DNA Methylation in Pituitary Adenomas: A Scoping Review
Abstract
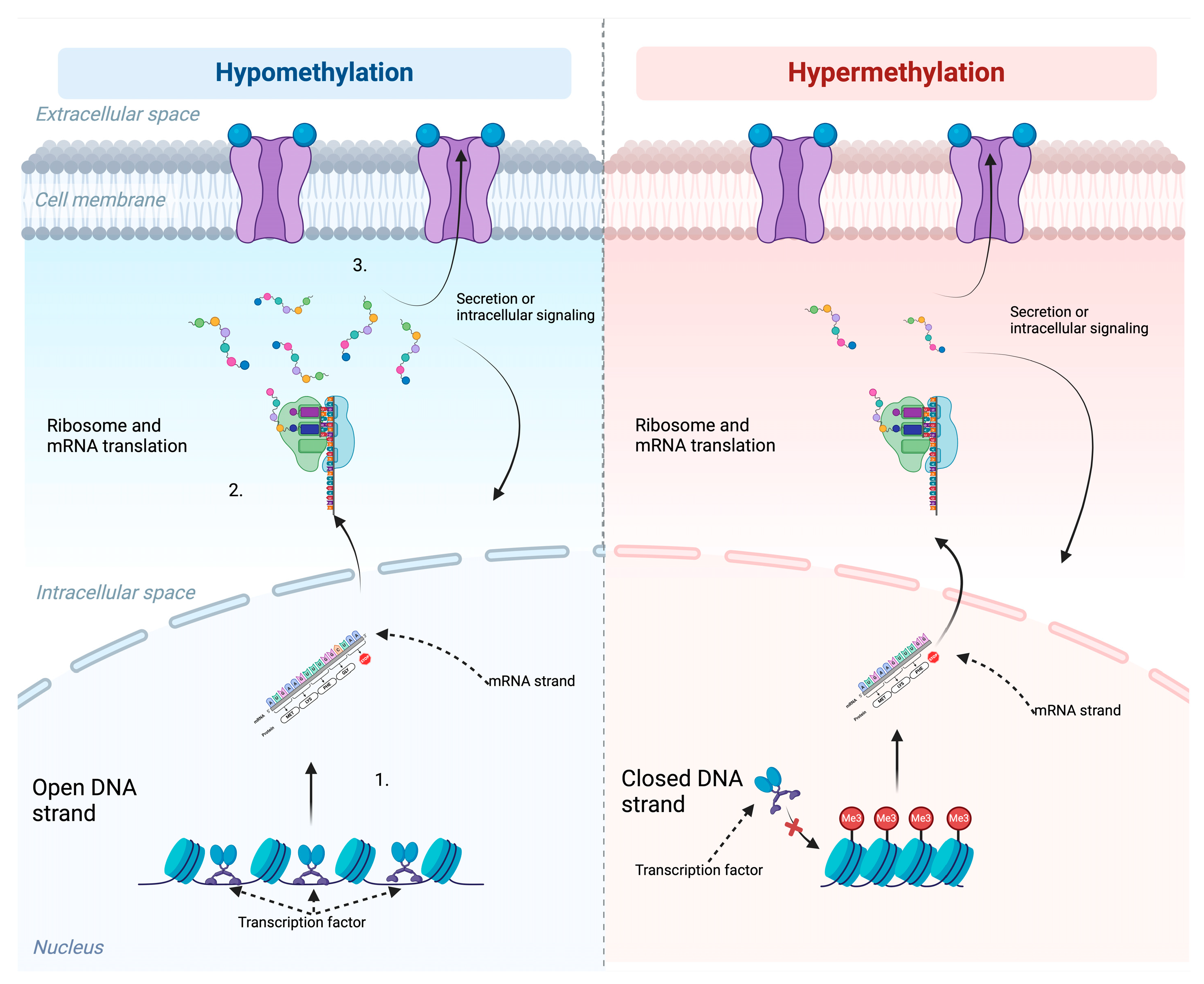
1. Introduction
1.1. Three Main Methods to Perform DNA Methylation
1.1.1. Methylation-Restricted Digestion
1.1.2. Methylation-Specific Polymerase Chain Reaction (MSP)
1.1.3. Chip-Based DNA Methylation Analysis
2. Results
2.1. Literature Screening and Eligibility
2.2. Study Characteristics
2.2.1. Methylation-Restricted Digestion
2.2.2. Methylation-Specific PCR
2.2.3. Chip-Based DNA Methylation Analysis
| Author/Year | Aim | Sample Size | Key Findings |
|---|---|---|---|
| Duong et al. [94]/2012 | Methylation profile of 27,578 CpG sites spanning more than 14,000 genes in each of the major pituitary adenoma subtypes | 7 GH-secreting tumors, 6 corticotrophinomas, 6 prolactinomas (PRL), and 13 non-functioning (NF) adenomas | First and unbiased survey of the pituitary tumor epigenome across different adenoma subtypes |
| Ling et al. [95]/2014 | DNA methylation alterations between invasive and noninvasive PAs subtypes | 24 patients with surgically resected PAs | DNA methylation analysis of key candidate genes may potentially complement histopathological classification systems for PA subtypes |
| Gu et al. [105]/2016 | DNA methylation differences between invasive and non-invasive non-functioning PAs | 12 adenomas were included in the discovery cohort; 7 adenomas were included in an independent cohort | Epigenetic modification of key gene substrates might partially account for the invasion of non-functioning PAs |
| Kober et al. [109]/2018 | The role of aberrant methylation at particular loci for gene expression in PAs | 31 gonadotroph NFPAs, 2 NFPAs that were positive for gonadotropins (FSH, LH, a-subunit) and TSH, and 1 null-cell adenoma | Invasive NFPAs showed invasiveness-related aberrant epigenetic upregulation of ITPKB and downregulation of CNKSR1 |
| Boresowicz [110]/2018 | Incidence of TERT abnormalities and to assess their role in telomere lengthening in PAs | Tissue samples from 101 patients | Telomerase abnormalities do not play any special role in pathogenesis of pituitary tumors |
| Johann et al. [96]/2018 | Characterize molecular alterations of sellar region ATRTs in adults as compared to pituitary adenomas | 47 pituitary adenomas were evaluated | Sellar region ATRTs in adults form a clinically distinct entity with a different mutational spectrum |
| Salomon et al. [97]/2018 | DNA methylation data were generated from the three major subtypes of pituitary adenomas | 48 patients | DNA methylation alterations play a major role in the disease etiology |
| Kober et al. [111]/2019 | DNA methylation in the misregulation of gene expression in gonadotroph NFPAs | 32 patients | Genes with aberrant methylation in pituitary tumors—STAT5A, RHOD, GALNT9, RASSF1, CDKN1A, TP73, STAT3 and HMGA2. FAM163A, HIF3A, and PRSS8—were hypermethylated in NFPAs |
| Cheng et al. [21]/2019 | Integrated analyses of paired whole-genome DNA methylation and gene expression in PA | Retrospectively enrolled 68 patients | Methylation and expression levels of PHYHD1, LTBR, MYBPHL, C22orf42, PRR5, ANKDD1A, RAB13, CAMKV, KIFC3, WNT4, and STAT6 play a pivotal role in the invasive behavior of NFPA |
| Neou et al. [98]/2019 | A molecularly unbiased classification, further deciphering the pathways responsible for tumorigenesis from a single set of PitNETs | The methylome of 86 PitNETs of all types | Identified three groups associated with tumor type and secretion; in particular, POU1F1/PIT1-lineage tumors showed global hypomethylation |
| Cheng et al. [112]/2020 | DNA methylation and expression parameters to evaluate the regrowth of NFPA | 71 patients diagnosed with NFPA | 6 of 13 genes (FAM90A1, ETS2, STAT6, MYT1L, ING2, and KCNK1) were considered potential biomarkers associated with the regrowth of NFPA |
| Boresowicz et al. [106]/2020 | CpGs located in miRNA genes that have differential methylation levels in gonadotroph PitNETs | 34 PitNETs and 5 samples of normal pituitary | Epigenetic regulation and changes in miRNA expression play a significant role in pathogenesis of PitNETs |
| Taniguchi-Ponciano et al. [14]/2020 | Identify the cellular pathways involved in their tumorigenesis | 6 non-tumoral pituitaries and 42 PAs | A divergent PA origin that segregates transcriptomically into three distinct clusters depending on the specific transcription factors |
| Wei et al. [11]/2020 | Activated and inhibited pathways and related key genes in hpNFPAs versus NFPAs | Eight snap-frozen NFPA specimens (four hpNF-PAs and four NFPAs) | The DNA methylation and gene expression patterns of two highly proliferative NFPAs occurring at young ages were noticeably distinct to those of six other NFPAs |
| Mosella et al. [15]/2021 | Identify, characterize, and validate methylation-based signatures that define PitNETs according to clinicopathological features | DNA methylation data from PitNETs from three independent institutions and from our cohort at the Hermelin Brain Tumor Center (n = 23) | Methylation signatures distinguished PitNETs by adenohypophyseal cell lineages |
| Schmid et al. [115]/2021 | Molecular differences between the three histologic types using DNA methylation analysis | 47 neoplasms of the posterior pituitary gland | Only subtle DNA methylation differences among tumors of the posterior pituitary |
| Nadaf et al. [99]/2021 | Use generated data to gain insights into the initiation and development of PitBs by identifying pathways differentially altered | A total of 64 tumor and normal FFPE tissues | PitB samples formed a distinct cluster separate from the various pituitary adenoma subtypes |
| Hagel et al. [107]/2021 | 12 double PAs (DPA) with diverse hormone profiles were investigated regarding DNA methylation profile | 12 cases were identified among 3654 surgical specimens of adenoma | Global DNA methylation profiling may yield additional information in lesions that appear as null-cell adenomas immunohistochemically |
| Asuzu et al. [117]/2022 | A mechanism of tumorigenesis with therapeutic implications for CD | Three tumors investigated for methylation (EPIC) | There may exist histone modifications that contribute to the pathogenesis of wild-type CD that were not captured by DNA methylation approaches |
| Dottermusch et al. [118]/2022 (Case report) | The role of epigenomic analyses in the diagnostic workup of a challenging sellar lesion | 57-year-old male | Exemplifies benefits and limitations of epigenomic analyses in molecular diagnostics of posterior pituitary neoplasms |
| Hickman et al. [119]/2022 (Case report) | A functioning corticotroph tumor with admixed adrenocortical cells, providing novel methylation profiling data | 33-year-old male | The methylation profile of this tumor was unique but was placed within the T-SNE plots adenohypophyseal entities, with the closest match being corticotroph tumors |
| Giuffrida et al. [114]/2022 | Correlate the methylation status of NFPAs and GH-omas with their epidemiological and clinicopathological features | 21 PA samples (11 GH-omas, 10 NFPAs) | C7orf50, GNG7, and BAHCC1 genes, which were found to be methylated in pituitary tumor biology |
| Hallén et al. [108]/2022 | Whether DNA methylation pattern differs in NFPAs between patients with residual adenoma with postoperative progression | 28 tumors from the reintervention group and 21 tumors from the radiologically stable group | Methylation patterns associated with clinically significant tumor growth requiring reintervention |
| Silva-Júnior et al. [100]/2022 | Methylome and transcriptome analysis of the three major subtypes of surgically resectable PitNETs | 77 patients (46 NFPT and 31 functioning pituitary tumors) | Methylome and transcriptome data resulted in three clusters that were associated with each other and with 2017 and 2022 WHO classifications |
| Aydin et al. [24]/2022 | Evaluate the molecular profiling of NF-PitNETs at three biological levels | 34 NF-PitNET samples and 6 normal pituitary glands | Proposed hub proteins, including DCC, DLG5, ETS2, FOXO1, HBP1, HMGA2, PCGF3, PSME4, RBPMS, RREB1, SMAD1, SOCS1, SOX2, YAP1, ZFHX3 |
| Herrgott et al. [116]/2022 | Differentiate PitNETs from OPD through analysis of LB specimens | PitNETs (n = 37) | PitNETs release DNA methylation markers in the serum/plasma |
| Santana-Santos et al. [101]/2022 | The Northwestern Medicine (NM) classifier of CNS tumors was developed and validated | 3905 central nervous tumor samples. 2801 samples were used in the original classifier training, and 1104 were used for validation. | Whole-genome methylation profiling of brain tumors for clinical testing has been developed and validated |
| Tucker et al. [113]/2023 | Validate the differential DNA methylation and related MAX protein expression profiles between NFPA and GHPA | 52 surgically resected tumors (37 NFPA, 15 GHPA) | MAX transcription factor binding sites are globally hypomethylated and demonstrate increased accessibility for transcription factor binding in GHPA compared to NFPA |
| Galbraith et al. [102]/2023 | The clinical utility of DNA methylation for primary diagnosis of brain tumors | 1921 primary CNS tumors | DNA methylation is of limited diagnostic and prognostic value in the diagnosis of meningioma, schwannoma, and pituitary adenoma |
| Kober et al. [103]/2023 | Genome-wide DNA methylation patterns in somatotroph tumors | Forty-eight tumor samples | Differences in DNA methylation profiles between three molecular subtypes are undeniable |
| Feng et al. [104]/2023 | DNA methylation analysis in PPETS tumors and the comparison cohort | 15 posterior pituitary tumors (PPT) | PPETS and PPT form a distinct molecular cluster |
2.2.4. Additional Papers
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mete, O.; Lopes, M.B. Overview of the 2017 WHO Classification of Pituitary Tumors. Endocr. Pathol. 2017, 28, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Burcea, I.F.; Năstase, V.N.; Poiană, C. Pituitary transcription factors in the immunohistochemical and molecular diagnosis of pituitary tumours—A systematic review. Endokrynol. Pol. 2021, 72, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Manojlovic-Gacic, E.; Engstrom, B.E.; Casar-Borota, O. Histopathological classification of non-functioning pituitary neuroendocrine tumors. Pituitary 2018, 21, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Trouillas, J.; Jaffrain-Rea, M.-L.; Vasiljevic, A.; Raverot, G.; Roncaroli, F.; Villa, C. How to Classify Pituitary Neuroendocrine Tumors (PitNET)s in 2020. Cancers 2020, 12, 514. [Google Scholar] [CrossRef]
- Mayr, B.; Apenberg, S.; Rothämel, T.; von zur Mühlen, A.; Brabant, G. Menin mutations in patients with multiple endocrine neoplasia type 1. Eur. J. Endocrinol. 1997, 137, 684–687. [Google Scholar] [CrossRef]
- Beckers, A.; Aaltonen, L.A.; Daly, A.F.; Karhu, A. Familial isolated pituitary adenomas (FIPA) and the pituitary adenoma predisposition due to mutations in the aryl hydrocarbon receptor interacting protein (AIP) gene. Endocr. Rev. 2013, 34, 239–277. [Google Scholar] [CrossRef]
- Mantovani, G.; Lania, A.G.; Spada, A. GNAS imprinting and pituitary tumors. Mol. Cell Endocrinol. 2010, 326, 15–18. [Google Scholar] [CrossRef]
- Reincke, M.; Sbiera, S.; Hayakawa, A.; Theodoropoulou, M.; Osswald, A.; Beuschlein, F.; Meitinger, T.; Mizuno-Yamasaki, E.; Kawaguchi, K.; Saeki, Y.; et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat. Genet. 2015, 47, 31–38. [Google Scholar] [CrossRef]
- Bi, W.L.; Horowitz, P.; Greenwald, N.F.; Abedalthagafi, M.; Agarwalla, P.K.; Gibson, W.J.; Mei, Y.; Schumacher, S.E.; Ben-David, U.; Chevalier, A.; et al. Landscape of Genomic Alterations in Pituitary Adenomas. Clin. Cancer Res. 2017, 23, 1841–1851. [Google Scholar] [CrossRef]
- Hauser, B.M.; Lau, A.; Gupta, S.; Bi, W.L.; Dunn, I.F. The Epigenomics of Pituitary Adenoma. Front. Endocrinol. 2019, 10, 290. [Google Scholar] [CrossRef]
- Wei, Z.; Zhou, C.; Li, M.; Huang, R.; Deng, H.; Shen, S.; Wang, R. Integrated multi-omics profiling of nonfunctioning pituitary adenomas. Pituitary 2021, 24, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Jiang, X.; Weisenthal, K.; Ma, J.; Botticelli, E.M.; Zhou, Y.; Hedley-Whyte, E.T.; Wang, B.; Swearingen, B.; Soberman, R.J.; et al. High Histone Deacetylase 2/3 Expression in Non-Functioning Pituitary Tumors. Front. Oncol. 2022, 12, 875122. [Google Scholar] [CrossRef]
- Taniguchi-Ponciano, K.; Andonegui-Elguera, S.; Peña-Martínez, E.; Silva-Román, G.; Vela-Patiño, S.; Gomez-Apo, E.; Chavez-Macias, L.; Vargas-Ortega, G.; Espinosa-de-los-Monteros, L.; Gonzalez-Virla, B.; et al. Transcriptome and methylome analysis reveals three cellular origins of pituitary tumors. Sci. Rep. 2020, 10, 19373. [Google Scholar] [CrossRef]
- Mosella, M.S.; Sabedot, T.S.; Silva, T.C.; Malta, T.M.; Dezem, F.S.; Asmaro, K.P.; Wells, M.; Mukherjee, A.; Poisson, L.M.; Snyder, J.; et al. DNA methylation-based signatures classify sporadic pituitary tumors according to clinicopathological features. Neuro Oncol. 2021, 23, 1292–1303. [Google Scholar] [CrossRef]
- Pease, M.; Ling, C.; Mack, W.J.; Wang, K.; Zada, G. The role of epigenetic modification in tumorigenesis and progression of pituitary adenomas: A systematic review of the literature. PLoS ONE 2013, 8, e82619. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Tycko, B. Epigenetic gene silencing in cancer. J. Clin. Investig. 2000, 105, 401–407. [Google Scholar] [CrossRef]
- Huttner, A.; Adams, E.F.; Buchfelder, M.; Fahlbusch, R. Growth hormone gene structure in human pituitary somatotrophinomas: Promoter region sequence and methylation studies. J. Mol. Endocrinol. 1994, 12, 167–172. [Google Scholar] [CrossRef]
- Asa, S.L.; Mete, O.; Perry, A.; Osamura, R.Y. Overview of the 2022 WHO Classification of Pituitary Tumors. Endocr. Pathol. 2022, 33, 6–26. [Google Scholar] [CrossRef]
- Cheng, S.; Xie, W.; Miao, Y.; Guo, J.; Wang, J.; Li, C.; Zhang, Y. Identification of key genes in invasive clinically non-functioning pituitary adenoma by integrating analysis of DNA methylation and mRNA expression profiles. J. Transl. Med. 2019, 17, 407. [Google Scholar] [CrossRef] [PubMed]
- Hallen, T.; Johannsson, G.; Dahlen, R.; Andersson-Assarsson, J.C.; Glad, C.A.; Orndal, C.; Engvall, A.; Caren, H.; Skoglund, T.; Olsson, D.S. Genome-Wide DNA Methylation Differences in Patients With Non-Functioning Pituitary Adenomas With or Without Postsurgical Intervention. J. Endocr. Soc. 2021, 5 (Suppl. S1), A643. [Google Scholar] [CrossRef]
- Ho, K.K.Y.; Fleseriu, M.; Wass, J.; Katznelson, L.; Raverot, G.; Little, A.S.; Castano, J.P.; Reincke, M.; Lopes, M.B.; Kaiser, U.B.; et al. A proposed clinical classification for pituitary neoplasms to guide therapy and prognosis. Lancet Diabetes Endocrinol. 2024, 12, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Aydin, B.; Beklen, H.; Arga, K.Y.; Bayrakli, F.; Turanli, B. Epigenomic and transcriptomic landscaping unraveled candidate repositioned therapeutics for non-functioning pituitary neuroendocrine tumors. J. Endocrinol. Investig. 2023, 46, 727–747. [Google Scholar] [CrossRef]
- Bello, M.J.; De Campos, J.M.; Isla, A.; Casartelli, C.; Rey, J.A. Promoter CpG methylation of multiple genes in pituitary adenomas: Frequent involvement of caspase-8. Oncol. Rep. 2006, 15, 443–448. [Google Scholar] [CrossRef]
- Gejman, R.; Batista, D.L.; Zhong, Y.; Zhou, Y.; Zhang, X.; Swearingen, B.; Stratakis, C.A.; Hedley-Whyte, E.T.; Klibanski, A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J. Clin. Endocrinol. Metab. 2008, 93, 4119–4125. [Google Scholar] [CrossRef]
- Stirzaker, C.; Taberlay, P.C.; Statham, A.L.; Clark, S.J. Mining cancer methylomes: Prospects and challenges. Trends Genet. 2014, 30, 75–84. [Google Scholar] [CrossRef]
- Ruebel, K.H.; Jin, L.; Zhang, S.; Scheithauer, B.W.; Lloyd, R.V. Inactivation of the p16 gene in human pituitary nonfunctioning tumors by hypermethylation is more common in null cell adenomas. Endocr. Pathol. 2001, 12, 281–289. [Google Scholar] [CrossRef]
- Miyata, K.; Naito, M.; Miyata, T.; Mokuda, S.; Asahara, H. Bisulfite Sequencing for DNA Methylation Analysis of Primary Muscle Stem Cells. Methods Mol. Biol. 2017, 1668, 3–13. [Google Scholar] [CrossRef]
- Newell-Price, J.; King, P.; Clark, A.J. The CpG island promoter of the human proopiomelanocortin gene is methylated in nonexpressing normal tissue and tumors and represses expression. Mol. Endocrinol. (Baltim. Md.) 2001, 15, 338–348. [Google Scholar] [CrossRef]
- Sahoo, K.; Sundararajan, V. Methods in DNA methylation array dataset analysis: A review. Comput. Struct. Biotechnol. J. 2024, 23, 2304–2325. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.J.; Hibberts, N.A.; McNicol, A.M.; Clayton, R.N.; Farrell, W.E. Loss of pRb expression in pituitary adenomas is associated with methylation of the RB1 CpG island. Cancer Res. 2000, 60, 1211–1216. [Google Scholar] [PubMed]
- Hoi, S.U.; Kelley, P.; Lee, W.H. Abnormalities of the human growth hormone gene and protooncogenes in some human pituitary adenomas. Mol. Endocrinol. 1988, 2, 85–89. [Google Scholar] [CrossRef]
- Woloschak, M.; Yu, A.; Post, K.D. Frequent inactivation of the p16 gene in human pituitary tumors by gene methylation. Mol. Carcinog. 1997, 19, 221–224. [Google Scholar] [CrossRef]
- Picard, C.; Silvy, M.; Gerard, C.; Buffat, C.; Lavaque, E.; Figarella-Branger, D.; Dufour, H.; Gabert, J.; Beckers, A.; Brue, T.; et al. Gsα overexpression and loss of Gsα imprinting in human somatotroph adenomas: Association with tumor size and response to pharmacologic treatment. Int. J. Cancer 2007, 121, 1245–1252. [Google Scholar] [CrossRef]
- Raverot, G.; Sturm, N.; De Fraipont, F.; Muller, M.; Salenave, S.; Caron, P.; Chabre, O.; Chanson, P.; Cortet-Rudelli, C.; Assaker, R.; et al. Temozolomide treatment in aggressive pituitary tumors and pituitary carcinomas: A French multicenter experience. J. Clin. Endocrinol. Metab. 2010, 95, 4592–4599. [Google Scholar] [CrossRef]
- Simpson, D.J.; Bicknell, J.E.; McNicol, A.M.; Clayton, R.N.; Farrell, W.E. Hypermethylation of the p16/CDKN2A/MTSI gene and loss of protein expression is associated with nonfunctional pituitary adenomas but not somatotrophinomas. Genes. Chromosomes Cancer 1999, 24, 328–336. [Google Scholar] [CrossRef]
- Kirsch, M.; Morz, M.; Pinzer, T.; Schackert, H.K.; Schackert, G. Frequent loss of the CDKN2C (p18INK4c) gene product in pituitary adenomas. Genes. Chromosomes Cancer 2009, 48, 143–154. [Google Scholar] [CrossRef]
- Simpson, D.J.; Clayton, R.N.; Farrell, W.E. Preferential loss of Death Associated Protein kinase expression in invasive pituitary tumours is associated with either CPG island methylation or homozygous deletion. Oncogene 2002, 21, 1217–1224. [Google Scholar] [CrossRef]
- Bahar, A.; Bicknell, J.E.; Simpson, D.J.; Clayton, R.N.; Farrell, W.E. Loss of expression of the growth inhibitory gene GADD45γ, in human pituitary adenomas, is associated with CpG island methylation. Oncogene 2004, 23, 936–944. [Google Scholar] [CrossRef]
- Qian, Z.R.; Sano, T.; Yoshimoto, K.; Yamada, S.; Ishizuka, A.; Mizusawa, N.; Horiguchi, H.; Hirokawa, M.; Asa, S.L. Inactivation of RASSF1A tumor suppressor gene by aberrant promoter hypermethylation in human pituitary adenomas. Lab. Investig. 2005, 85, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Liu, H.; Zhao, S.; Wu, J.; Fan, J.; Liao, J. Silencing of RASSF3 by DNA Hypermethylation Is Associated with Tumorigenesis in Somatotroph Adenomas. PLoS ONE 2013, 8, e59024. [Google Scholar] [CrossRef] [PubMed]
- Ruebel, K.H.; Jin, L.; Qian, X.; Scheithauer, B.W.; Kovacs, K.; Nakamura, N.; Zhang, H.; Raz, A.; Lloyd, R.V. Effects of DNA methylation on galectin-3 expression in pituitary tumors. Cancer Res. 2005, 65, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Buslei, R.; Kreutzer, J.; Hofmann, B.; Schmidt, V.; Siebzehnrul, F.; Hahnen, E.; Eyupoglu, I.Y.; Fahlbusch, R.; Blumcke, I. Abundant hypermethylation of SOCS-1 in clinically silent pituitary adenomas. Acta Neuropathol. 2006, 111, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Asa, S.L.; Ezzat, S. Fibroblast growth factor 2 and estrogen control the balance of histone 3 modifications targeting MAGE-A3 in pituitary neoplasia. Clin. Cancer Res. 2008, 14, 1984–1996. [Google Scholar] [CrossRef]
- Yuan, Y.; Qian, Z.R.; Sano, T.; Asa, S.L.; Yamada, S.; Kagawa, N.; Kudo, E. Reduction of GSTP1 expression by DNA methylation correlates with clinicopathological features in pituitary adenomas. Mod. Pathol. 2008, 21, 856–865. [Google Scholar] [CrossRef]
- Ferrau, F.; Romeo, P.D.; Puglisi, S.; Ragonese, M.; Spagnolo, F.; Salpietro, C.; Ientile, R.; Curro, M.; Visalli, G.; Alibrandi, A.; et al. GSTP1 gene methylation and AHR rs2066853 variant predict resistance to first generation somatostatin analogs in patients with acromegaly. J. Endocrinol. Investig. 2019, 42, 825–831. [Google Scholar] [CrossRef]
- Revill, K.; Dudley, K.J.; Clayton, R.N.; McNicol, A.M.; Farrell, W.E. Loss of neuronatin expression is associated with promoter hypermethylation in pituitary adenoma. Endocr.-Relat. Cancer 2009, 16, 537–548. [Google Scholar] [CrossRef]
- Duong, C.V.; Yacqub-Usman, K.; Emes, R.D.; Clayton, R.N.; Farrell, W.E. The EFEMP1 gene: A frequent target for epigenetic silencing in multiple human pituitary adenoma subtypes. Neuroendocrinology 2013, 98, 200–211. [Google Scholar] [CrossRef]
- Wu, Y.; Bai, J.; Li, Z.; Wang, F.; Cao, L.; Liu, C.; Yu, S.; Yu, G.; Zhang, Y. Low expression of secreted frizzled-related protein 4 in aggressive pituitary adenoma. Pituitary 2015, 18, 335–342. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, C.; Yu, S.; Gao, H.; Li, Z.; Li, C.; Zhang, Y. Assessment of sFRP4 as a bio-marker for predicting aggressiveness and recurrence of growth hormone-secreting pituitary adenomas. Oncol. Rep. 2016, 35, 2991–2999. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Qian, L.; Jing, G.; Jie, F.; Xiaosong, S.; Chunhui, L.; Yangfang, L.; Guilin, L.; Gao, H.; Yazhuo, Z. Aberrant expression of the sFRP and WIF1 genes in invasive non-functioning pituitary adenomas. Mol. Cell. Endocrinol. 2018, 474, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Li, J.; Liu, Q.; Liu, C.; Li, C.; Song, G.; Zhu, H.; Gao, H.; Zhang, Y. P21Waf1/Cip1 and p27Kip1 are correlated with the development and invasion of prolactinoma. J. Neuro-Oncol. 2018, 136, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Hage, M.; Chaligne, R.; Viengchareun, S.; Villa, C.; Salenave, S.; Bouligand, J.; Letouze, E.; Tosca, L.; Rouquette, A.; Tachdjian, G.; et al. Hypermethylator Phenotype and Ectopic GIP Receptor in GNAS Mutation-Negative Somatotropinomas. J. Clin. Endocrinol. Metab. 2019, 104, 1777–1787. [Google Scholar] [CrossRef] [PubMed]
- Dalle Nogare, M.; D’Annunzio, S.; Vazza, G.; Regazzo, D.; Picello, L.; Denaro, L.; Voltan, G.; Scaroni, C.; Ceccato, F.; Occhi, G. The Methylation Analysis of the Glucose-Dependent Insulinotropic Polypeptide Receptor (GIPR) Locus in GH-Secreting Pituitary Adenomas. Int. J. Mol. Sci. 2023, 24, 9264. [Google Scholar] [CrossRef]
- Ma, H.S.; Wang, E.L.; Xu, W.F.; Yamada, S.; Yoshimoto, K.; Qian, Z.R.; Shi, L.; Liu, L.L.; Li, X.H. Overexpression of dna (Cytosine-5)-methyltransferase 1 (dnmt1) and dna (cytosine-5)-methyltransferase 3a (dnmt3a) is associated with aggressive behavior and hypermethylation of tumor suppressor genes in human pituitary adenomas. Med. Sci. Monit. 2018, 24, 4841–4850. [Google Scholar] [CrossRef]
- Wang, R.Q.; Lan, Y.L.; Lou, J.C.; Lyu, Y.Z.; Hao, Y.C.; Su, Q.F.; Ma, B.B.; Yuan, Z.B.; Yu, Z.K.; Zhang, H.Q.; et al. Expression and methylation status of LAMA2 are associated with the invasiveness of nonfunctioning PitNET. Ther. Adv. Endocrinol. Metab. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Miyake, Y.; Adachi, J.I.; Suzuki, T.; Mishima, K.; Araki, R.; Mizuno, R.; Nishikawa, R. TERT promoter methylation is significantly associated with TERT upregulation and disease progression in pituitary adenomas. J. Neuro-Oncol. 2019, 141, 131–138. [Google Scholar] [CrossRef]
- Alzoubi, H.; Minasi, S.; Gianno, F.; Antonelli, M.; Belardinilli, F.; Giangaspero, F.; Jaffrain-Rea, M.-L.; Buttarelli, F.R. Alternative Lengthening of Telomeres (ALT) and Telomerase Reverse Transcriptase Promoter Methylation in Recurrent Adult and Primary Pediatric Pituitary Neuroendocrine Tumors. Endocr. Pathol. 2022, 33, 494–505. [Google Scholar] [CrossRef]
- Endo, M.; Adachi, J.I.; Murakami, C.; Inomoto, C.; Komatsu, M.; Hanakita, S.; Oyama, K.I.; Matsuno, A.; Nishikawa, R.; Oya, S. A case of aggressive pituitary neuroendocrine tumour with extremely rapid progression: Possible diagnostic value of TERT promoter methylation. Br. J. Neurosurg. 2022, 1–7. [Google Scholar] [CrossRef]
- Valimaki, N.; Schalin-Jantti, C.; Karppinen, A.; Paetau, A.; Kivipelto, L.; Aaltonen, L.A.; Karhu, A. Genetic and epigenetic characterization of growth hormone-secreting pituitary tumors. Mol. Cancer Res. 2019, 17, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Romanet, P.; Galluso, J.; Kamenicky, P.; Hage, M.; Theodoropoulou, M.; Roche, C.; Graillon, T.; Etchevers, H.C.; De Murat, D.; Mougel, G.; et al. Somatotroph Tumors and the Epigenetic Status of the GNAS Locus. Int. J. Mol. Sci. 2021, 22, 7570. [Google Scholar] [CrossRef] [PubMed]
- Araki, T.; Tone, Y.; Kameda, H.; Ben-Shlomo, A.; Yamada, S.; Takeshita, A.; Yamamoto, M.; Kawakami, Y.; Tone, M.; Melmed, S. Two Distinctive POMC Promoters Modify Gene Expression in Cushing Disease. J. Clin. Endocrinol. Metab. 2021, 106, E3346–E3363. [Google Scholar] [CrossRef]
- Xu, B.; Sano, T.; Yoshimoto, K.; Yamada, S. Downregulation of E-cadherin and its undercoat proteins in pituitary growth hormone cell adenomas with prominent fibrous bodies. Endocr. Pathol. 2002, 13, 341–351. [Google Scholar] [CrossRef]
- Bahar, A.; Simpson, D.J.; Cutty, S.J.; Bicknell, J.E.; Hoban, P.R.; Holley, S.; Mourtada-Maarabouni, M.; Williams, G.T.; Clayton, R.N.; Farrell, W.E. Isolation and characterization of a novel pituitary tumor apoptosis gene. Mol. Endocrinol. 2004, 18, 1827–1839. [Google Scholar] [CrossRef]
- Zhao, J.; Dahle, D.; Zhou, Y.; Zhang, X.; Klibanski, A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J. Clin. Endocrinol. Metab. 2005, 90, 2179–2186. [Google Scholar] [CrossRef]
- Ogino, A.; Yoshino, A.; Katayama, Y.; Watanabe, T.; Ota, T.; Komine, C.; Yokoyama, T.; Fukushima, T. The p15(INK4b)/p16(INK4a)/RB1 pathway is frequently deregulated in human pituitary adenomas. J. Neuropathol. Exp. Neurol. 2005, 64, 398–403. [Google Scholar] [CrossRef]
- Yoshino, A.; Katayama, Y.; Ogino, A.; Watanabe, T.; Yachi, K.; Ohta, T.; Komine, C.; Yokoyama, T.; Fukushima, T. Promoter hypermethylation profile of cell cycle regulator genes in pituitary adenomas. J. Neuro-Oncol. 2007, 83, 153–162. [Google Scholar] [CrossRef]
- Qian, Z.R.; Sano, T.; Yoshimoto, K.; Asa, S.L.; Yamada, S.; Mizusawa, N.; Kudo, E. Tumor-specific downregulation and methylation of the CDH13 (H-cadherin) and CDH1 (E-cadherin) genes correlate with aggressiveness of human pituitary adenomas. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc. 2007, 20, 1269–1277. [Google Scholar] [CrossRef]
- Zhu, X.; Lee, K.; Asa, S.L.; Ezzat, S. Epigenetic silencing through DNA and histone methylation of fibroblast growth factor receptor 2 in neoplastic pituitary cells. Am. J. Pathol. 2007, 170, 1618–1628. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, R.; Du, W.; Yang, F.; Wei, X. RIZ1 and histone methylation status in pituitary adenomas. Tumour Biol. 2017, 39, 1010428317711794. [Google Scholar] [CrossRef] [PubMed]
- Pedraza-Arevalo, S.; Ibanez-Costa, A.; Blazquez-Encinas, R.; Branco, M.R.; Vazquez-Borrego, M.C.; Herrera-Martinez, A.D.; Venegas-Moreno, E.; Serrano-Blanch, R.; Arjona-Sanchez, A.; Galvez-Moreno, M.A.; et al. Epigenetic and post-transcriptional regulation of somatostatin receptor subtype 5 (SST5) in pituitary and pancreatic neuroendocrine tumors. Mol. Oncol. 2022, 16, 764–779. [Google Scholar] [CrossRef] [PubMed]
- Szabo, B.; Meszaros, K.; Krokker, L.; Liko, I.; Saskoi, E.; Nemeth, K.; Szabo, P.T.; Szucs, N.; Czirjak, S.; Szaloki, G.; et al. Aspirin Mediates Its Antitumoral Effect Through Inhibiting PTTG1 in Pituitary Adenoma. J. Clin. Endocrinol. Metab. 2022, 107, 3066–3079. [Google Scholar] [CrossRef] [PubMed]
- Guaraldi, F.; Morandi, L.; Zoli, M.; Mazzatenta, D.; Righi, A.; Evangelisti, S.; Ambrosi, F.; Tonon, C.; Giannini, C.; Lloyd, R.V.; et al. Epigenomic and somatic mutations of pituitary tumors with clinical and pathological correlations in 111 patients. Clin. Endocrinol. (Oxf.) 2022, 97, 763–772. [Google Scholar] [CrossRef]
- Dong, W.; Shi, W.; Liu, Y.; Li, J.; Zhang, Y.; Dong, G.; Dong, X.; Gao, H. CHST7 Methylation Status Related to the Proliferation and Differentiation of Pituitary Adenomas. Cells 2022, 11, 2400. [Google Scholar] [CrossRef]
- Jaffrain-Rea, M.L.; Ferretti, E.; Toniato, E.; Cannita, K.; Santoro, A.; Di Stefano, D.; Ricevuto, E.; Maroder, M.; Tamburrano, G.; Cantore, G.; et al. p16 (INK4a, MTS-1) gene polymorphism and methylation status in human pituitary tumours. Clin. Endocrinol. 1999, 51, 317–325. [Google Scholar] [CrossRef]
- Seemann, N.; Kuhn, D.; Wrocklage, C.; Keyvani, K.; Hackl, W.; Buchfelder, M.; Fahlbusch, R.; Paulus, W. CDKN2A/p 16 inactivation is related to pituitary adenoma type and size. J. Pathol. 2001, 193, 491–497. [Google Scholar] [CrossRef]
- Simpson, D.J.; McNicol, A.M.; Murray, D.C.; Bahar, A.; Turner, H.E.; Wass, J.A.H.; Esiri, M.M.; Clayton, R.N.; Farrell, W.E. Molecular Pathology Shows p16 Methylation in Nonadenomatous Pituitaries from Patients with Cushing’s Disease. Clin. Cancer Res. 2004, 10, 1780–1788. [Google Scholar] [CrossRef]
- Evang, J.A.; Berg, J.P.; Casar-Borota, O.; Lekva, T.; Kringen, M.K.; Ramm-Pettersen, J.; Bollerslev, J. Reduced levels of E-cadherin correlate with progression of corticotroph pituitary tumours. Clin. Endocrinol. 2011, 75, 811–818. [Google Scholar] [CrossRef]
- Hossain Md, G.; Iwata, T.; Mizusawa, N.; Qian, Z.R.; Shima, S.W.N.; Okutsu, T.; Yamada, S.; Sano, T.; Yoshimoto, K. Expression of p18INK4C is down-regulated in human pituitary adenomas. Endocr. Pathol. 2009, 20, 114–121. [Google Scholar] [CrossRef]
- Vaitkiene, P.; Valiulyte, I.; Glebauskiene, B.; Liutkeviciene, R. N-myc downstream-regulated gene 2 (NDRG2) promoter methylation and expression in pituitary adenoma. Diagn. Pathol. 2017, 12, 33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Michaelis, K.A.; Knox, A.J.; Xu, M.; Kiseljak-Vassiliades, K.; Edwards, M.G.; Geraci, M.; Kleinschmidt-DeMasters, B.K.; Lillehei, K.O.; Wierman, M.E. Identification of growth arrest and DNA-damage- inducible gene β (GADD45β) as a novel tumor suppressor in pituitary gonadotrope tumors. Endocrinology 2011, 152, 3603–3613. [Google Scholar] [CrossRef] [PubMed]
- Raef, H.; Zou, M.; Baitei, E.Y.; Al-Rijjal, R.A.; Kaya, N.; Al-Hamed, M.; Monies, D.; Abu-Dheim, N.N.; Al-Hindi, H.; Al-Ghamdi, M.H.; et al. A novel deletion of the MEN1 gene in a large family of multiple endocrine neoplasia type 1 (MEN1) with aggressive phenotype. Clin. Endocrinol. 2011, 75, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Kochling, M.; Ewelt, C.; Furtjes, G.; Peetz-Dienhart, S.; Koos, B.; Hasselblatt, M.; Paulus, W.; Stummer, W.; Brokinkel, B. hTERT promoter methylation in pituitary adenomas. Brain Tumor Pathol. 2016, 33, 27–34. [Google Scholar] [CrossRef]
- Valiulyte, I.; Steponaitis, G.; Skiriute, D.; Tamasauskas, A.; Vaitkiene, P. Signal transducer and activator of transcription 3 (STAT3) promoter methylation and expression in pituitary adenoma. BMC Med. Genet. 2017, 18, 72. [Google Scholar] [CrossRef]
- McCormack, A.I.; McDonald, K.L.; Gill, A.J.; Clark, S.J.; Burt, M.G.; Campbell, K.A.; Braund, W.J.; Little, N.S.; Cook, R.J.; Grossman, A.B.; et al. Low O6-methylguanine-DNA methyltransferase (MGMT) expression and response to temozolomide in aggressive pituitary tumours. Clin. Endocrinol. 2009, 71, 226–233. [Google Scholar] [CrossRef]
- Losa, M.; Mazza, E.; Terreni, M.R.; McCormack, A.; Gill, A.J.; Motta, M.; Cangi, M.G.; Talarico, A.; Mortini, P.; Reni, M. Salvage therapy with temozolomide in patients with aggressive or metastatic pituitary adenomas: Experience in six cases. Eur. J. Endocrinol. 2010, 163, 843–851. [Google Scholar] [CrossRef]
- Bush, Z.M.; Longtine, J.A.; Cunningham, T.; Schiff, D.; Jane Jr, J.A.; Vance, M.L.; Thorner, M.O.; Laws Jr, E.R.; Lopes, M.B.S. Temozolomide treatment for aggressive pituitary tumors: Correlation of clinical outcome with O6-methylguanine methyltransferase (MGMT) promoter methylation and expression. J. Clin. Endocrinol. Metab. 2010, 95, E280–E290. [Google Scholar] [CrossRef][Green Version]
- Arya, S.; Majaid, M.A.; Shwetha, S.D.; Sravani, K.; Arivazhagan, A.; Sampath, S.; Santosh, V. Implications of MGMT methylation status in pituitary adenoma. Pathol. Res. Pract. 2014, 210, 407–411. [Google Scholar] [CrossRef]
- Ceccato, F.; Lombardi, G.; Manara, R.; Emanuelli, E.; Denaro, L.; Milanese, L.; Gardiman, M.P.; Bertorelle, R.; Scanarini, M.; D’Avella, D.; et al. Temozolomide and pasireotide treatment for aggressive pituitary adenoma: Expertise at a tertiary care center. J. Neuro-Oncol. 2015, 122, 189–196. [Google Scholar] [CrossRef]
- Jiang, X.B.; Hu, B.; He, D.S.; Mao, Z.G.; Wang, X.; Song, B.B.; Zhu, Y.H.; Wang, H.J. Expression profiling of O6 methylguanine-DNA-methyl transferase in prolactinomas: A correlative study of promoter methylation and pathological features in 136 cases. BMC Cancer 2015, 15, 644. [Google Scholar] [CrossRef] [PubMed]
- Micko, A.S.G.; Hoftberger, R.; Wohrer, A.; Millesi, M.; Knosp, E.; Wolfsberger, S. MGMT assessment in pituitary adenomas: Comparison of different immunohistochemistry fixation chemicals. Pituitary 2018, 21, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chang, M.; Zhou, G.; Liu, P.; Lou, J.; Zhang, Y.; Guo, X.; Bao, X.; Lian, W.; Wang, Y.; et al. Multi-Omics Investigations Revealed Underlying Molecular Mechanisms Associated With Tumor Stiffness and Identified Sunitinib as a Potential Therapy for Reducing Stiffness in Pituitary Adenomas. Front. Cell Dev. Biol. 2022, 10, 820562. [Google Scholar] [CrossRef] [PubMed]
- Duong, C.V.; Emes, R.D.; Wessely, F.; Yacqub-Usman, K.; Clayton, R.N.; Farrell, W.E. Quantitative, genome-wide analysis of the DNA methylome in sporadic pituitary adenomas. Endocr.-Relat. Cancer 2012, 19, 805–816. [Google Scholar] [CrossRef]
- Ling, C.; Pease, M.; Shi, L.; Punj, V.; Shiroishi, M.S.; Commins, D.; Weisenberger, D.J.; Wang, K.; Zada, G. A pilot genome-scale profiling of DNA methylation in sporadic pituitary macroadenomas: Association with Tumor invasion and histopathological subtype. PLoS ONE 2014, 9, e96178. [Google Scholar] [CrossRef]
- Johann, P.D.; Bens, S.; Oyen, F.; Wagener, R.; Giannini, C.; Perry, A.; Raisanen, J.M.; Reis, G.F.; Nobusawa, S.; Arita, K.; et al. Sellar Region Atypical Teratoid/Rhabdoid Tumors (ATRT) in Adults Display DNA Methylation Profiles of the ATRT-MYC Subgroup. Am. J. Surg. Pathol. 2018, 42, 506–511. [Google Scholar] [CrossRef]
- Salomon, M.P.; Wang, X.; Marzese, D.M.; Hsu, S.C.; Nelson, N.; Zhang, X.; Matsuba, C.; Takasumi, Y.; Ballesteros-Merino, C.; Fox, B.A.; et al. The Epigenomic Landscape of Pituitary Adenomas Reveals Specific Alterations and Differentiates Among Acromegaly, Cushing’s Disease and Endocrine-Inactive Subtypes. Clin. Cancer Res. 2018, 24, 4126–4136. [Google Scholar] [CrossRef]
- Neou, M.; Villa, C.; Armignacco, R.; Jouinot, A.; Raffin-Sanson, M.L.; Septier, A.; Letourneur, F.; Diry, S.; Diedisheim, M.; Izac, B.; et al. Pangenomic Classification of Pituitary Neuroendocrine Tumors. Cancer Cell 2020, 37, 123–134.e125. [Google Scholar] [CrossRef]
- Nadaf, J.; de Kock, L.; Chong, A.S.; Korbonits, M.; Thorner, P.; Benlimame, N.; Fu, L.; Peet, A.; Warner, J.; Ploner, O.; et al. Molecular characterization of DICER1-mutated pituitary blastoma. Acta Neuropathol. 2021, 141, 929–944. [Google Scholar] [CrossRef]
- Silva-Júnior, R.; Bueno, A.C.; Martins, C.S.; Coelli-Lacchini, F.; Ozaki, J.G.O.; Almeida, E.S.D.C.; Marrero-Gutierrez, J.; Santos, A.C.D.; Garcia-Peral, C.; Machado, H.R.; et al. Integrating Methylome and Transcriptome Signatures Expands the Molecular Classification of the Pituitary Tumors. J. Clin. Endocrinol. Metab. 2023, 108, 1452–1463. [Google Scholar] [CrossRef]
- Santana-Santos, L.; Kam, K.L.; Dittmann, D.; De Vito, S.; McCord, M.; Jamshidi, P.; Fowler, H.; Wang, X.; Aalsburg, A.M.; Brat, D.J.; et al. Validation of Whole Genome Methylation Profiling Classifier for Central Nervous System Tumors. J. Mol. Diagn. 2022, 24, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, K.; Vasudevaraja, V.; Serrano, J.; Shen, G.; Tran, I.; Abdallat, N.; Wen, M.; Patel, S.; Movahed-Ezazi, M.; Faustin, A.; et al. Clinical utility of whole-genome DNA methylation profiling as a primary molecular diagnostic assay for central nervous system tumors—A prospective study and guidelines for clinical testing. Neuro-Oncol. Adv. 2023, 5, vdad076. [Google Scholar] [CrossRef] [PubMed]
- Kober, P.; Rymuza, J.; Baluszek, S.; Maksymowicz, M.; Nyc, A.; Mossakowska, B.J.; Zielinski, G.; Kunicki, J.; Bujko, M. DNA Methylation Pattern in Somatotroph Pituitary Neuroendocrine Tumors. Neuroendocrinology 2024, 114, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Duan, Z.; Yao, K.; Gui, Q.; Liu, X.; Wang, X.; Du, Z.; Shao, L.; Zhang, B.; Cai, S.; et al. Primary papillary epithelial tumor of the sella and posterior pituitary tumor show similar (epi)genetic features and constitute a single neuro-oncological entity. Neuro-Oncol. 2023, 25, 1487–1497. [Google Scholar] [CrossRef]
- Gu, Y.; Zhou, X.; Hu, F.; Yu, Y.; Xie, T.; Huang, Y.; Zhao, X.; Zhang, X. Differential DNA methylome profiling of nonfunctioning pituitary adenomas suggesting tumour invasion is correlated with cell adhesion. J. Neuro-Oncol. 2016, 129, 23–31. [Google Scholar] [CrossRef]
- Boresowicz, J.; Kober, P.; Rusetska, N.; Maksymowicz, M.; Paziewska, A.; Dabrowska, M.; Zeber-Lubecka, N.; Kunicki, J.; Bonicki, W.; Ostrowski, J.; et al. DNA Methylation Influences miRNA Expression in Gonadotroph Pituitary Tumors. Life 2020, 10, 59. [Google Scholar] [CrossRef]
- Hagel, C.; Schuller, U.; Flitsch, J.; Knappe, U.J.; Kellner, U.; Bergmann, M.; Buslei, R.; Buchfelder, M.; Rudiger, T.; Herms, J.; et al. Double adenomas of the pituitary reveal distinct lineage markers, copy number alterations, and epigenetic profiles. Pituitary 2021, 24, 904–913. [Google Scholar] [CrossRef]
- Hallén, T.; Johannsson, G.; Dahlén, R.; Glad, C.A.M.; Örndal, C.; Engvall, A.; Carén, H.; Skoglund, T.; Olsson, D.S. Genome-wide DNA Methylation Differences in Nonfunctioning Pituitary Adenomas With and Without Postsurgical Progression. J. Clin. Endocrinol. Metab. 2022, 107, 2318–2328. [Google Scholar] [CrossRef]
- Kober, P.; Boresowicz, J.; Rusetska, N.; Maksymowicz, M.; Goryca, K.; Kunicki, J.; Bonicki, W.; Siedlecki, J.A.; Bujko, M. DNA methylation profiling in nonfunctioning pituitary adenomas. Mol. Cell. Endocrinol. 2018, 473, 194–204. [Google Scholar] [CrossRef]
- Boresowicz, J.; Kober, P.; Rusetska, N.; Maksymowicz, M.; Goryca, K.; Kunicki, J.; Bonicki, W.; Bujko, M. Telomere Length and TERT abnormalities in pituitary adenomas. Neuroendocrinol. Lett. 2018, 39, 49–55. [Google Scholar]
- Kober, P.; Boresowicz, J.; Rusetska, N.; Maksymowicz, M.; Paziewska, A.; Dabrowska, M.; Kunicki, J.; Bonicki, W.; Ostrowski, J.; Siedlecki, J.A.; et al. The role of aberrant DNA methylation in misregulation of gene expression in gonadotroph nonfunctioning pituitary tumors. Cancers 2019, 11, 1650. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Li, C.; Xie, W.; Miao, Y.; Guo, J.; Wang, J.; Zhang, Y. Integrated analysis of DNA methylation and mRNA expression profiles to identify key genes involved in the regrowth of clinically non-functioning pituitary adenoma. Aging 2020, 12, 2408–2427. [Google Scholar] [CrossRef] [PubMed]
- Tucker, D.W.; Pangal, D.J.; Du, R.; Gogia, A.S.; Tafreshi, A.; Ruzevick, J.; Hurth, K.T.; Triche, T.; Micko, A.; Carpten, J.D.; et al. Validation of Myc-Associated Protein X (MAX) regulation in growth hormone secreting and nonfunctional pituitary adenoma. PLoS ONE 2023, 18, e0284949. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, G.; D’Argenio, V.; Ferrau, F.; Lasorsa, V.A.; Polito, F.; Aliquo, F.; Ragonese, M.; Cotta, O.R.; Alessi, Y.; Oteri, R.; et al. Methylome Analysis in Nonfunctioning and GH-Secreting Pituitary Adenomas. Front. Endocrinol. 2022, 13, 841118. [Google Scholar] [CrossRef]
- Schmid, S.; Solomon, D.A.; Perez, E.; Thieme, A.; Kleinschmidt-DeMasters, B.K.; Giannini, C.; Reinhardt, A.; Asa, S.L.; Mete, O.; Stichel, D.; et al. Genetic and epigenetic characterization of posterior pituitary tumors. Acta Neuropathol. 2021, 142, 1025–1043. [Google Scholar] [CrossRef]
- Herrgott, G.A.; Asmaro, K.P.; Wells, M.; Sabedot, T.S.; Malta, T.M.; Mosella, M.S.; Nelson, K.; Scarpace, L.; Barnholtz-Sloan, J.S.; Sloan, A.E.; et al. Detection of tumor-specific DNA methylation markers in the blood of patients with pituitary neuroendocrine tumors. Neuro Oncol. 2022, 24, 1126–1139. [Google Scholar] [CrossRef]
- Asuzu, D.T.; Alvarez, R.; Fletcher, P.A.; Mandal, D.; Johnson, K.; Wu, W.; Elkahloun, A.; Clavijo, P.; Allen, C.; Maric, D.; et al. Pituitary adenomas evade apoptosis via noxa deregulation in Cushing’s disease. Cell Rep. 2022, 40, 111223. [Google Scholar] [CrossRef]
- Dottermusch, M.; Rotermund, R.; Ricklefs, F.L.; Wefers, A.K.; Saeger, W.; Flitsch, J.; Glatzel, M.; Matschke, J. The Diagnostic Impact of Epigenomics in Pituicyte-derived Tumors: Report of an Unusual Sellar Lesion with Extensive Hemorrhage and Necrotic Debris. Endocr. Pathol. 2022, 33, 411–413. [Google Scholar] [CrossRef]
- Hickman, R.A.; Gionco, J.T.; Faust, P.L.; Miller, M.L.; Bruce, J.; Page-Wilson, G.; Rosenblum, M.K.; Asa, S.L. Pituitary corticotroph tumour with adrenocortical cells: A distinct clinicopathologic entity with unique morphology and methylation profile. Neuropathol. Appl. Neurobiol. 2022, 48, e12754. [Google Scholar] [CrossRef]
- Batisse, M.; Raverot, G.; Maqdasy, S.; Durando, X.; Sturm, N.; Montoriol, P.F.; Kemeny, J.L.; Chazal, J.; Trouillas, J.; Tauveron, I. Aggressive silent GH pituitary tumor resistant to multiple treatments, including temozolomide. Cancer Investig. 2013, 31, 190–196. [Google Scholar] [CrossRef]
- Ronsley, R.; Boue, D.R.; Venkata, L.P.R.; Scott, S.; Shaikhouni, A.; Jones, J.; Schieffer, K.M.; Cottrell, C.E.; Mardis, E.R.; Olshefski, R.; et al. An unusual case of atypical teratoid/rhabdoid tumor, initially diagnosed as atypical pituitary adenoma in a 13-year-old male patient. Neuro-Oncol. Adv. 2022, 4, vdac121. [Google Scholar] [CrossRef] [PubMed]
- Barrantes-Freer, A.; Braune, M.; Sandner, B.; Dottermusch, M.; Lindner, D. Comparative epigenomics indicate a common origin of ectopic and intrasellar corticotroph pituitary neuroendocrine tumors/adenomas: A case report. Virchows Arch. 2024, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Yachi, K.; Ohta, T.; Fukushima, T.; Yoshino, A.; Katayama, Y.; Shinojima, Y.; Terui, T.; Nagase, H. Aberrant hypermethylation of non-promoter zygote arrest 1 (Zar1) in human brain tumors. Neurol. Med.-Chir. 2010, 50, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Salehi, F.; Scheithauer, B.W.; Kros, J.M.; Lau, Q.; Fealey, M.; Erickson, D.; Kovacs, K.; Horvath, E.; Lloyd, R.V. MGMT promoter methylation and immunoexpression in aggressive pituitary adenomas and carcinomas. Neuropathol. Appl. Neurobiol. 2011, 37 (Suppl. S1), 17–18. [Google Scholar] [CrossRef]
- Rusetska, N.; Kober, P.; Krol, S.K.; Boresowicz, J.; Maksymowicz, M.; Kunicki, J.; Bonicki, W.; Bujko, M. Invasive and noninvasive nonfunctioning gonadotroph pituitary tumors differ in dna methylation level of line-1 repetitive elements. J. Clin. Med. 2021, 10, 560. [Google Scholar] [CrossRef]
- Chang, M.; Wang, Z.; Gao, J.; Yang, C.; Feng, M.; Niu, Y.; Tong, W.M.; Bao, X.; Wang, R. METTL3-mediated RNA m6A Hypermethylation Promotes Tumorigenesis and GH Secretion of Pituitary Somatotroph Adenomas. J. Clin. Endocrinol. Metab. 2022, 107, 136–149. [Google Scholar] [CrossRef]
- Garcia-Martinez, A.; Sottile, J.; Sanchez-Tejada, L.; Fajardo, C.; Camara, R.; Lamas, C.; Barbera, V.M.; Pico, A. DNA Methylation of Tumor Suppressor Genes in Pituitary Neuroendocrine Tumors. J. Clin. Endocrinol. Metab. 2019, 104, 1272–1282. [Google Scholar] [CrossRef]
- Szabo, B.; Nemeth, K.; Meszaros, K.; Szucs, N.; Czirjak, S.; Reiniger, L.; Rajnai, H.; Krencz, I.; Karaszi, K.; Krokker, L.; et al. Demethylation Status of Somatic DNA Extracted From Pituitary Neuroendocrine Tumors Indicates Proliferative Behavior. J. Clin. Endocrinol. Metab. 2020, 105, 2015–2026. [Google Scholar] [CrossRef]
- Ilangumaran, S.; Gui, Y.; Shukla, A.; Ramanathan, S. SOCS1 expression in cancer cells: Potential roles in promoting antitumor immunity. Front. Immunol. 2024, 15, 1362224. [Google Scholar] [CrossRef]
- Dubois, F.; Bergot, E.; Zalcman, G.; Levallet, G. RASSF1A, puppeteer of cellular homeostasis, fights tumorigenesis, and metastasis-an updated review. Cell Death Dis. 2019, 10, 928. [Google Scholar] [CrossRef]
- Chinnam, M.; Goodrich, D.W. RB1, development, and cancer. Curr. Top. Dev. Biol. 2011, 94, 129–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cai, F.; Cao, J.; Gao, F.; Lv, Y.; Tang, Y.; Zhang, A.; Yan, W.; Wang, Y.; Hu, X.; et al. Analysis of Related Factors of Tumor Recurrence or Progression After Transnasal Sphenoidal Surgical Treatment of Large and Giant Pituitary Adenomas and Establish a Nomogram to Predict Tumor Prognosis. Front. Endocrinol. 2021, 12, 793337. [Google Scholar] [CrossRef] [PubMed]
- Noguera-Castells, A.; García-Prieto, C.A.; Álvarez-Errico, D.; Esteller, M. Validation of the new EPIC DNA methylation microarray (900K EPIC v2) for high-throughput profiling of the human DNA methylome. Epigenetics 2023, 18, 2185742. [Google Scholar] [CrossRef] [PubMed]
- Livoreil, B.; Glanville, J.; Haddaway, N.R.; Bayliss, H.; Bethel, A.; de Lachapelle, F.F.; Robalino, S.; Savilaakso, S.; Zhou, W.; Petrokofsky, G.; et al. Systematic searching for environmental evidence using multiple tools and sources. Environ. Evid. 2017, 6, 23. [Google Scholar] [CrossRef]
- Bramer, W.M.; Rethlefsen, M.L.; Kleijnen, J.; Franco, O.H. Optimal database combinations for literature searches in systematic reviews: A prospective exploratory study. Syst. Rev. 2017, 6, 245. [Google Scholar] [CrossRef]
- Møller, M.W.; Nortvig, M.J.; Andersen, M.S.; Poulsen, F.R. DNA methylation in pituitary adenomas: Scoping review protocol. medRxiv 2024. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]

| Methylation Association | Method | Genes/Pathways | Clinical Outcomes | Key Findings |
|---|---|---|---|---|
| Correlation with methylation and expression/clinical behavior | Methylation-restricted digestion | GH-gene, p16, RB1 promoter, POMC promoter, 1A DMR | POMC promoter methylation repressed expression. Partial methylation of 1A DMR induced higher expression | No clinical function of these findings |
| Methylation-specific PCR | CDKN2A, DAP kinase, GADD45g, RASSF1A, RASSF3, Gal-3, SOCS-1, FGFR2-IIIb, GSTP1, NNAT, EFEMP1, sFRP4, p21, p27, WIF1, GIPR, DNMT1, DNMT3A, DNMT3B, LAMA2, TERT, GNAS, POMC promoter | RASSF1A, RASSF3—somatotroph tumorigensis, SOCS-1—NFPAs, FGFR2-IIIb—oncogenic signals, GSTP1—invasiveness and response to SSA, sFRP4—aggresiveness, p21 and p27—invasiveness, DNMT1, DNMT3A, and DNMT3B—oncogenic factors, TERT—disease progression, POMC promoter | Related primarily to invasive behavior | |
| Chip-based | STAT5A, RHOD, GALNT9, RASSF1, CDKN1A, TP73, STAT3, HMGA2, FAM163A, HIF3A, PRSS8 | Hypermethylated in NFPA | Different significant sites for NFPA, minimal overlap between studies | |
| PHYHD1, LTBR, MYBPHL, C22orf42, PRR5, ANKDD1A, RAB13, CAMKV, KIFC3, WNT4, and STAT6 | Invasive behavior of NFPA | |||
| FAM90A1, ETS2, STAT6, MYT1L, ING2, and KCNK1 | Regrowth of NFPA | |||
| C7orf50, GNG7, and BAHCC1 | Methylated in pituitary adenomas | |||
| DCC, DLG5, ETS2, FOXO1, HBP1, HMGA2, PCGF3, PSME4, RBPMS, RREB1, SMAD1, SOCS1, SOX2, YAP1, ZFHX3 | Hub proteins for NFPA | |||
| Myc-associated protein X transcription factor binding sites | Globally hypomethylated in NFPA | |||
| Other methods | METTL3 | Regulation of cell growth or hormone secretion of GH-PA | ||
| Probable association (no significant findings) | Methylation-specific PCR | E-cadherin, C22orf3, MEG3, p15INK4b, RB1, CDH13, FGFR2, CDKN2C, RIZ1, SSTR5, Pttg1, autosomal and X-linked genes, CHST7 promoter | E-cadherin—aggressive behavior, MEG3—pathogenesis of NFPAs, CDH13—pathogenesis, Autosomal and X-linked genes—aggressiveness and response to treatment, CHST7—promoter for lineage | Related to pathogenesis |
| No association with clinical differences | Methylation-restricted digestion | MGMT promoter | Poor predictor of outcome of treatment with Temozolomide | No significant association between methylation status of MGMT and clinical outcomes |
| Methylation-specific PCR | p16, CDH1, p18INK4C, NDRG2, MGMT, GADD45b, CDH1, MEN1, hTERT promoter, STAT3 promoter | p16—aggressiveness, MGMT—expression, NDRG2—invasiveness, STAT3 promoter—prognostic marker | Genes otherwise proven related to aggressive pathologies showed no association with aggressive behavior in pituitary adenomas | |
| Chip-based | TERT, Myc-associated protein X transcription factor binding sites | TERT—pathogenesis, Myc-associated protein X transcription factor binding sites | ||
| Other methods | ZAR1, MGMT promoter, LINE-1 | MGMT promoter—immunoreactivity, LINE-1—aggresiveness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Møller, M.W.; Nortvig, M.J.; Andersen, M.S.; Poulsen, F.R. DNA Methylation in Pituitary Adenomas: A Scoping Review. Int. J. Mol. Sci. 2025, 26, 531. https://doi.org/10.3390/ijms26020531
Møller MW, Nortvig MJ, Andersen MS, Poulsen FR. DNA Methylation in Pituitary Adenomas: A Scoping Review. International Journal of Molecular Sciences. 2025; 26(2):531. https://doi.org/10.3390/ijms26020531
Chicago/Turabian StyleMøller, Morten Winkler, Mathias Just Nortvig, Mikkel Schou Andersen, and Frantz Rom Poulsen. 2025. "DNA Methylation in Pituitary Adenomas: A Scoping Review" International Journal of Molecular Sciences 26, no. 2: 531. https://doi.org/10.3390/ijms26020531
APA StyleMøller, M. W., Nortvig, M. J., Andersen, M. S., & Poulsen, F. R. (2025). DNA Methylation in Pituitary Adenomas: A Scoping Review. International Journal of Molecular Sciences, 26(2), 531. https://doi.org/10.3390/ijms26020531





