sexual reproduction in rice. Plant Physiol. 2021, 188, 1189–1209. [Google Scholar] [CrossRef]
- Ren, D.; Ding, C.; Qian, Q. Molecular bases of rice grain size and quality for optimized productivity. Sci. Bull. 2023, 68, 314–350. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Pan, T.; Zhu, Y.; Jiang, X.; Yu, M.; Wang, R.; Zhang, F.; Luo, S.; Bao, X.; Chen, Y.; et al. FLOURY ENDOSPERM20 encoding SHMT4 is required for rice endosperm development. Plant Biotechnol. J. 2022, 20, 1438–1440. [Google Scholar] [CrossRef]
- Guo, H.; Wu, T.; Li, S.; He, Q.; Yang, Z.; Zhang, W.; Gan, Y.; Sun, P.; Xiang, G.; Zhang, H.; et al. The methylation patterns and transcriptional responses to chilling stress at the seedling stage in rice. Int. J. Mol. Sci. 2019, 20, 5089. [Google Scholar] [CrossRef]
- Kim, W.; Iizumi, T.; Nishimori, M. Global patterns of crop production losses associated with droughts from 1983 to 2009. J. Appl. Meteorol. Climatol. 2019, 58, 1233–1244. [Google Scholar] [CrossRef]
- Kou, S.; Gu, Q.; Duan, L.; Liu, G.; Yuan, P.; Li, H.; Wu, Z.; Liu, W.; Huang, P.; Liu, L. Genome-wide bisulphite sequencing uncovered the contribution of DNA methylation to rice short-term drought memory formation. J. Plant Growth Regul. 2022, 41, 2903–2917. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Xia, H.; Wei, H.; Lou, Q.; Li, M.; Li, T.; Luo, L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017, 7, 39843. [Google Scholar] [CrossRef]
- Wang, J.; Nan, N.; Li, N.; Liu, Y.; Wang, T.J.; Hwang, I.; Liu, B.; Xu, Z.Y. A DNA methylation reader–chaperone regulator–transcription factor complex activates OsHKT1;5 expression during salinity stress. Plant Cell 2020, 32, 3535–3558. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Fahad, M.; Zhang, L.; Wu, L.; Wu, X. Microrchidia OsMORC6 positively regulates cadmium tolerance and uptake by mediating DNA methylation in rice. Rice 2025, 18, 25. [Google Scholar] [CrossRef]
- Cong, W.; Miao, Y.; Xu, L.; Zhang, Y.; Yuan, C.; Wang, J.; Zhuang, T.; Lin, X.; Jiang, L.; Wang, N.; et al. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 282. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhu, J.; Guo, R.; Whelan, J.; Shou, H. DNA methylation is involved in acclimation to iron-deficiency in rice (Oryza sativa). Plant J. 2021, 107, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xing, L.; Li, Z.; Jiang, D. Epigenetic control of plant abiotic stress responses. J. Genet. Genom. 2025, 52, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Wang, L.; Han, T.; Ye, W.; Liu, Y.; Sun, Y.; Moose, S.P.; Song, Q.; Chen, Z.J. Small RNAs mediate transgenerational inheritance of genome-wide trans-acting epialleles in maize. Genome Biol. 2022, 23, 53. [Google Scholar] [CrossRef] [PubMed]
- Andreoni, I.; Coughlin, M.W.; Perley, D.A.; Yao, Y.; Lu, W.; Cenko, S.B.; Kumar, H.; Anand, S.; Ho, A.Y.Q.; Kasliwal, M.M.; et al. A very luminous jet from the disruption of a star by a massive black hole. Nature 2022, 612, 430–434. [Google Scholar] [CrossRef]
- Li, Y.; Xia, Q.; Kou, H.; Wang, D.; Lin, X.; Wu, Y.; Xu, C.; Xing, S.; Liu, B. Induced Pib expression and resistance to Magnaporthe grisea are compromised by cytosine demethylation at critical promoter regions in rice. J. Integr. Plant Biol. 2011, 53, 814–823. [Google Scholar] [CrossRef]
- Sheng, C.; Li, X.; Xia, S.; Zhang, Y.; Yu, Z.; Tang, C.; Xu, L.; Wang, Z.; Zhang, X.; Zhou, T.; et al. An OsPRMT5-OsAGO2/miR1875-OsHXK1 module regulates rice immunity to blast disease. J. Integr. Plant Biol. 2023, 65, 1077–1095. [Google Scholar] [CrossRef]
- Campo, S.; Sánchez-Sanuy, F.; Camargo-Ramírez, R.; Gómez-Ariza, J.; Baldrich, P.; Campos-Soriano, L.; Soto-Suárez, M.; San Segundo, B. A novel Transposable element-derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol. J. 2021, 19, 1798–1811. [Google Scholar] [CrossRef]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef]
- Zhang, H.; Tao, Z.; Hong, H.; Chen, Z.; Wu, C.; Li, X.; Xiao, J.; Wang, S. Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants 2016, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhou, Y.; Mao, L.; Ye, C.; Wang, W.; Zhang, J.; Yu, Y.; Fu, F.; Wang, Y.; Qian, F.; et al. Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat. Commun. 2017, 8, 15323. [Google Scholar] [CrossRef]
- Yan, T.; Kuang, L.; Gao, F.; Chen, J.; Li, L.; Wu, D. Differentiation of genome-wide DNA methylation between japonica and indica rice. Plant J. 2025, 121, e17218. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, Y.; Gui, Y.; An, D.; Liu, J.; Xu, X.; Li, Q.; Wang, J.; Wang, W.; Shi, C.; et al. Elimination of a retrotransposon for quenching genome instability in modern rice. Mol. Plant 2019, 12, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Xing, M.; Zhao, Z.; Gu, Y.; Xiao, Y.; Liu, Q.; Xue, H. DNA methylation modification in heterosis initiation through analyzing rice hybrid contemporary seeds. Crop J. 2021, 9, 1179–1190. [Google Scholar] [CrossRef]
- Ma, X.; Xing, F.; Jia, Q.; Zhang, Q.; Hu, T.; Wu, B.; Shao, L.; Zhao, Y.; Zhang, Q.; Zhou, D.X. Parental variation in CHG methylation is associated with allelic-specific expression in elite hybrid rice. Plant Physiol. 2021, 186, 1025–1041. [Google Scholar] [CrossRef]
- Sigurpalsdottir, B.D.; Stefansson, O.A.; Holley, G.; Beyter, D.; Zink, F.; Hardarson, M.Þ.; Sverrisson, S.Þ.; Kristinsdottir, N.; Magnusdottir, D.N.; Magnusson, O.Þ.; et al. A comparison of methods for detecting DNA methylation from long-read sequencing of human genomes. Genome Biol. 2024, 25, 69. [Google Scholar] [CrossRef]
- Liu, J.; Zhong, X. Population epigenetics: DNA methylation in the plant omics era. Plant Physiol. 2024, 194, 2039–2048. [Google Scholar] [CrossRef]
- Rozov, S.M.; Permyakova, N.V.; Deineko, E.V. The problem of the low rates of CRISPR/Cas9-mediated knock-ins in plants: Approaches and solutions. Int. J. Mol. Sci. 2019, 20, 3371. [Google Scholar] [CrossRef]
- Yan, N.; Feng, H.; Sun, Y.; Xin, Y.; Zhang, H.; Lu, H.; Zheng, J.; He, C.; Zuo, Z.; Yuan, T.; et al. Cytosine base editors induce off-target mutations and adverse phenotypic effects in transgenic mice. Nat. Commun. 2023, 14, 1784. [Google Scholar] [CrossRef]
- Shankar, S.; Sreekumar, A.; Prasad, D.; Das, A.V.; Pillai, M.R. Genome editing of oncogenes with ZFNs and TALENs: Caveats in nuclease design. Cancer Cell Int. 2018, 18, 169. [Google Scholar] [CrossRef]
- Gong, Z.; Previtera, D.A.; Wang, Y.; Botella, J.R. Geminiviral-induced genome editing using miniature CRISPR/Cas12j (CasΦ) and Cas12f variants in plants. Plant Cell Rep. 2024, 43, 71. [Google Scholar] [CrossRef]
- Tang, X.; Ren, Q.; Yan, X.; Zhang, R.; Liu, L.; Han, Q.; Zheng, X.; Qi, Y.; Song, H.; Zhang, Y. Boosting genome editing in plants with single transcript unit surrogate reporter systems. Plant Commun. 2024, 5, 100921. [Google Scholar] [CrossRef]
- Přibylová, A.; Fischer, L.; Pyott, D.; Bassett, A.; Molnar, A. DNA methylation can alter CRISPR/Cas9 editing frequency and DNA repair outcome in a target-specific manner. New Phytol. 2022, 235, 2285–2299. [Google Scholar] [CrossRef]
- Aussel, C.; Cathomen, T.; Fuster-García, C. The hidden risks of CRISPR/Cas: Structural variations and genome integrity. Nat. Commun. 2025, 16, 7208. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Yang, C.; Wang, D.; Deng, X.; Cao, X.; Song, X. Targeted DNA demethylation produces heritable epialleles in rice. Sci. China Life Sci. 2022, 65, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Cao, P.; Wang, C.; Lai, J.; Deng, Y.; Li, C.; Hao, Y.; Wu, Z.; Chen, R.; Qiang, Q.; et al. Population analysis reveals the roles of DNA methylation in tomato domestication and metabolic diversity. Sci. China Life Sci. 2023, 66, 1888–1902. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Yu, J.; Li, X.; Li, J.; Fan, J.; Liu, H.; Wei, W.; Zhang, L.; Gu, K.; Liu, D.; et al. Increased DNA methylation contributes to the early ripening of pear fruits during domestication and improvement. Genome Biol. 2024, 25, 87. [Google Scholar] [CrossRef]
| No. | Gene in Rice | Function Analysis | Reference |
|---|---|---|---|
| 1 | OsDMR2 | Plant height, fertility, number of tillers | [57] |
| 2 | OsCMT3a | Plant height, heading time, grain filling rate, zygote development | [58,59] |
| 3 | OsCMT3b | Zygote development | [59] |
| 4 | OsMET1a | / | [60] |
| 5 | OsMET1b | / | [60] |
| 6 | OsDDM1a | Plant height, grain filling rate | [57] |
| 7 | OsDDM1b | Internode length, fertility, spike length, fruit set rate, grain shape, organ size | [61] |
| 8 | OsROS1a | Endosperm components, germ cell development, root development, zygote development | [62,63,64] |
| 9 | OsROS1b | Zygote development | [63] |
| 10 | OsROS1c | Zygote development, embryo development | [63] |
| 11 | OsDML4 | Heat resistance, endosperm components | [65,66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Li, W.-J.; Xu, J.-H. DNA Methylation in Rice: Mechanisms, Regulatory Roles, and Beyond. Int. J. Mol. Sci. 2025, 26, 8454. https://doi.org/10.3390/ijms26178454
Li T, Li W-J, Xu J-H. DNA Methylation in Rice: Mechanisms, Regulatory Roles, and Beyond. International Journal of Molecular Sciences. 2025; 26(17):8454. https://doi.org/10.3390/ijms26178454
Chicago/Turabian StyleLi, Ting, Wen-Jing Li, and Jian-Hong Xu. 2025. "DNA Methylation in Rice: Mechanisms, Regulatory Roles, and Beyond" International Journal of Molecular Sciences 26, no. 17: 8454. https://doi.org/10.3390/ijms26178454
APA StyleLi, T., Li, W.-J., & Xu, J.-H. (2025). DNA Methylation in Rice: Mechanisms, Regulatory Roles, and Beyond. International Journal of Molecular Sciences, 26(17), 8454. https://doi.org/10.3390/ijms26178454






