Abstract
Buchloe dactyloides is one of the typical ecological grass species, characterized by its strong salt tolerance. Hexokinase (HXK) plays a crucial role in plant growth, development, and resistance to abiotic stresses. To understand the function of HXKs in the salt tolerance of B. dactyloides, this study identified and analyzed the HXK gene family members using the whole-genome data of B. dactyloides. Additionally, transcriptomic methods were employed to investigate the expression levels and stress response patterns of the HXK family genes under salt stress. The results showed that 25 HXK genes were identified in the B. dactyloides HXK gene family, which were classified into three subfamilies based on the phylogenetic tree. Members within the same subfamily exhibited similar gene structures and conserved motifs. The promoter regions of BdHXKs contained numerous cis-regulatory elements associated with plant hormone responses, plant growth and development, and resistance to abiotic stresses. Quantitative real-time PCR analysis provided preliminary evidence that the BdHXK5, BdHXK7, and BdHXK23 genes might play important roles in the salt tolerance regulation of B. dactyloides. These findings offer a theoretical foundation for further elucidating the functions and molecular regulatory mechanisms of BdHXKs under salt stress. This study has provided a theoretical basis for the breeding of new varieties of ecological restoration grasses with stronger salt tolerance and better growth and development. This is of great significance for the improvement and ecological restoration of saline–alkali land.
1. Introduction
Buchloe dactyloides (Nutt.) Engelm., also known as buffalo grass or buffalograss, is a perennial herbaceous plant belonging to the genus Buchloe in the subfamily Eragrostoideae of the family Gramineae. It is native to the semi-arid temperate and subtropical regions of central North America [1]. Compared to most turfgrasses, B. dactyloides exhibits remarkable traits such as tolerance to poor soil, cold resistance, drought resistance, and salt–alkali tolerance, along with extremely low maintenance costs, making it an important ecological grass species for improving saline–alkali soils and a highly promising candidate for development [2].
In recent years, global climate change and the frequent occurrence of natural disasters have increasingly drawn attention to the problem of soil salinization. According to statistics from UNESCO and the Food and Agriculture Organization of the United Nations, the global area of saline–alkali land is approximately 954.38 million hectares [3]. In China, the scale of saline–alkali land is also substantial, covering about 99.13 million hectares, and these figures continue to grow [4]. Soil salt stress can disrupt the normal physiological metabolism of plants through osmotic stress, ion stress, and oxidative stress, thereby affecting plant growth and development and even causing plant death [5]. To resist salt stress, plants employ a series of complex physiological activities and metabolic processes as defense mechanisms.
Hexokinase (HXK) is ubiquitously present in various plants and primarily participates in the phosphorylation of hexoses. By regulating glycolysis, it provides substrates for other physiological processes, thereby influencing plant growth and development [6]. HXK genes have been cloned in several plant species, including Arabidopsis thaliana [7], rice [8], corn [9], tomato [10], and Sorghum bicolor [11], and all findings indicate their significant role in sugar metabolism [12]. Furthermore, HXK can function as a regulatory signal. For instance, PsHXK1 and PsHXK2 can enhance glucose sensitivity, leading to the accumulation of anthocyanins [13]; GcHXK in citrus can induce stomatal closure, reducing the photosynthetic rate and affecting seed germination [14]. Many studies have confirmed that HXK is also involved in biotic and abiotic stress responses. Under salt stress, hexokinase catalyzes the phosphorylation of hexoses, providing energy and products that participate in osmotic regulation and antioxidant defense, helping plants withstand the damage caused by salt stress. PrunusHXK3 can enhance the salt and drought tolerance of Arabidopsis transgenic plants by altering primary C-metabolism [15]; the overexpression of GmHXK15 significantly enhances the tolerance of transgenic soybeans to alkaline stress [16]; CsHXKs genes in tea plants play a critical role in responding to four types of abiotic stresses: drought, salinity, cold, and exogenous abscisic acid (ABA) [17]. Therefore, HXK is considered a bifunctional enzyme, not only catalyzing metabolic reactions such as glucose phosphorylation but also serving as a key connecting element between sugar signaling and plant hormone signaling, playing a vital role in plant growth and development and environmental adaptation [18].
Despite the important functions of HXK genes, there are no reported studies on the HXK gene family in B. dactyloides. This study, based on the whole-genome data of B. dactyloides (the genome of the cultivar homologous tetraploid was made available in high quality in 2024), identified and analyzed 25 members of the HXK gene family. It included predictions of protein physicochemical properties, chromosomal localization, cis-acting element analysis, motif prediction, and analysis of conservation motifs. Additionally, the expression patterns of these genes in different tissues under salt stress were investigated. The aim is to provide a theoretical foundation for subsequent research on the functions of HXK genes and for the genetic improvement of salt tolerance using these genes.
2. Results
2.1. Identification and Physicochemical Property Analysis of B. dactyloides HXK Family Members
Based on the HXK sequences from Arabidopsis thaliana, rice (Oryza sativa), and Brachypodium distachyon, a Blast search was conducted on the full-length genome data of B. dactyloides. A total of 25 HXK genes were identified and named BdHXK1 to BdHXK25 (Supplementary Table S1). The physicochemical properties of these 25 BdHXKs genes were analyzed using the MEME online tool. The amino acid sequence lengths ranged from 441 residues (BdHXK1) to 531 residues (BdHXK23). The molecular weights varied from 47,372.95 Da (BdHXK1) to 58,056.96 Da (BdHXK23). As plants adapt to their environment, they develop unique metabolic requirements. This leads to the mutation and duplication of encoding genes, which in turn alters the protein structure and thus affects the molecular weight. The theoretical isoelectric points (pI) ranged from 5.03 to 6.02, indicating that all BdHXKs family proteins are acidic and rich in acidic amino acids. This is somewhat similar to what is observed in some grasses, such as Setaria italica, where many hexokinases also have acidic pI values [19]. The instability index of BdHXKs genes ranged from 33.41 to 46.38, suggesting that these genes are relatively stable. The aliphatic index was between 89.65 and 103.84, and the hydropathy index of BdHXKs genes ranged from -0.172 to 0.167. BdHXK2, BdHXK3, BdHXK8 to BdHXK13, and BdHXK22 to BdHXK25 had positive hydropathy indices, indicating that they are likely hydrophobic proteins. In contrast, BdHXK1, BdHXK4 to BdHXK7, and BdHXK14 to BdHXK21 had negative hydropathy indices, suggesting that they are hydrophilic proteins. Subcellular localization predictions indicated that BdHXK6, BdHXK7, BdHXK18, and BdHXK21 are located in the chloroplast, while BdHXK2, BdHXK3, BdHXK8 to BdHXK10, BdHXK19 to BdHXK20, and BdHXK22 to BdHXK25 are located in the mitochondria. The remaining proteins were found to be predicted in both chloroplasts and mitochondria. Environmental factors drive the evolution of HXK in plants toward different physicochemical properties to respond to diverse ecological niches. For example, under salt stress, B. dactyloides generates HXK with enhanced stability. This ensures that effective glycolysis can provide energy to counteract ion toxicity. Meanwhile, as salt stress causes water efflux from cells, the hydrophilicity of HXK increases. This promotes sufficient substrate–enzyme interactions, thus helping the plant maintain growth under osmotic stress.
2.2. Chromosomal Localization and Gene Structure Analysis of the B. dactyloides HXK Gene Family
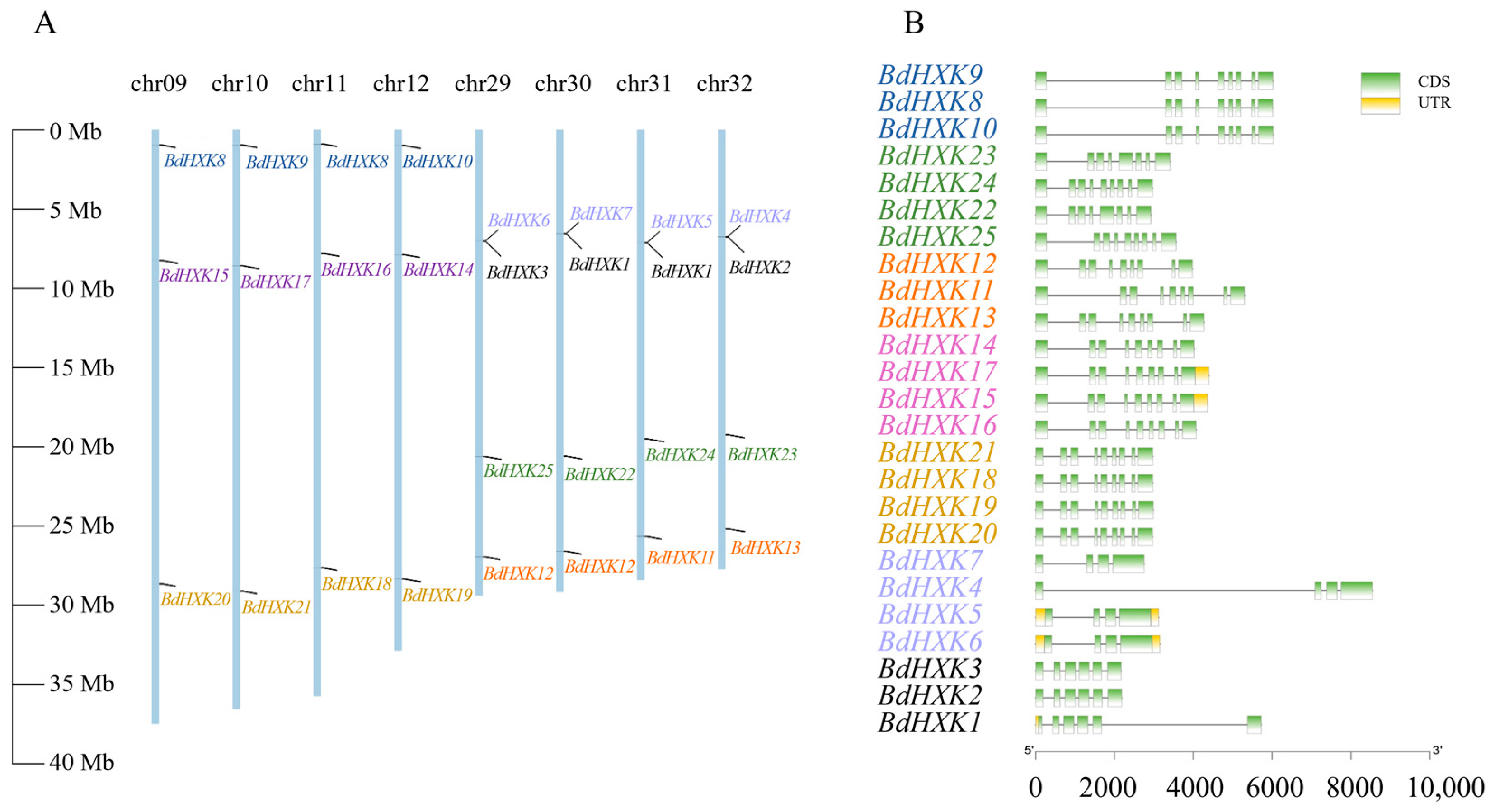
The chromosomal positions of BdHXKs genes were determined using TBtools software. Buchloe dactyloides has a total of 40 chromosomes. Chromosomes 9, 10, 11, and 12 each contained three BdHXK genes, and their distribution positions were similar. Chromosomes 29, 30, 31, and 32 each contained four BdHXKs genes, with similarly distributed positions. BdHXK3 and BdHXK6, BdHXK1 and BdHXK7, BdHXK1 and BdHXK5, and BdHXK2 and BdHXK4 were clustered in small regions (Figure 1A). These results indicate that BdHXK gene family members are unevenly distributed across the chromosomes, suggesting that gene tandem duplication events may have occurred during evolution, which could facilitate the rapid response of B. dactyloides to salt stress and enhance its salt tolerance. Our gene structure analysis of BdHXK genes using TBtools software (Figure 1B) showed that the structures of genes at the same positions were highly similar. Each gene contained three to eight introns. The number of exons in different genes ranged from 4 to 9, with 4 genes (16% of the total) containing 4 exons, 3 genes (12% of the total) containing 6 exons, 2 genes (8% of the total) containing 8 exons, and 16 genes (64% of the total) containing 9 exons.
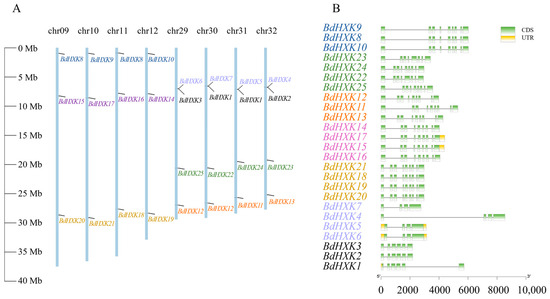
Figure 1.
Chromosome distribution and gene structure of HXK family genes in Buchloe dactyloides. (A) A total of 8 chromosomes with varying lengths are shown in relation to the Mb (million base pair) scale on the left, and individual chromosomes (bars) are labeled with respective BdHXK genes. (B) Genetic structure analysis of BdHXKs gene family members, with exons represented by yellow rectangles and introns represented by black lines.
2.3. Phylogenetic Tree and Conserved Motif Analysis of the B. dactyloides HXK Gene Family
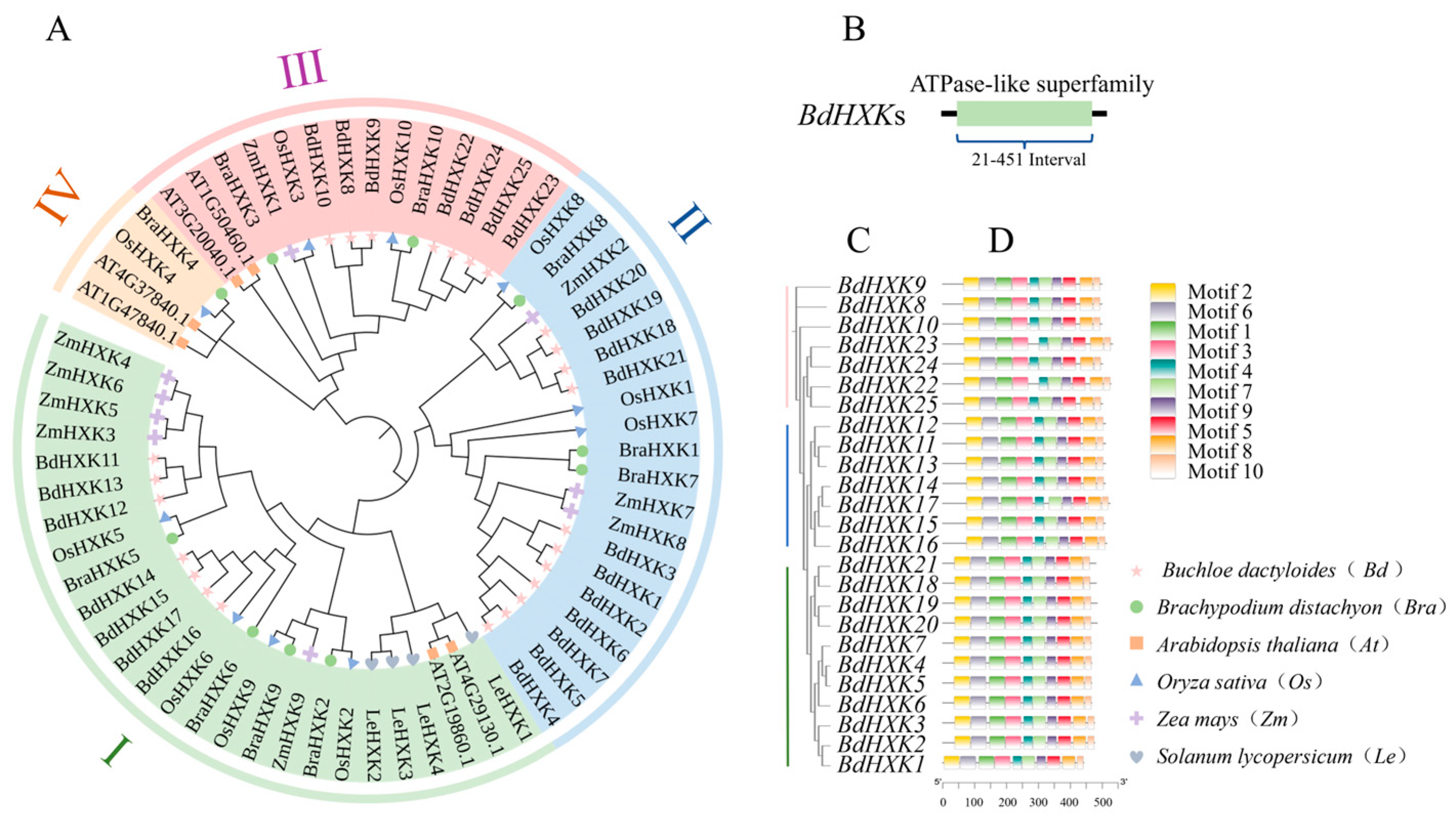
To further understand the evolutionary relationships of BdHXKs genes with HXK genes in other species, a phylogenetic tree was constructed by aligning 6 HXK genes from Arabidopsis thaliana, 10 HXK genes from rice (Oryza sativa), 9 HXK genes from Zea mays, 4 HXK genes from Solanum lycopersicum, and 10 HXK genes from Brachypodium distachyon with the 25 HXK genes identified in B. dactyloides (Figure 2A). The results indicate that the 25 BdHXKs genes have high homology with those from rice and Brachypodium distachyon, but a relatively distant relationship with those from Arabidopsis thaliana. Based on their homology with rice HXK genes, the BdHXKs genes were divided into three phylogenetic clusters [20]: Cluster I: BdHXK11 to BdHXK17 and OsHXK2, OsHXK5, OsHXK6, and OsHXK9; Cluster II: BdHXK1 to BdHXK7 and BdHXK18 to BdHXK21 with OsHXK1, OsHXK7, and OsHXK8; Cluster III: The remaining BdHXK genes and OsHXK3 and OsHXK10; Cluster IV: None of the BdHXK genes were clustered with OsHXK4.
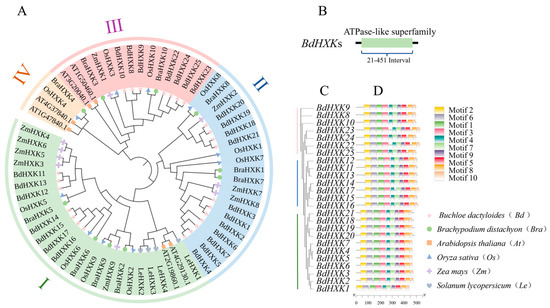
Figure 2.
Phylogenetic trees, conserved domain, and conserved motif of HXK family members for Buchloe dactyloides. (A) Phylogenetic tree was constructed based on HXK sequences of Buchloe dactyloides, Arabidopsis thaliana, Oryza sativa, Zea mays, Solanum lycopersicum, and Brachypodium distachyon. The tree was then categorized into four groups, each represented by a distinct color. (B) Conserved domain analysis of Buchloe dactyloides HXK gene family members. (C) Evolutionary relationship of HXK gene family in Buchloe dactyloides. (D) A total of 10 motifs were identified, represented by rectangles of different colors.
Furthermore, the conserved domains of the BdHXKs gene family members were analyzed using the NCBI conserved domain search program (Figure 2B). The results show that BdHXK proteins contain the ASKHA ATPase-like superfamily domain, which is generally located between residues 21 and 451. Proteins within the same subfamily share similar conserved domains, indicating that genes in the same subfamily have a relatively conserved evolution and may have similar regulatory mechanisms and functions. Studies have shown that these proteins can accelerate sugar metabolism by catalyzing the phosphorylation of glucose, fructose, and sorbitol, producing more ATP to meet cellular energy requirements and enhance salt tolerance [21]. In some plants, hexokinases can also regulate the activity of sodium-potassium ion exchange factors, promoting the efflux of sodium ions from cells and maintaining ion balance, thereby enhancing plant salt tolerance [21].
To further identify the conserved motifs of the BdHXKs proteins, the MEME website was used to predict the conserved motifs of the encoded proteins, and 10 conserved motifs were identified (Figure 2D). Combining the phylogenetic results of the BdHXKs genes (Figure 2C), it is evident that most family members contain all 10 motifs, and their arrangement order is also largely the same. This suggests that the BdHXK gene family members have similar and conserved structures and may play similar roles in salt tolerance regulation in B. dactyloides.
2.4. qRT-PCR Validation Analysis of the B. dactyloides HXK Family Under Salt Stress
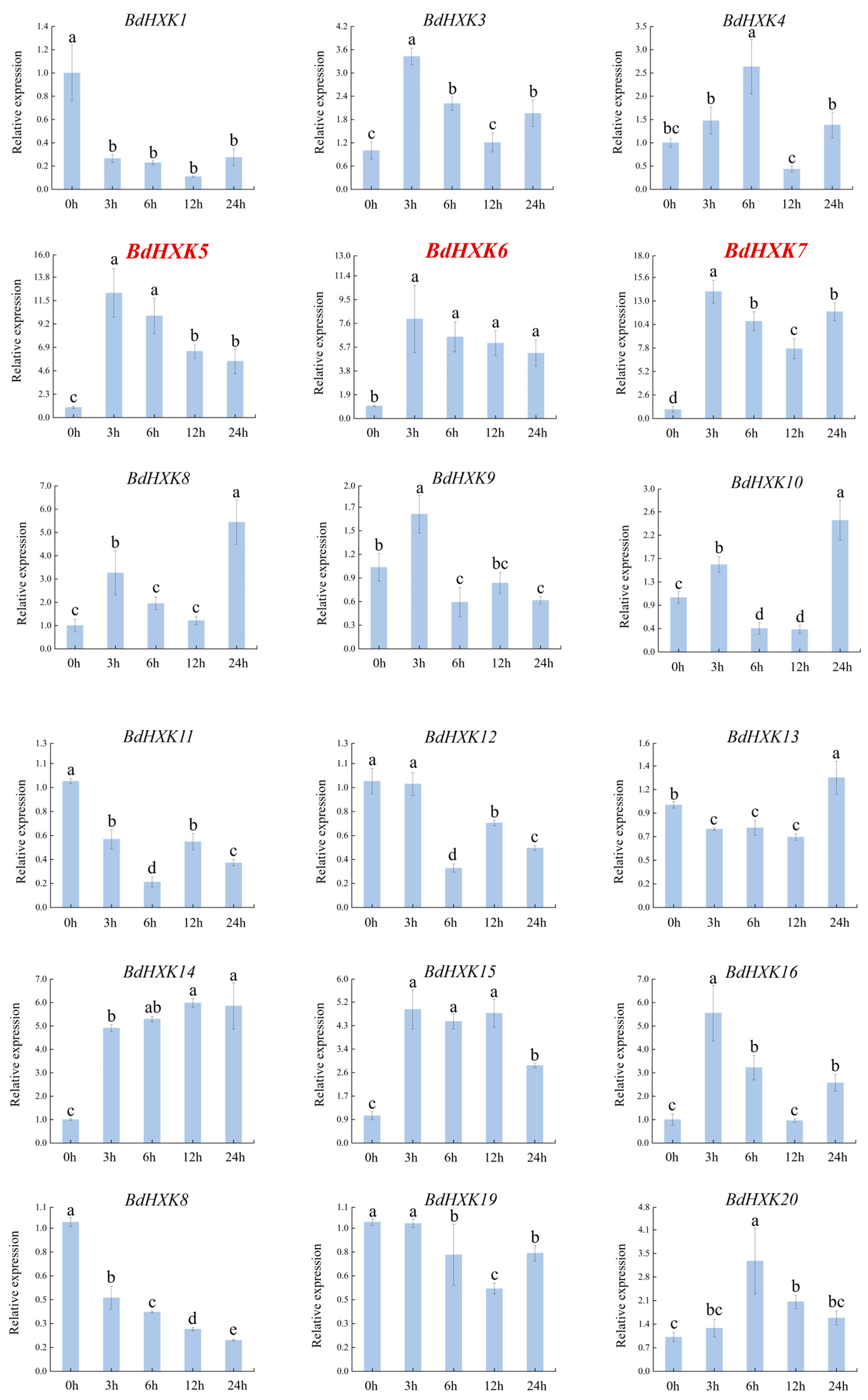
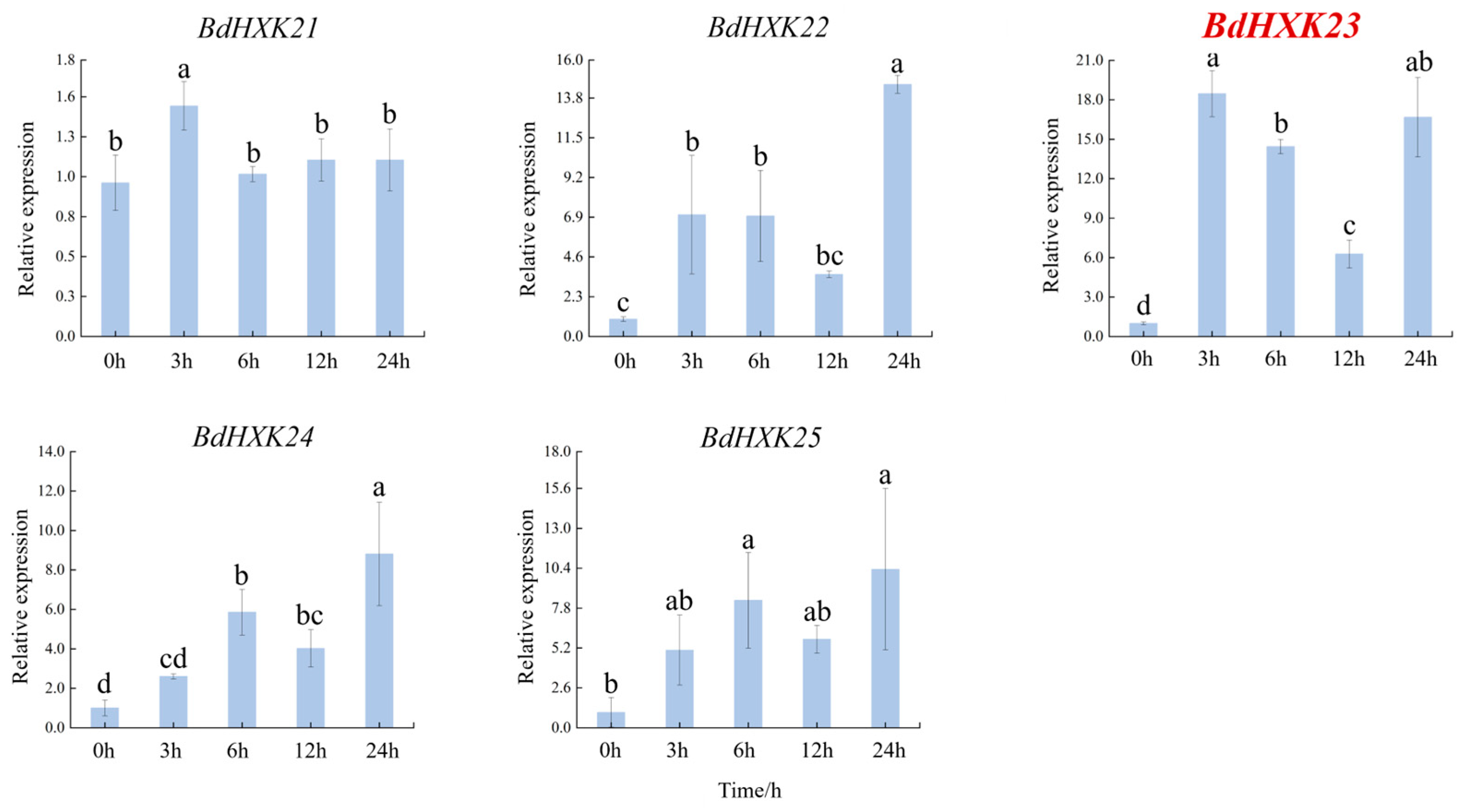
To investigate the response of HXK genes to salt stress, the differential expression of BdHXKs genes of B. dactyloides under 600 mM NaCl stress was analyzed using real-time quantitative PCR (RT-qPCR). The RT-qPCR results (Figure 3) show the following expression patterns: BdHXK5, BdHXK6, BdHXK14, and BdHXK20: their relative expression levels first increased and then decreased; BdHXK9, BdHXK15, and BdHXK21: their relative expression levels first increased, then decreased, and showed a slight recovery at 12 h; BdHXK3, BdHXK4, BdHXK7, BdHXK8, BdHXK10, BdHXK16, BdHXK22, BdHXK23, BdHXK24, and BdHXK25: their relative expression levels first increased and then decreased, with a recovery at 24 h; BdHXK1, BdHXK13, and BdHXK19: their relative expression levels first decreased and then increased; BdHXK11 and BdHXK12: their relative expression levels first decreased, then increased, and showed a decline again at 24 h; BdHXK18: it exhibited a unique pattern, showing continuous decline without any recovery; BdHXK2 and BdHXK17: they did not show significant expression changes under salt stress.
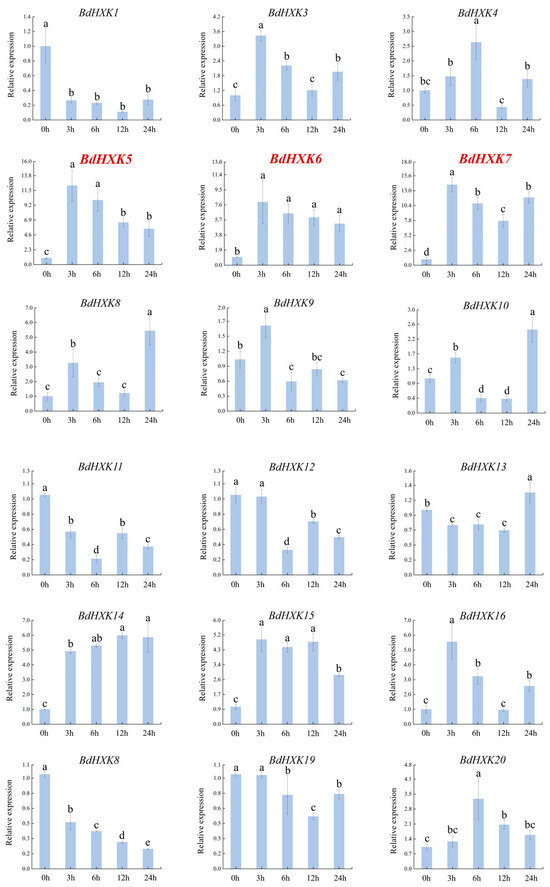
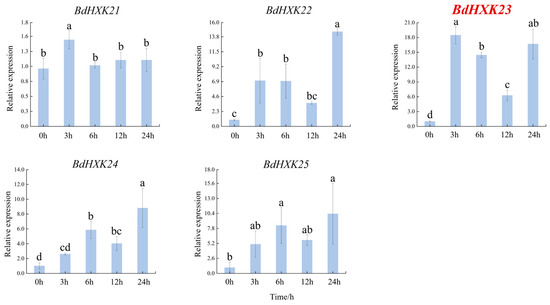
Figure 3.
Expression pattern of HXK gene in Buchloe dactyloides under salt stress. The horizontal coordinate is the processing time, and the vertical coordinate is the relative expression level. Duncan’s test of SPSS was used to determine the significance of the differences. Different letters indicate significant differences between groups (p < 0.05).
Among these, BdHXK5, BdHXK6, BdHXK7, and BdHXK23 were more sensitive to salt stress, and their relative expression levels were significantly upregulated, making them potential candidates for further research on salt tolerance in B. dactyloides.
2.5. Tissue-Specific Expression Analysis of Key B. dactyloides HXK Genes
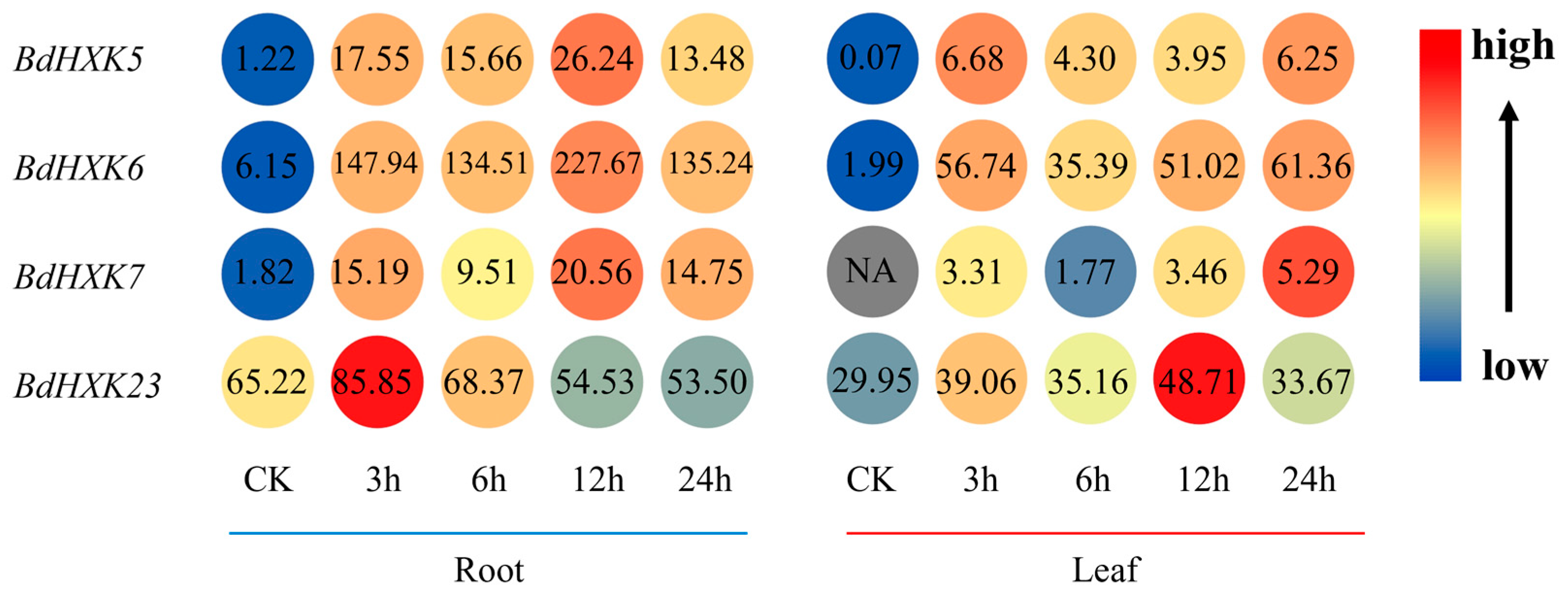
Transcriptome data were used to analyze the expression of HXK genes in the roots and leaves of B. dactyloides (Figure 4). The results show that the expression levels of these genes vary across different tissues. HXK genes exhibit higher expression in the roots compared to the leaves, where the expression levels are significantly reduced. Among the four key genes, their expression levels were significantly upregulated at 3 h under salt stress. In the roots, the expression level of BdHXK6 was notably higher than that of the other genes, suggesting that it may play a crucial role in salt stress tolerance.
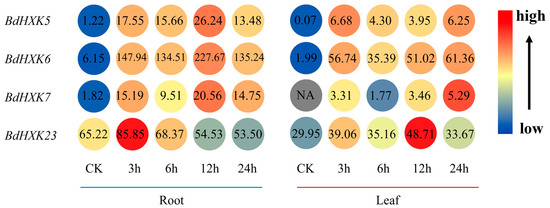
Figure 4.
Expression differential of Buchloe dactyloides HXK key gene in different tissues. The expression profiles of the BdHXK5, BdHXK6, BdHXK7, and BdHXK23 genes under different tissues and different durations of salt stress treatment are presented in a heatmap. In the heatmap, the color ranging from blue to red indicates the expression levels from low to high, with the specific expression amounts shown as numerical values. On the left side is the expression level of the key genes in roots at different periods, and on the right side is the expression level in leaves at different periods.
2.6. Promoter Analysis of Key Members of the B. dactyloides HXK Gene Family
Analysis of the cis-acting elements in the promoters of B. dactyloides can provide insights into the tissue-specific or stress-responsive expression patterns of these genes. As shown in Supplementary Figure S1, the promoters of each BdHXK gene generally contain different types of plant hormone response elements, abiotic stress response elements, and plant growth and development response elements. The plant hormone response elements include 10 types: abscisic acid response elements (ABRE), methyl jasmonate response elements (CGTCA-motif and TGACG-motif), auxin response elements (TGA-element and AuxRR-core), gibberellin response elements (P-box and TATC-box), zein metabolism regulation elements (O2-site), and salicylic acid response elements (TCA-element). The abiotic stress response elements include six types: anoxic-inducible cis-elements (ARE), hypoxia-specific induction enhancers (GC-motif), MYB drought-induced binding sites (CCAAT-box and MBS), low-temperature response cis-elements (LTR), and cis-acting factors involved in salt stress defense and response (TC-rich repeats). Finally, the plant growth and development response elements include 15 types: light response elements (G-box, AAAC-motif, ATCT-motif, Box4, Box II, GA-motif, GT1-motif, I-box, Pc-CMA2c, Sp1, TCCC-motif, and TCT-motif), meristematic tissue expression regulation elements (CAT-box), and seed-specific regulation cis-elements (RY-element).
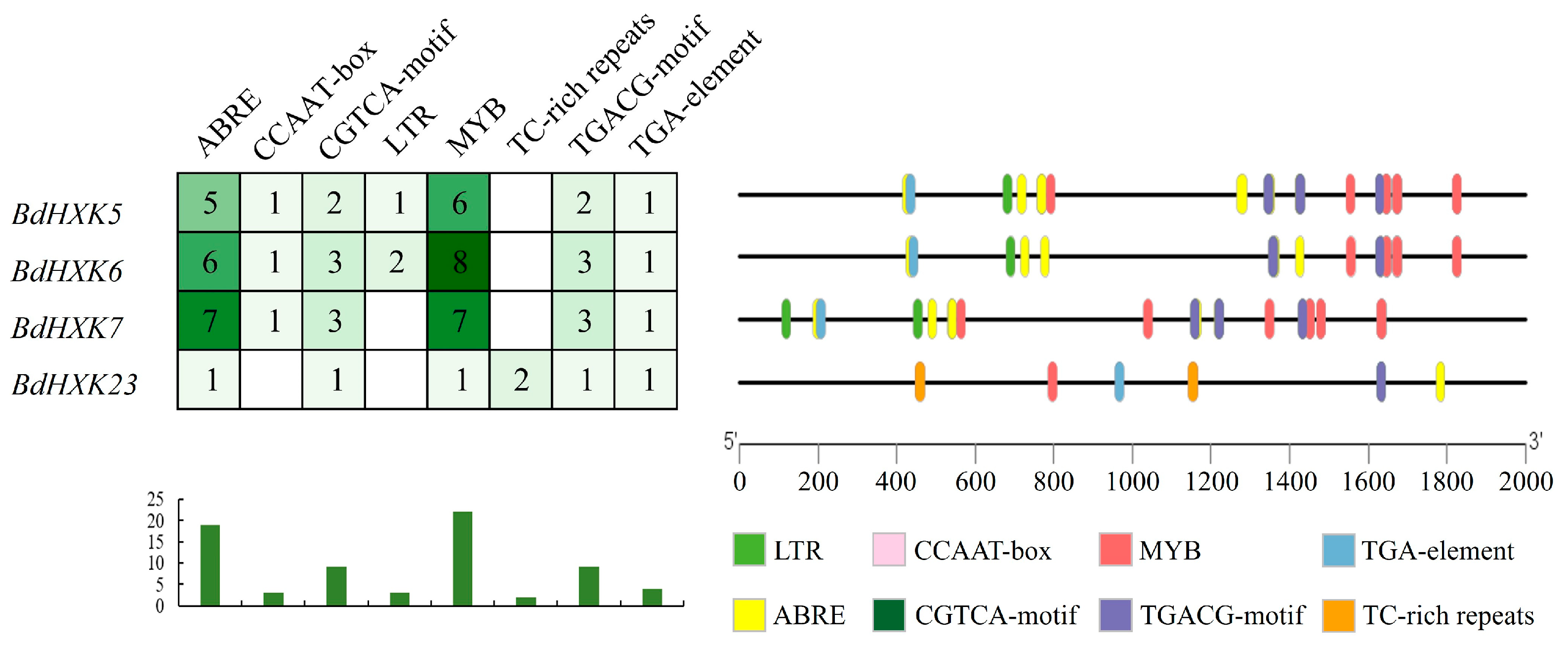
Each of the 25 BdHXKs genes contains 11 to 20 different cis-acting elements, with significant variations in the types and quantities of these elements. All members of the BdHXKs gene family contain light response, abscisic acid, and methyl jasmonate-related cis-acting elements, which are significantly associated with plant abiotic stress tolerance. This suggests that the BdHXKs gene family in B. dactyloides has diverse functions and may play important roles in plant growth and development as well as in resistance to salt, drought, low temperature, and anoxic stresses. Notably, some members, particularly BdHXK6, contain unique cis-acting elements such as TC-rich repeats, TCA-element, and ARE, which can interact with specific genes to regulate the expression of downstream genes, thereby influencing the defense and stress response of B. dactyloides to salt stress.
Promoter analysis of the key genes BdHXK5, BdHXK6, BdHXK7, and BdHXK23 was conducted, and the results are shown in Figure 5. BdHXK6 contains the highest number of various cis-acting elements, while BdHXK23 has the fewest, lacking the CCAAT-box and LTR elements but containing the unique TC-rich repeats element. Statistical analysis revealed that abscisic acid, MYB drought-induced binding sites, and methyl jasmonate-related cis-acting elements are the most prevalent. Research has shown that abscisic acid is significantly associated with plant abiotic stress tolerance, and methyl jasmonate is related to plant growth and development, as well as abiotic stress tolerance. This indicates that BdHXK5, BdHXK6, BdHXK7, and BdHXK23 may play strong roles in plant growth and development and in responses to abiotic stress.
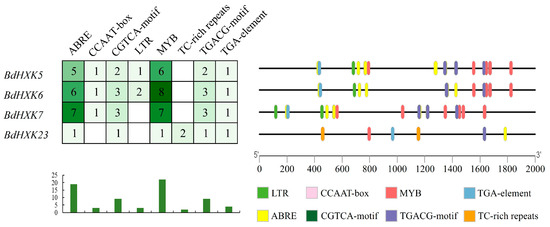
Figure 5.
Buchloe dactyloides HXK key gene cis-acting element. Different cis-acting elements are represented in different colors. These cis-acting elements may be related to plant stress resistance and hormone regulation.
2.7. Correlation Network Analysis
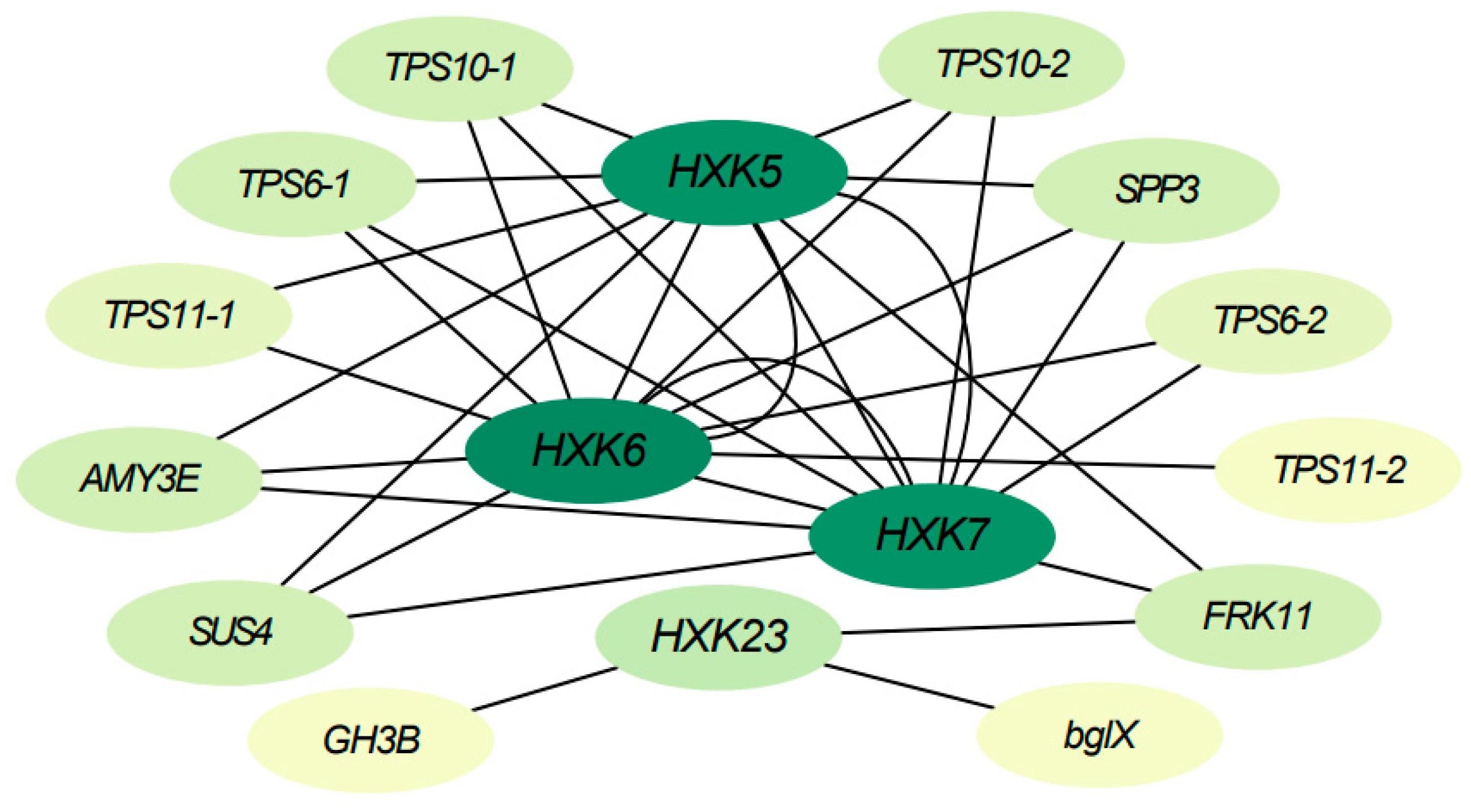
To further understand the functional mechanisms of BdHXKs genes, a correlation network analysis was performed based on transcriptome data for BdHXK5, BdHXK6, BdHXK7, and BdHXK23. The results are presented in Figure 6, and detailed information on the interacting genes is provided in Supplementary Table S3. The interaction gene information shows that BdHXK5, BdHXK6, BdHXK7, and BdHXK23 are significantly positively correlated with genes from families such as trehalose-6-phosphate synthase (TPS), sucrose phosphate synthase (SPP), α-amylase (AMY), and sucrose synthase (SUS). This suggests the possibility of upstream and downstream interactions between these genes. Moreover, the same BdHXK gene can interact with multiple other BdHXK genes, indicating a tight interaction network within the BdHXK gene family. These findings suggest that HXK family genes in B. dactyloides may regulate various metabolic processes through interactions with genes involved in carbohydrate supply and conversion, providing more energy and carbon sources to the plant, helping to maintain osmotic balance in plant cells, and reducing cell dehydration caused by salt stress, thereby enabling the plant to better cope with salt stress conditions.
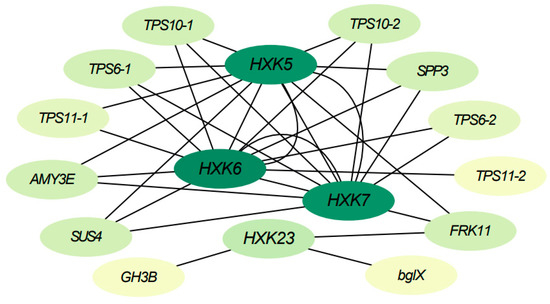
Figure 6.
Buchloe dactyloides BdHXK5, BdHXK6, BdHXK7, and BdHXK23 genes’ correlation network analysis in leaves and roots. The ovals represent each gene. The darker the color, the higher the number of interactions with other genes. Edges represent interactions between genes.
3. Discussion
Hexokinase (HXK) not only plays a crucial role in plant metabolism but is also actively involved in the response to various abiotic stresses such as salinity, alkalinity, and drought. To date, 6 HXK members have been reported in Arabidopsis thaliana [7], 10 members in rice (Oryza sativa) [8], 9 members in Zea mays [9], 4 members in Solanum lycopersicum [10], and 10 members in Brachypodium distachyon. However, no studies have reported HXKs in Buchloe dactyloides. In this study, based on the B. dactyloides genome database, 25 members of the HXK gene family were identified, which is significantly more than in other plants. The HXK gene family plays an important role in the growth, development, and environmental adaptation of B. dactyloides. In the long-term evolutionary process, B. dactyloides may face specific natural selection pressure, making the expansion of the HXK gene family a favorable evolutionary strategy. Therefore, we speculate that the number of HXK gene family members may increase in order to adapt to the environment such as salt stress, so as to better regulate various physiological processes.
In previous research, we comprehensively applied flow cytometry and the chromosome squash method and found that B. dactyloides had multiple ploidies [22]. We selected the most representative tetraploid materials for in-depth study. As a homologous tetraploid plant, B. dactyloides presents a unique gene distribution pattern in its genetic architecture. Specifically, on the homologous chromosomes numbered 9, 10, 11, and 12, each chromosome carries three members of the HXK gene family, and on the homologous chromosomes numbered 29, 30, 31, and 32, each chromosome has four members of the HXK gene family. Through in-depth research, it was found that there were differences in the sequences of alleles on homologous chromosomes. Therefore, the proteins encoded by them were also different, and it was further speculated that there would also be differences in their functions. Hence, we determined that these genes were different gene individuals. This phenomenon was most likely the result of natural selection during the long evolutionary process. Previous studies have shown that most HXK genes in model plants such as rice, Arabidopsis, foxtail millet, and sorghum contain nine exons [23]. In our study, 16 of the 25 BdHXKs genes contained 9 exons, while the remaining members had fewer exons due to intron loss, containing 4, 6, or 8 exons. This indicates that the gene structures of different BdHXK members are highly variable. By studying the structural changes, we can infer the evolutionary history of the gene family. Current research on HXK genes in rice and Arabidopsis suggests that they originated from a common ancestor [20]. Given the close evolutionary relationships between B. dactyloides and rice, Arabidopsis, and B. distachyon, it is reasonable to hypothesize that B. dactyloides may have evolved from the same ancestor.
Through the construction and analysis of a phylogenetic tree for BdHXKs gene in B. dactyloides, Arabidopsis thaliana, Oryza sativa, Zea mays, Solanum lycopersicum, and Brachypodium distachyon, BdHXKs can be clearly divided into three subfamilies, consistent with previous findings in rice [20]. Each BdHXK gene in B. dactyloides has at least one homologous gene in rice and B. distachyon. For example, BdHXK1 to BdHXK7 share the closest evolutionary relationship with OsHXK1, suggesting that they may regulate sugar signal transduction and interact with other signaling molecules to maintain ion balance inside and outside cells [24]. BdHXK14 is most closely related to OsHXK6 in rice and BdHXK6 in B. distachyon, indicating that it may be light-induced and have similar functions, potentially contributing to the plant’s response to abiotic stress, such as by regulating sugar metabolism and energy supply to help B. dactyloides adapt to high-salinity environments and maintain normal cellular physiological functions [25].
Subcellular localization analysis of BdHXKs family members revealed that BdHXK6, BdHXK7, BdHXK18, and BdHXK21 are specifically located in chloroplasts, where they can phosphorylate hexoses or bind to the chloroplast membrane to phosphorylate glucose [26]. Notably, BdHXK7 showed a significant upregulation under salt stress, with its expression level increasing by up to 14-fold. This suggests that BdHXK7 may accelerate the catalysis of sugar metabolism under salt stress, providing energy for the synthesis of osmotic regulatory substances such as proline and antioxidant enzymes [27]. BdHXK2, BdHXK3, BdHXK8, BdHXK9, BdHXK10, BdHXK19, BdHXK20, and BdHXK22 to BdHXK25 are specifically located in mitochondria, possibly through membrane-anchored structures, and may contribute to the regulation of plant sugar signaling [26]. Other BdHXK proteins are expressed in both chloroplasts and mitochondria, collectively participating in the regulation of sugar metabolism and plant growth and development.
HXK plays an irreplaceable role in the normal growth, development, and metabolic activities of organisms [28]. Multiple studies have shown that HXK, in response to salt stress, primarily catalyzes the phosphorylation of hexoses, converting them into products like glucose-6-phosphate, thereby initiating metabolic pathways such as glycolysis to provide cells with sufficient energy to cope with the stress caused by salinity [29]. Additionally, HXK can regulate the activity of sodium and hydrogen ion exchangers through phosphorylation, promoting the efflux of sodium ions from cells to maintain the intracellular ion balance [30]. For example, MdHXK1 in apples interacts with the Na+/H+ exchanger and phosphorylates the Ser275 residue, enhancing the stability and transport activity of MdNHX1, thus improving the salt tolerance of apple plants [31].
The differential expression analysis revealed that the expression levels of the remaining BdHXK members varied, indicating that different members of the HXK gene family play distinct roles in the plant’s response to salt stress. Notably, BdHXK5, BdHXK6, BdHXK7, and BdHXK23 showed a significant upregulation of up to 14–18-fold under salt stress, making them potential candidate genes for further functional studies on salt stress response in B. dactyloides. Subsequently, we analyzed the expression patterns of BdHXK5, BdHXK6, BdHXK7, and BdHXK23 genes in the roots and leaves of B. dactyloides through transcriptome data. The research results revealed that the expression levels of these genes in the roots were significantly higher than those in the leaves. The HXK gene family demonstrated the characteristic of high expression in root tissues. As the root is the main salt-sensing organ of plants, this finding implies that the HXK genes may play a significant role in the process of B. dactyloides responding to salt stress.
Further analysis of the cis-acting elements in the promoter regions of key BdHXKs genes in B. dactyloides revealed that most of them contain stress-responsive elements such as ABRE, ARE, and TC-rich repeats [32]. This suggests that the BdHXKs gene family may play important roles in plant growth and development, as well as in resistance to abiotic stresses like salinity, drought, low temperature, and anoxia.
Transcriptome correlation network analysis revealed a significant positive correlation between key BdHXK members and genes from families such as TPS, SPP, AMY, and SUS, suggesting potential upstream and downstream interaction relationships. For instance, TPS is involved in the synthesis of trehalose-6-phosphate. The interaction with HXK can promote the production of trehalose within cells, ensuring adequate energy supply and helping B. dactyloides cells maintain osmotic balance and reduce water loss and turgor pressure decline caused by salt stress [33]. SPP primarily converts sucrose-6-phosphate to sucrose, and its interaction with HXK can increase sucrose synthesis, thereby activating certain antioxidant enzymes and helping plants mitigate oxidative damage [34]. AMY degrades starch to produce glucose, and its interaction with HXK under salt stress can facilitate the rapid entry of glucose into glycolysis and other metabolic pathways, providing more energy to the plant [35]. SUS, which is involved in the synthesis and degradation of sucrose, can accelerate the breakdown of sucrose into glucose and fructose under salt stress [36]. After phosphorylation by HXK, these sugars release energy, affecting hormone levels such as auxin and cytokinin, and thus maintaining basic cellular physiological functions, enabling plants to better adapt to salt stress environments and sustain growth [37].
This study is the first to elucidate the number, distribution, and structural characteristics of the HXK gene family in the B. dactyloides genome under salt stress and to analyze their expression patterns over different time periods. These findings can provide important genetic information for future research on gene functions and molecular mechanisms and for the targeted improvement of salt tolerance in B. dactyloides.
4. Materials and Methods
4.1. Experimental Materials and Salt Stress Treatment
The experimental material was Buchloe dactyloides (Nutt.) Engelm., provided by the Chinese Academy of Forestry Sciences. Plump B. dactyloides seeds were surface-sterilized by soaking in 0.5% sodium hypochlorite for 5 min with occasional stirring to ensure thorough disinfection. After rinsing thoroughly, the seeds were soaked in 0.5% potassium nitrate for 24 h. Following another thorough rinse, the seeds were soaked in 2000 mg/L gibberellic acid for 24 h. After rinsing again and soaking in distilled water for 18 h, the seeds were washed and sown in seedling pots. The pots had an inner diameter of 12 cm and a depth of 11 cm, and the seedling substrate was nutrient soil. The plant growth conditions were maintained at a day/night temperature of 28 °C/24 °C. Twenty days after sowing, uniformly growing seedlings were selected and treated with 600 mM NaCl solution. Samples were collected at 0, 3, 6, 12, and 24 h after treatment, washed thoroughly, and divided into aboveground and underground parts. Each part was rapidly frozen in liquid nitrogen and stored at −80 °C for further analysis.
4.2. Identification and Physicochemical Property Analysis of B. dactyloides HXK Gene Family Members
The genome data of B. dactyloides was downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/). The protein sequences of the known hexokinase (HXK) gene family members from Arabidopsis thaliana (GenBank accession number: GCA_000001735.2), Oryza sativa (GenBank accession number: GCA_034140825.1), Zea mays (GenBank accession number: GCA_902167145.1), Solanum lycopersicum (GenBank accession number: GCA_036512215.2), and Brachypodium distachyon (GenBank accession number: GCA_000005505.4) were used as queries to perform Blastp comparisons against the full-length genome data of B. dactyloides to identify the HXK gene family members in B. dactyloides. All the websites were accessed on 2 November 2024. The physicochemical properties of the identified B. dactyloides HXK proteins, including the number of amino acids, relative molecular weight, isoelectric point (PI), and instability index, were predicted using the TBtools software. In order to ensure the accuracy and reliability of the results, we used the Plant—mPLoc subcellular localization prediction website (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/, accessed on 11 November 2024) and the WoLF PSORT (https://wolfpsort.hgc.jp/, accessed on 13 November 2024) to predict B. dactyloides HXK proteins’ subcellular localization.
4.3. Phylogenetic Tree Construction and Salt Tolerance Expression Analysis of the B. dactyloides HXK Gene Family
To investigate the phylogenetic relationships within the B. dactyloides HXK gene family, multiple sequence alignments of the HXK protein sequences from B. dactyloides, Arabidopsis thaliana, Oryza sativa, Zea mays, Solanum lycopersicum, and Brachypodium distachyon were performed using MEGA X. The aligned sequences were used to construct a phylogenetic tree using the maximum likelihood (ML) method in TBtools software. The tree parameters were submitted to the ITOL website (https://itol.embl.de/, accessed on 14 November 2024) for beautification. The classification of the B. dactyloides HXK gene family was based on the classification method used for the rice HXK gene family [20]. This comparative analysis provides deeper insights into the evolutionary relationships and potential functional similarities of HXK genes across different species.
To understand the expression patterns of BdHXK genes in response to salt stress, the expression data of BdHXK genes under different salt stress treatments were extracted from the B. dactyloides transcriptome data. The TBtools software was used to generate a heatmap of the expression levels of key BdHXK genes in the roots at different time points of salt stress treatment.
4.4. Chromosomal Localization, Structure Analysis, Conserved Domain Analysis, Conserved Motif Analysis, and Cis-Acting Element Analysis of the B. dactyloides HXK Gene Family
The chromosomal positions of HXK genes in B. dactyloides were visualized using TBtools software. The coding sequence (CDS) of B. dactyloides HXK genes was used to obtain gene structure diagrams via TBtools software. The amino acid sequences of B. dactyloides HXK proteins were input into the NCBI conserved domain search program to identify the conserved domains they contain. The amino acid sequences were also uploaded to the MEME online tool (http://meme-suite.org/tools/meme accessed on 16 November 2024) to predict conserved motifs in the BdHXK gene family, with the number of motifs set to 10 [38]. The results were visualized using TBtools software.
The promoter sequences, 2000 bp upstream of the start codon of B. dactyloides HXK genes, were analyzed for cis-acting elements using the Plant CARE online tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/, accessed on 17 November 2024) [39]. The analysis results were visualized using TBtools software.
4.5. Real-Time Quantitative PCR (RT-qPCR) Detection
The RNA was extracted using the TaKaRa RNA extraction kit, and cDNA was synthesized by reverse transcription(The kit was provided by Takara Bio Technology, Beijing Co., Ltd.). Primers were designed using Primer Premier 5.0 software (Premier Biosoft International is located in Palo Alto, CA, USA) and synthesized by Beijing RiboBio Tech Co., Ltd. (Beijing, China) (Supplementary Table S2). The PCR amplification was performed using 2× M5 HiPer SYBR Premix Es Taq (with Tli RNaseH) (Beijing PolyMedGene Biotech Co., Ltd., MF787). The reaction mixture was prepared as follows: 0.8 μL of cDNA template, 0.4 μL each of forward and reverse primers, 5 μL of 2× M5 HiPer SYBR Premix Es Taq, 0.2 μL of 50× ROX Reference Dye II, and 3.2 μL of ddH2O, bringing the total volume to 10 μL. The PCR reaction conditions were as follows: initial denaturation at 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 30 s; followed by 95 °C for 5 s, 60 °C for 60 s, and 95 °C for 15 s; and a final extension at 8 °C for an indefinite period. Each gene was analyzed in triplicate. DNAJ was used as the reference gene [40]. The expression levels of the target genes were quantified using the 2−ΔΔCt method [41], with the expression level of DNAJ at 0 h set to 1. The expression levels of other genes and at different time points were compared to this reference for quantification.
4.6. WGCNA Analysis of the B. dactyloides HXK Gene Family
To determine the functions and evolutionary relationships of the BdHXKs genes, we analyzed the transcriptome data of the key genes. Firstly, the WGCNA network was constructed using the one-step method [42]. The FPKN values were input, and the genes with similar expression patterns in the output were grouped into one module. The key BdHXK genes were selected as the hub genes to construct a plant co-expression network. The network was visualized using Cytoscape v3.10.0 [43].
4.7. Data Analysis
Data analysis was performed using Excel 2010. One-way ANOVA was conducted using IBM SPSS Statistics 27 to assess the significance of differences (p < 0.05). The Duncan test was used to evaluate the significance of the differences. Bar charts were created using Origin 2021 for visual representation of the data.
5. Conclusions
In this study, 25 hexokinase (HXK) genes, designated BdHXK1 to BdHXK25, were identified from the Buchloe dactyloides genome. The 25 BdHXK genes are unevenly distributed across 8 chromosomes.
Phylogenetic analysis divided the BdHXK family into three subfamilies, with the closest evolutionary relationships to rice HXKs. Promoter sequence analysis revealed the presence of stress-responsive elements, indicating their involvement in responses to abiotic stresses and hormone signaling. Relative expression analysis showed significant upregulation of BdHXK5, BdHXK6, BdHXK7, and BdHXK23 under salt stress, suggesting their potential as key regulators of salt tolerance in B. dactyloides. This provides a potential gene target for the screening of salt-tolerant crops, and the subsequent focus on crop varieties with these key regulatory factors or their related expression characteristics can help the screening of salt-tolerant crops.
These results enhance our understanding of the BdHXK gene family’s role in salt stress response and provide a foundation for further research on sugar metabolism, energy supply, and signal transduction. Future studies can focus on elucidating the specific functions and interactions of individual BdHXK members to gain more comprehensive insight into the growth and development of B. dactyloides and its resistance to abiotic stresses. It is expected to promote the cultivation of new strains of ecological restoration grass with more salt resistance and better growth and development and help the improvement and ecological restoration of saline–alkali land.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms26020838/s1.
Author Contributions
H.Q. and S.W. conceived and designed the experiments. H.Q., Y.L., and X.W. performed the experiments. H.Q., Y.L., and S.W. analyzed the data. X.L. and F.S. provided suggestions on experiments. H.Q. wrote and revised the manuscript. S.W., Y.L., X.L., and F.S. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundamental Research Funds of the Chinese Academy of Forestry, Project number: CAFYBB2022XA002; Open Competition Project to Select the Best Candidates of the National Forestry and Grassland Administration “Breeding of Excellent Grass Varieties” (202201).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, 2 November 2024, GCA_000001735.2, GCA_034140825.1, GCA_902167145.1, GCA_036512215.2, GCA_000005505.4.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guo, L.; Meng, H.; Teng, K.; Fan, X.; Zhang, H.; Teng, W.; Yue, Y.; Wu, J. Effects of Nitrogen Forms on the Growth and Nitrogen Accumulation in Buchloe dactyloides Seedlings. Plants 2022, 11, 2086. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.R.; John, J. The life history of buffalo grass. Trans. Kans. Acad. Sci. 1941, 44, 58–75. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. World Soil Day: The Food and Agriculture Organization of the United Nations Emphasizes That Soil Salinization Poses a Threat to Global Food Security. Available online: https://www.fao.org/newsroom/detail/world-soil-day-fao-highlights-threat-of-soil-salinization-to-food-security-031221/ (accessed on 1 January 2025).
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The threat of soil salinity: A Europeans calere view. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Birhanie, Z.M.; Yang, D.; Luan, M.; Xiao, A.; Liu, L.; Zhang, C.; Biswas, A.; Dey, S.; Deng, Y.; Li, D. Salt Stress Induces Changes in Physiological Characteristics, Bioactive Constituents, and Antioxidants in Kenaf (Hibiscus cannabinus L.). Antioxidants 2022, 11, 2005. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Cho, J.I.; Ryoo, N.; Shin, D.H.; Park, Y.I.; Hwang, Y.S.; Lee, S.K.; An, G.; Jeon, J.S. Role of rice cytosolic hexokinase OsHXK7 in sugar signaling and metabolism. J. Integr. Plant Biol. 2016, 58, 127–135. [Google Scholar] [CrossRef]
- Karve, A.; Rauh, B.L.; Xia, X.; Kandasamy, M.; Meagher, R.B.; Sheen, J.; Moore, B.D. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 2008, 228, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.I.; Ryoo, N.; Eom, J.S.; Lee, D.W.; Kim, H.B.; Jeong, S.W.; Lee, Y.H.; Kwon, Y.K.; Cho, M.H.; Bhoo, S.H.; et al. Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 2008, 149, 745–759. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Zhang, J.W.; Chen, Y.J.; Li, R.F.; Wang, H.Z.; Ding, L.P.; Wei, J.H. Isolation, structural analysis, and expression characteristics of the maize (Zea mays L.) hexokinase gene family. Mol. Biol. Rep. 2014, 41, 6157–6166. [Google Scholar] [CrossRef] [PubMed]
- Granot, D. Role of tomato hexose kinases. Funct. Plant Biol. 2007, 34, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, Y.; Qin, X.; He, X.; Han, S.; Lv, Y.; Deng, Z.; Zeng, G.; Gao, X.; Hu, Y.; et al. Isolation, structure analysis and expression characterization of the Hexokinase gene family in Sorghum bicolor. 3 Biotech 2025, 15, 20. [Google Scholar] [CrossRef]
- Wang, P.; Huo, J.; Liu, T.; Liang, G.; Mao, J. Identification and Expression Analysis of the HXK Gene Family in Grapes. Acta Agric. Boreali-Sin. 2020, 35, 57–64. [Google Scholar]
- Zhang, C.; Zhang, L.; Fu, J.; Dong, L. Isolation and characterization of hexokinase genes PsHXK1 and PsHXK2 from tree peony (Paeonia suffruticosa Andrews). Mol. Biol. Rep. 2020, 47, 327–336. [Google Scholar] [CrossRef]
- Lugassi, N.; Kelly, G.; Arad, T.; Farkash, C.; Yaniv, Y.; Yeselson, Y.; Schaffer, A.A.; Raveh, E.; Granot, D.; Carmi, N. Expression of Hexokinase in Stomata of Citrus Fruit Reduces Fruit Transpiration and Affects Seed Development. Front. Plant Sci. 2020, 11, 255. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Díaz, J.; Batista-Silva, W.; Almada, R.; Medeiros, D.B.; Arrivault, S.; Correa, F.; Bastías, A.; Rojas, P.; Beltrán, M.F.; Pozo, M.F.; et al. Prunus Hexokinase 3 genes alter primary C-metabolism and promote drought and salt stress tolerance in Arabidopsis transgenic plants. Sci. Rep. 2021, 11, 7098. [Google Scholar] [CrossRef]
- Jiao, F.; Chen, Y.; Zhang, D.; Wu, J. Genome-Wide Characterization of Soybean Hexokinase Genes Reveals a Positive Role of GmHXK15 in Alkali Stress Response. Plants 2023, 12, 3121. [Google Scholar] [CrossRef]
- Li, N.N.; Qian, W.J.; Wang, L.; Cao, H.L.; Hao, X.Y.; Yang, Y.J.; Wang, X.C. Isolation and expression features of hexose kinase genes under various abiotic stresses in the tea plant (Camellia sinensis). Plant Physiol. 2017, 209, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhao, S.; Chen, X.; Wang, W.; Dong, W.; Chen, J.; Shen, J.R.; Liu, L.; Kuang, T. Biochemical and structural study of Arabidopsis hexokinase 1. Acta Crystallogr. D Biol. Crystallogr. 2015, 71 Pt 2, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, M.; Hu, D.; Zhang, D. Identification and Expression Analysis of Setaria italica Hexokinase Gene Family. Mol. Plant Breed. 2021, 1–11. [Google Scholar]
- Cho, J.I.; Ryoo, N. Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 2006, 224, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tian, Z.; Guo, Y. Characterization of hexokinase gene family members in Glycine max and functional analysis of GmHXK2 under salt stress. Front. Genet. 2023, 14, 1135290. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C. Correlation Analysis of Different Ploidy and Phenotypic Characteristics of Buchloe dactyloides. Master’s Thesis, Chinese Academy of Forestry, Beijing, China, 2014. [Google Scholar]
- Granot, D.; David-Schwartz, R.; Kelly, G. Hexose kinases and their role in sugar-sensing and plant development. Front. Plant Sci. 2013, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, L.H.; Tun, W.; Jeon, J.S.; An, G. Sucrose signaling in higher plants. Plant Sci. 2021, 302, 110703. [Google Scholar] [CrossRef]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, Y.; Zhang, Q.; Wu, R.; Wang, X.; Feng, S.; Chen, S.; Lu, C.; Du, L. Genome-Wide Identification and Characterization of Hexokinase Genes in Moso Bamboo (Phyllostachys edulis). Front. Plant Sci. 2020, 11, 600. [Google Scholar] [CrossRef]
- Dai, N.; Kandel-Kfir, M.; Petreikov, M. The tomato hexokinase LeHXK1 cloning, mapping, expression pattern and phylogenetic relationships. Plant Sci. 2022, 163, 581–590. [Google Scholar] [CrossRef]
- Granot, D.; Kelly, G.; Stein, O.; David-Schwartz, R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 2014, 65, 809–819. [Google Scholar] [CrossRef]
- Geng, M.T.; Yao, Y. Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava. Int. J. Mol. Sci. 2017, 18, 1041. [Google Scholar] [CrossRef] [PubMed]
- Pilic, J.; Gottschalk, B. Hexokinase 1 forms rings that regulate mitochondrial fission during energy stress. Mol. Cell 2024, 84, 2732–2746. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.H.; Ma, Q.J. The Glucose Sensor MdHXK1 Phosphorylates a Tonoplast Na+/H+ Exchanger to Improve Salt Tolerance. Plant Physiol. 2018, 176, 2977–2990. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chen, Y. MicroRNA2871b of Dongxiang Wild Rice (Oryza rufipogon Griff.) Negatively Regulates Cold and Salt Stress Tolerance in Transgenic Rice Plants. Int. J. Mol. Sci. 2023, 24, 14502. [Google Scholar] [CrossRef] [PubMed]
- Iturriaga, G.; Suárez, R.; Nova-Franco, B. Trehalose Metabolism: From Osmoprotection to Signaling. Int. J. Mol. Sci. 2009, 10, 3793–3810. [Google Scholar] [CrossRef] [PubMed]
- Maloney, V.J.; Park, J.Y.; Unda, F.; Mansfield, S.D. Sucrose phosphate synthase and sucrose phosphate phosphatase interact in planta and promote plant growth and biomass accumulation. J. Exp. Bot. 2015, 66, 4383–4394. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Jin, L. Genome-Wide Identification and Expression Profiling of the α-Amylase (AMY) Gene Family in Potato. Genes 2024, 15, 793. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, H. Genome-Wide Identification and Expression Analysis of the Sucrose Synthase Gene Family in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2023, 24, 12493. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.; Harter, K.; Rohr, L.; Mishra, S.K.; Chauhan, P.S. Elucidation of PGPR-responsive OsNAM2 regulates salt tolerance in Arabidopsis by AFP2 and SUS protein interaction. Microbiol. Res. 2024, 289, 127890. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qian, Y.Q.; Han, L.; Liu, J.X.; Sun, Z.Y. Identification of suitable reference genes in buffalo grass for accurate transcript normalization under various abiotic stress conditions. Gene 2014, 547, 55–62. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, X.B.; Shi, Y.F. OsCDC48/48E complex is required for plant survival in rice (Oryza sativa L.). Plant Mol. Biol. 2019, 100, 163. [Google Scholar] [CrossRef]
- Li, F.; Hu, Q.; Chen, F.; Jiang, J.F. Transcriptome analysis reveals Vernalization is independent of cold acclimation in Arabidopsis. BMC Genom. 2021, 22, 462. [Google Scholar] [CrossRef]
- Xuan, W.; Dai, M.; Buckner, J.; Mirel, B.; Song, J.; Athey, B.; Watson, S.J.; Meng, F. Cross-domain neurobiology data integration and exploration. BMC Genom. 2010, 11 (Suppl. 3), S6. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).