Functional Characterization of OsCSN1 in the Agronomic Trait Control of Rice Seedlings Under Far-Red Light
Abstract
1. Introduction
2. Results
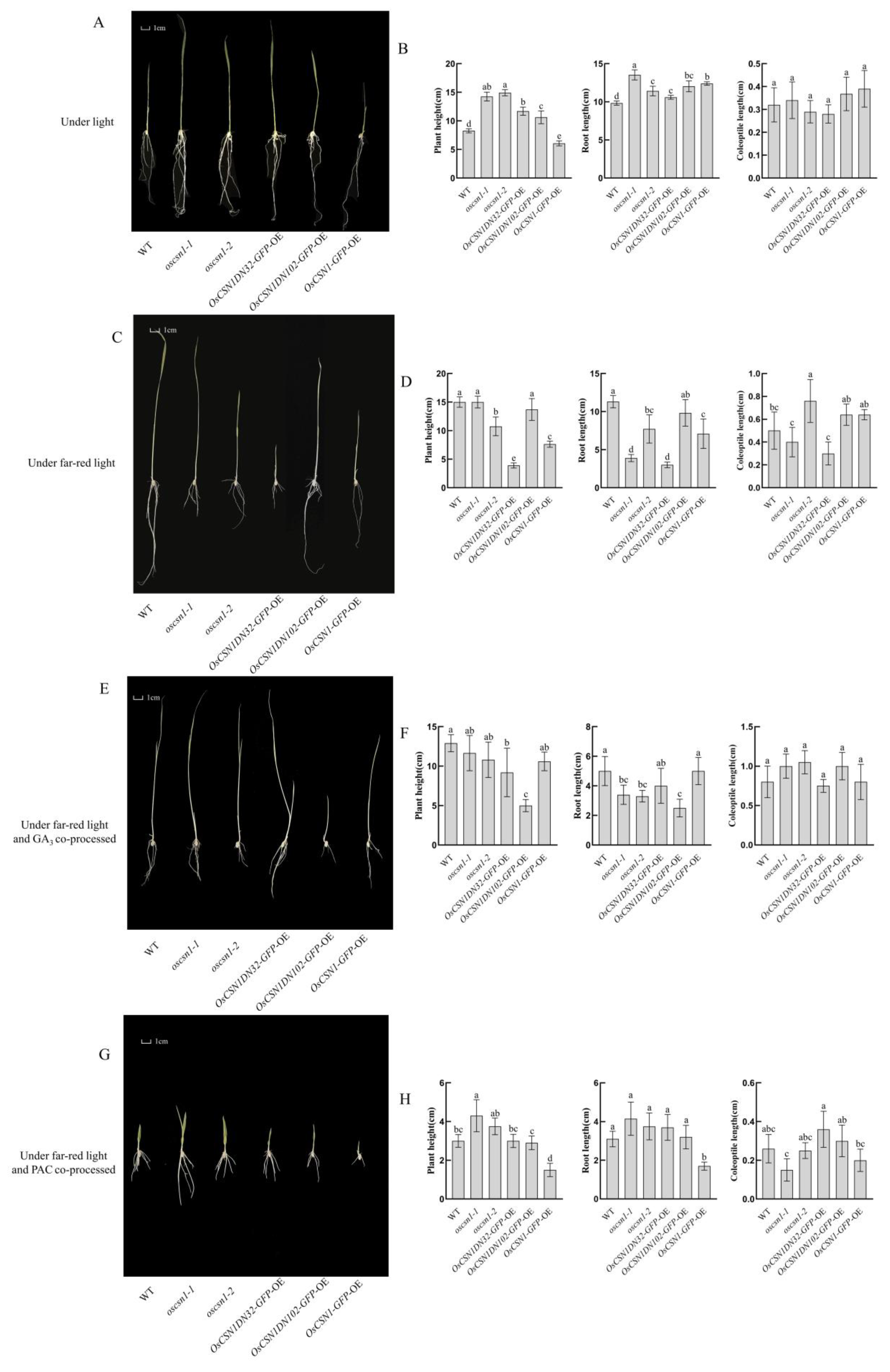
2.1. OsCSN1 Is a Positive Regulator of Plant Height in Rice Seedlings Under Far-Red Light
2.2. OsCSN1 Affects the Length of the Coleoptile by Far-Red Light Through the N-Terminal
2.3. OsCSN1 Inhibits Root Length Elongation Mediated by Far-Red Light Through the N-Terminal
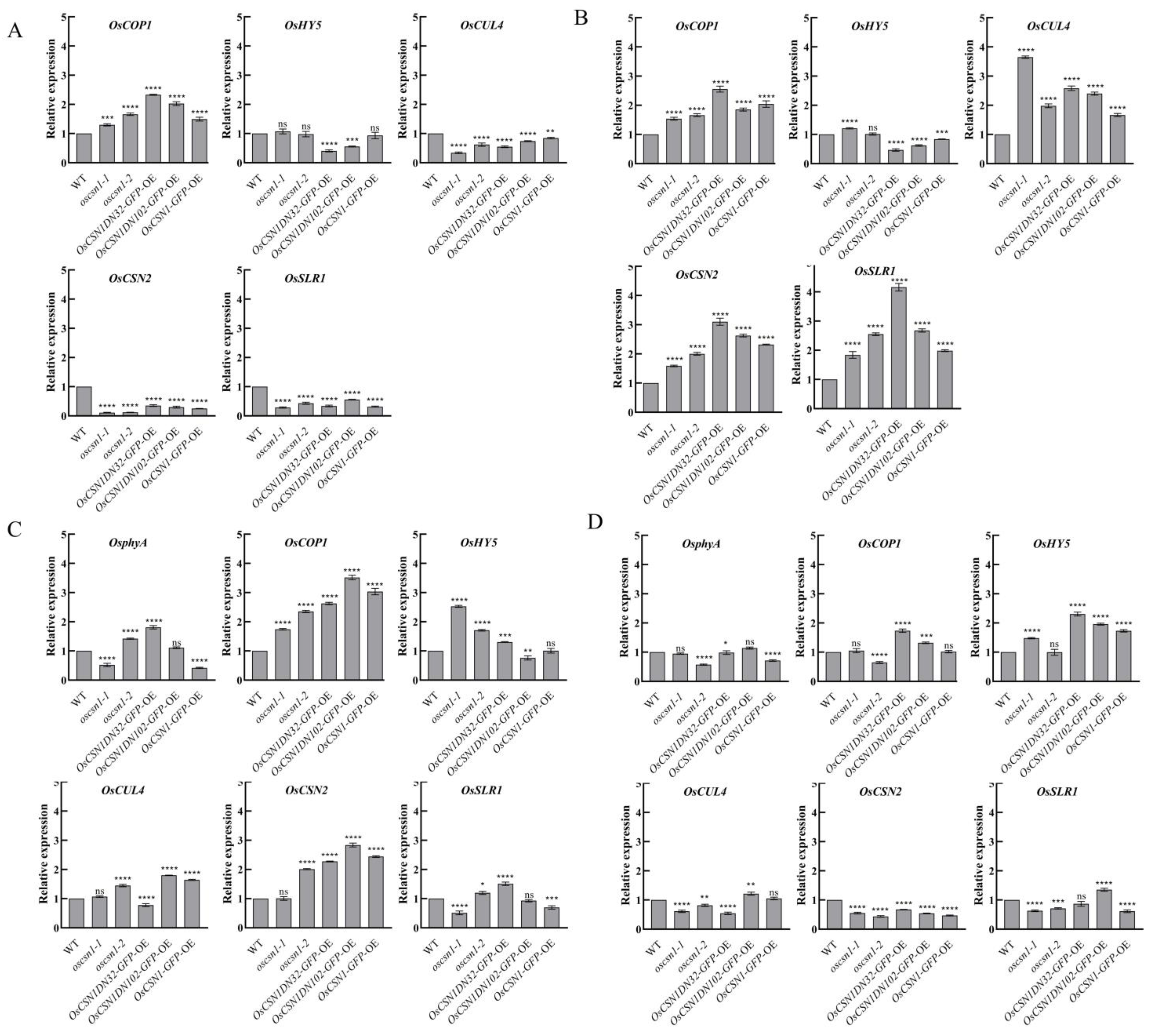
2.4. Far-Red Light Signals and Hormone Signals Affect the Expression of Proteins and Genes in Mutants
3. Discussion
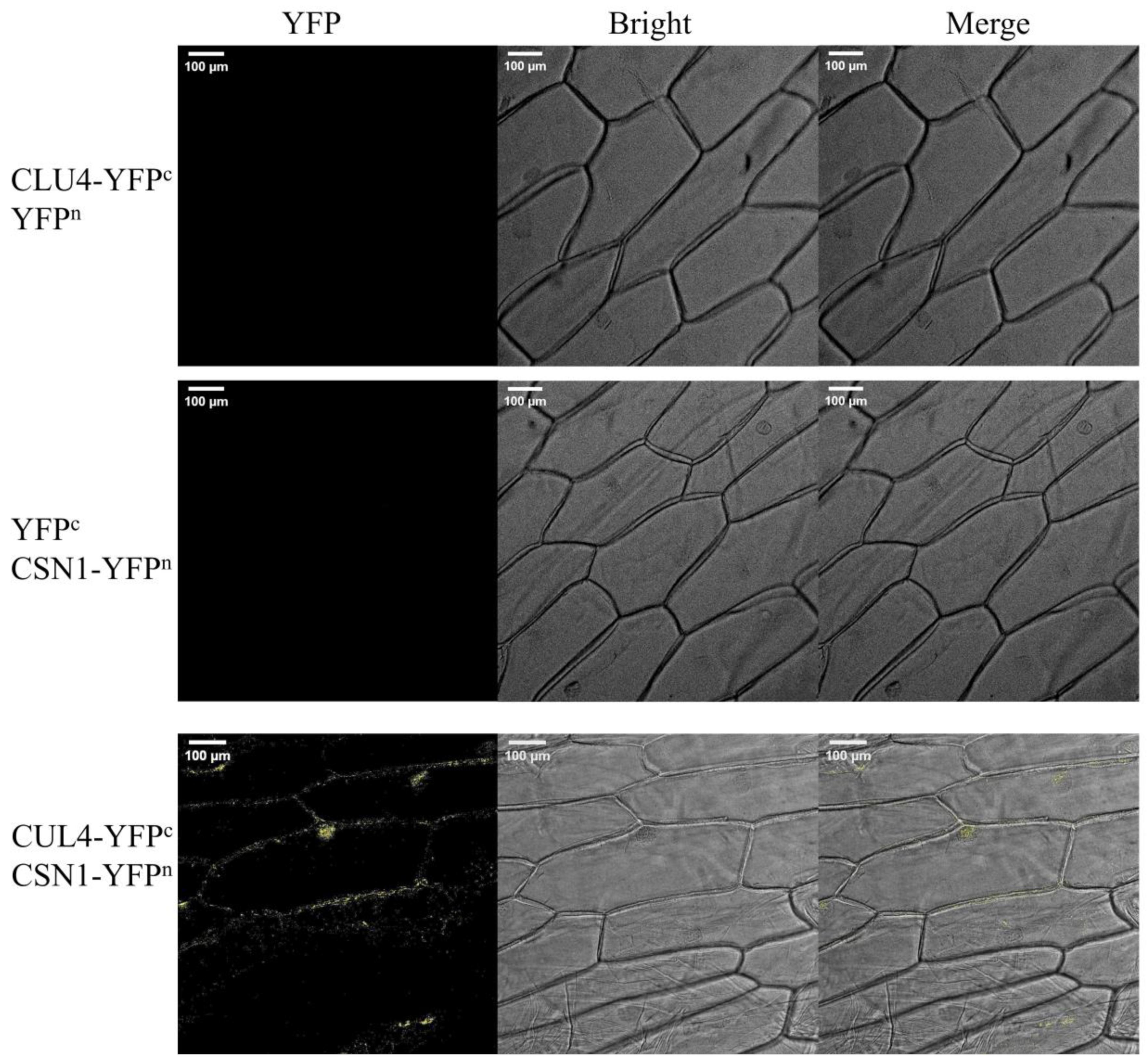
3.1. OsCSN1 Localization Affects Rice Seedling Growth
3.2. OsCSN1 Is Involved in the Regulation of Rice Plant Phenotype Through N-Terminal Under Far-Red Light and There Are Different Sensitivities and Synergistic Effects with Hormone Signals
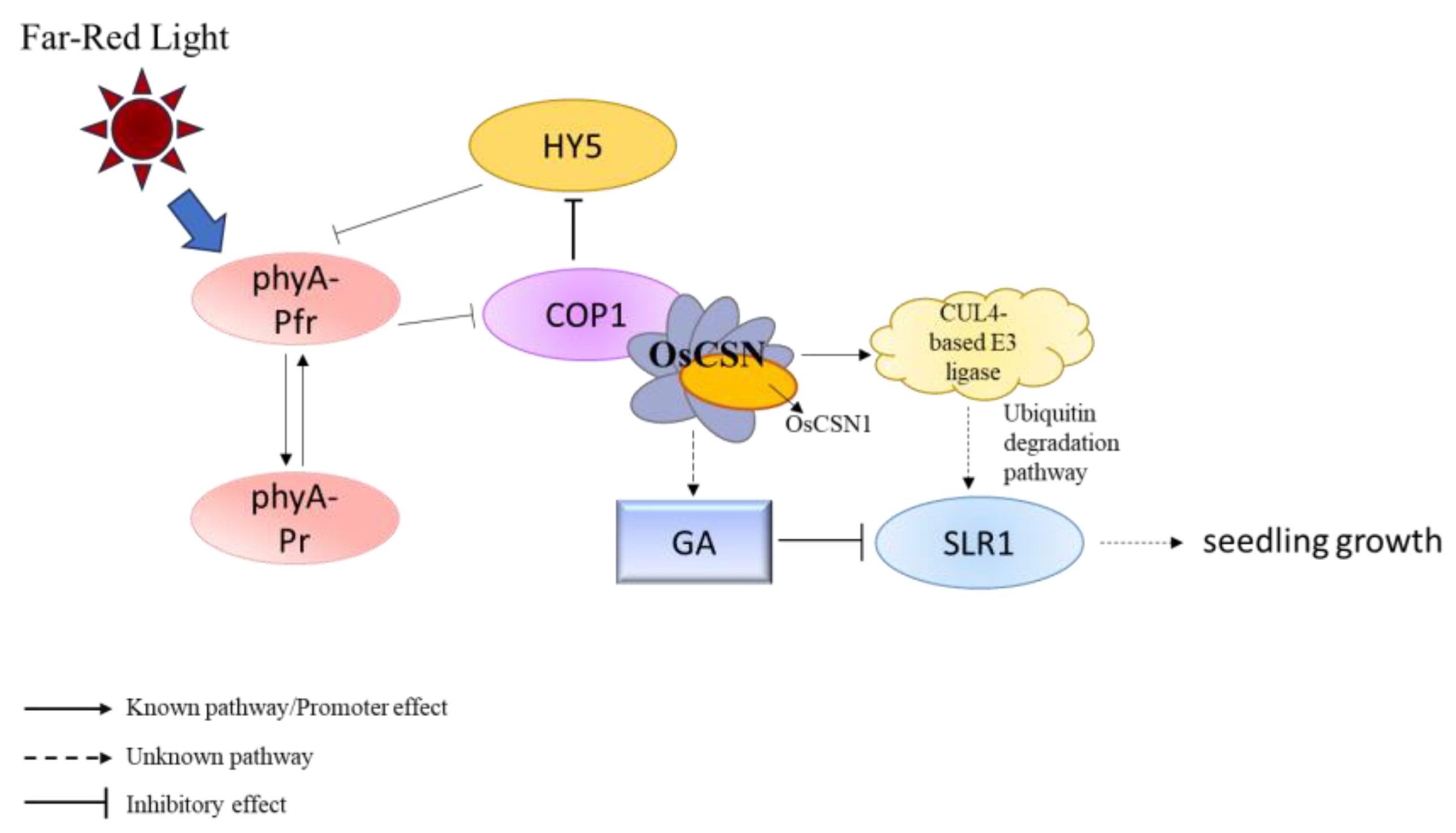
3.3. OsCSN1 Responds to Far-Red Light by Regulating the SLR1 Factor in the GA Pathway
3.4. OsCSN1 Senses Far-Red Light Signals Through phyA to Regulate Seedling Growth
3.5. Limitations and Potential of Far-Red Light in Regulating Rice Growth
4. Materials and Methods
4.1. Plant Material and Culture Conditions
4.2. Phenotypic Analysis of Rice Seedlings
4.3. Protein Extraction and Western Blot Analysis
4.4. Total RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction
4.5. Spatiotemporal Positioning
4.6. Bimolecular Fluorescence Complementation
4.7. Data Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wei, N.; Chamovitz, D.A.; Deng, X.W. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 1994, 78, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Wei, N.; Deng, X. COP9: A new genetic locus involved in light-regulated development and gene expression in arabidopsis. Plant Cell 1992, 4, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Bech-Otschir, D.; Seeger, M.; Dubiel, W. The COP9 signalosome: At the interface between signal transduction and ubiquitin-dependent proteolysis. J. Cell Sci. 2002, 115 Pt 3, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Deng, X. COP9 signalosome revisited: A novel mediator of protein degradation. Trends Cell Biol. 2001, 11, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Serino, G.; Deng, X. The COP9 signalosome: Regulating plant development through the control of proteolysis. Annu. Rev. Plant Biol. 2003, 54, 165–182. [Google Scholar] [CrossRef]
- Sharon, M.; Mao, H.; Boeri Erba, E.; Stephens, E.; Zheng, N.; Robinson, C. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure 2009, 17, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Kotiguda, G.; Weinberg, D.; Dessau, M.; Salvi, C.; Serino, G.; Chamovitz, D.; Hirsch, J. The organization of a CSN5-containing subcomplex of the COP9 signalosome. J. Biol. Chem. 2012, 287, 42031–42041. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Bucher, P. The PCI domain: A common theme in three multiprotein complexes. Trends Biochem. Sci. 1998, 23, 204–205. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Huang, X.; Gusmaroli, G.; Terzaghi, W.; Lau, O.S.; Yanagawa, Y.; Zhang, Y.; Li, J.; Lee, J.H.; Zhu, D.; et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 2010, 22, 108–123. [Google Scholar] [CrossRef]
- Tsuge, T.; Matsui, M.; Wei, N. The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J. Mol. Biol. 2001, 305, 1–9. [Google Scholar] [CrossRef]
- Wang, X.; Kang, D.; Feng, S.; Serino, G.; Schwechheimer, C.; Wei, N. CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: A structure-function study of CSN1 subunit of COP9 signalosome. Mol. Biol. Cell 2002, 13, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Wu, M.; Li, B.; Bücker, B.; Keil, P.; Zhang, S.; Li, J.; Kang, D.; Liu, J.; Dong, J.; et al. The COP9 Signalosome regulates seed germination by facilitating protein degradation of RGL2 and ABI5. PLoS Genet. 2018, 14, e1007237. [Google Scholar] [CrossRef] [PubMed]
- Wei, N.; Deng, X. The role of the COP/DET/FUS genes in light control of arabidopsis seedling development. Plant Physiol. 1996, 112, 871–878. [Google Scholar] [CrossRef]
- Osterlund, M.; Hardtke, C.; Wei, N.; Deng, X. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lopez-Molina, L. Control of seed germination in the shade. Cell Cycle 2012, 11, 4489–4490. [Google Scholar] [CrossRef][Green Version]
- Bae, G.; Choi, G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef] [PubMed]
- Takano, M.; Inagaki, N.; Xie, X.; Yuzurihara, N.; Hihara, F.; Ishizuka, T.; Yano, M.; Nishimura, M.; Miyao, A.; Hirochika, H.; et al. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 2005, 17, 3311–3325. [Google Scholar] [CrossRef] [PubMed]
- Kneissl, J.; Shinomura, T.; Furuya, M.; Bolle, C. A rice phytochrome A in Arabidopsis: The Role of the N-terminus under red and far-red light. Mol. Plant 2008, 1, 84–102. [Google Scholar] [CrossRef] [PubMed]
- Kircher, S.; Kozma-Bognar, L.; Kim, L.; Adam, E.; Harter, K.; Schafer, E.; Nagy, F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 1999, 11, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Sharrock, R.; Clack, T. Heterodimerization of type II phytochromes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 11500–11505. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Watanabe, E.; Tokutomi, S.; Nagatani, A.; Chua, N. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004, 18, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Clough, R.; Jordan-Beebe, E.; Lohman, K.; Marita, J.; Walker, J.; Gatz, C.; Vierstra, R. Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 1999, 17, 155–167. [Google Scholar] [CrossRef]
- Hoecker, U.; Xu, Y.; Quail, P. SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 1998, 10, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Matsui, M.; Wei, N.; Wagner, D.; Chu, A.; Feldmann, K.; Quail, P. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 1992, 71, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Sheerin, D.; Menon, C.; zur Oven-Krockhaus, S.; Enderle, B.; Zhu, L.; Johnen, P.; Schleifenbaum, F.; Stierhof, Y.; Huq, E.; Hiltbrunner, A. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 2015, 27, 189–201. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.; Tan, G.; Zhou, W.; Wang, G. Gibberellin and the plant growth retardant Paclobutrazol altered fruit shape and ripening in tomato. Protoplasma 2020, 257, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Dill, A.; Thomas, S.; Hu, J.; Steber, C.; Sun, T. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 2004, 16, 1392–1405. [Google Scholar] [CrossRef]
- Peng, J.; Harberd, N. The role of GA-mediated signalling in the control of seed germination. Curr. Opin. Plant Biol. 2002, 5, 376–381. [Google Scholar] [CrossRef]
- Dai, C.; Xue, H. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Bao, A.; Zeng, H.; Huang, G.; Geng, M.; Zhang, C.; Zhang, Q.; Lu, J.; Wu, M.; et al. OsCSN1 regulates the growth of rice seedlings through the GA signaling pathway in blue light. J. Plant Physiol. 2023, 280, 153904. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Liu, Y.; Bao, A.; Jiao, T.; Zeng, H.; Yue, W.; Yin, L.; Xu, M.; Lu, J.; Wu, M.; et al. A Characterization of the Functions of OsCSN1 in the Control of Sheath Elongation and Height in Rice Plants under Red Light. Agronomy 2024, 14, 572. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C.; Chien, C.; Hsieh, H. FAR-RED INSENSITIVE219 modulates CONSTITUTIVE PHOTOMORPHOGENIC1 activity via physical interaction to regulate hypocotyl elongation in Arabidopsis. Plant Physiol. 2011, 156, 631–646. [Google Scholar] [CrossRef]
- Takano, M.; Inagaki, N.; Xie, X.; Kiyota, S.; Baba-Kasai, A.; Tanabata, T.; Shinomura, T. Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. Proc. Natl. Acad. Sci. USA 2009, 106, 14705–14710. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Tanabata, T.; Xie, X.; Inagaki, N.; Takano, M.; Shinomura, T.; Yamamoto, K.T. Phytochrome-mediated growth inhibition of seminal roots in rice seedlings. Physiol. Plant. 2009, 137, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Andel, F.; Lagarias, J.; Mathies, R. Resonance raman analysis of chromophore structure in the lumi-R photoproduct of phytochrome. Biochemistry 1996, 35, 15997–16008. [Google Scholar] [CrossRef]
- Hiltbrunner, A.; Tscheuschler, A.; Viczián, A.; Kunkel, T.; Kircher, S.; Schäfer, E. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006, 47, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Jang, I.; Lee, J.; Shin., E.; Nam, S. Evaluation of Growth, Flowering, and Chlorophyll Fluorescence Responses of Viola cornuta cv. Penny Red Wing according to Spectral Power Distributions. J. People Plants Environ. 2023, 26, 335–349. [Google Scholar] [CrossRef]
- Musazade, E.; Liu, Y.; Ren, Y.; Wu, M.; Zeng, H.; Han, S.; Gao, X.; Chen, S.; Guo, L. OsCSN1 Regulates the Growth and Development of Rice Seedlings through the Degradation of SLR1 in the GA Signaling Pathway. Agronomy 2022, 12, 2946. [Google Scholar] [CrossRef]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar] [CrossRef] [PubMed]

| Primer | Sequence (5′-3′) |
|---|---|
| GAPDHF | AAGCCAGCATCCTATGATCAGATT |
| GAPDHR | CGTAACCCAGAATACCCTTGAGTTT |
| Dye-SLR1F | CATGCTTTCCGAGCTCAACG |
| Dye-SLR1R | TGACAGTGGACGAGGTGGAA |
| Dye-CUL4F | AGGACAGACAGTATCAGGTGGATGC |
| Dye-CUL4R | TCCGATGGCTTGATTGGGAACTTG |
| Dye-COP1F | CATCTCAGCCACAAGAGCGACTG |
| Dye-COP1R | GGTCTATCGGTGATGCTGTCTTCG |
| Dye-CSN2F | GAGCAGCTCTTGGTCTCACTCATTC |
| Dye-CSN2R | CGACCTGTCACCACGTTCTAGTAAC |
| Dye-phyAF | GATGGTGCTCTGAGTGGAATGC |
| Dye-phyAR | ACAGGAGGCGTTGGTGCTATC |
| Dye-HY5F | AGGTGAAGGTGAAGGACTTGGAG |
| Dye-HY5R | GAGCATCTGGTTCTCATTCTGTAGG |
| Primer | Sequence (5′-3′) |
|---|---|
| CSN1-c/nEYFP-F | GATCTCGAGCTCAAGCTTCGAATTCATGGACGTCGAGGGCGAGGTCCCGG |
| CSN1-nEYFP-R | TCGCCCTTGCTCACCATCAGGATCCCATCTTCCTTTGTCCAGCTCTTTGG |
| CUL4-c/nEYFP-F | GATCTCGAGCTCAAGCTTCGAATTCATGCACAAAAACTAAGCTTC |
| CUL4-cEYFP-R | GCGAGCTGCACGCTGCCCAGGATCCAGCCAGGTAATTGTAGATCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zeng, H.; Shang, Y.; Zhang, H.; Jiao, T.; Yin, L.; Yang, J.; Xu, M.; Lu, J.; Wu, M.; et al. Functional Characterization of OsCSN1 in the Agronomic Trait Control of Rice Seedlings Under Far-Red Light. Int. J. Mol. Sci. 2025, 26, 522. https://doi.org/10.3390/ijms26020522
Liu Y, Zeng H, Shang Y, Zhang H, Jiao T, Yin L, Yang J, Xu M, Lu J, Wu M, et al. Functional Characterization of OsCSN1 in the Agronomic Trait Control of Rice Seedlings Under Far-Red Light. International Journal of Molecular Sciences. 2025; 26(2):522. https://doi.org/10.3390/ijms26020522
Chicago/Turabian StyleLiu, Yanxi, Hua Zeng, Yuqing Shang, Hexin Zhang, Tongtong Jiao, Le Yin, Jinyuan Yang, Miao Xu, Jingmei Lu, Ming Wu, and et al. 2025. "Functional Characterization of OsCSN1 in the Agronomic Trait Control of Rice Seedlings Under Far-Red Light" International Journal of Molecular Sciences 26, no. 2: 522. https://doi.org/10.3390/ijms26020522
APA StyleLiu, Y., Zeng, H., Shang, Y., Zhang, H., Jiao, T., Yin, L., Yang, J., Xu, M., Lu, J., Wu, M., & Guo, L. (2025). Functional Characterization of OsCSN1 in the Agronomic Trait Control of Rice Seedlings Under Far-Red Light. International Journal of Molecular Sciences, 26(2), 522. https://doi.org/10.3390/ijms26020522





