The Role of Alternative Splicing in Polyploids in Response to Abiotic Stress
Abstract
1. Introduction
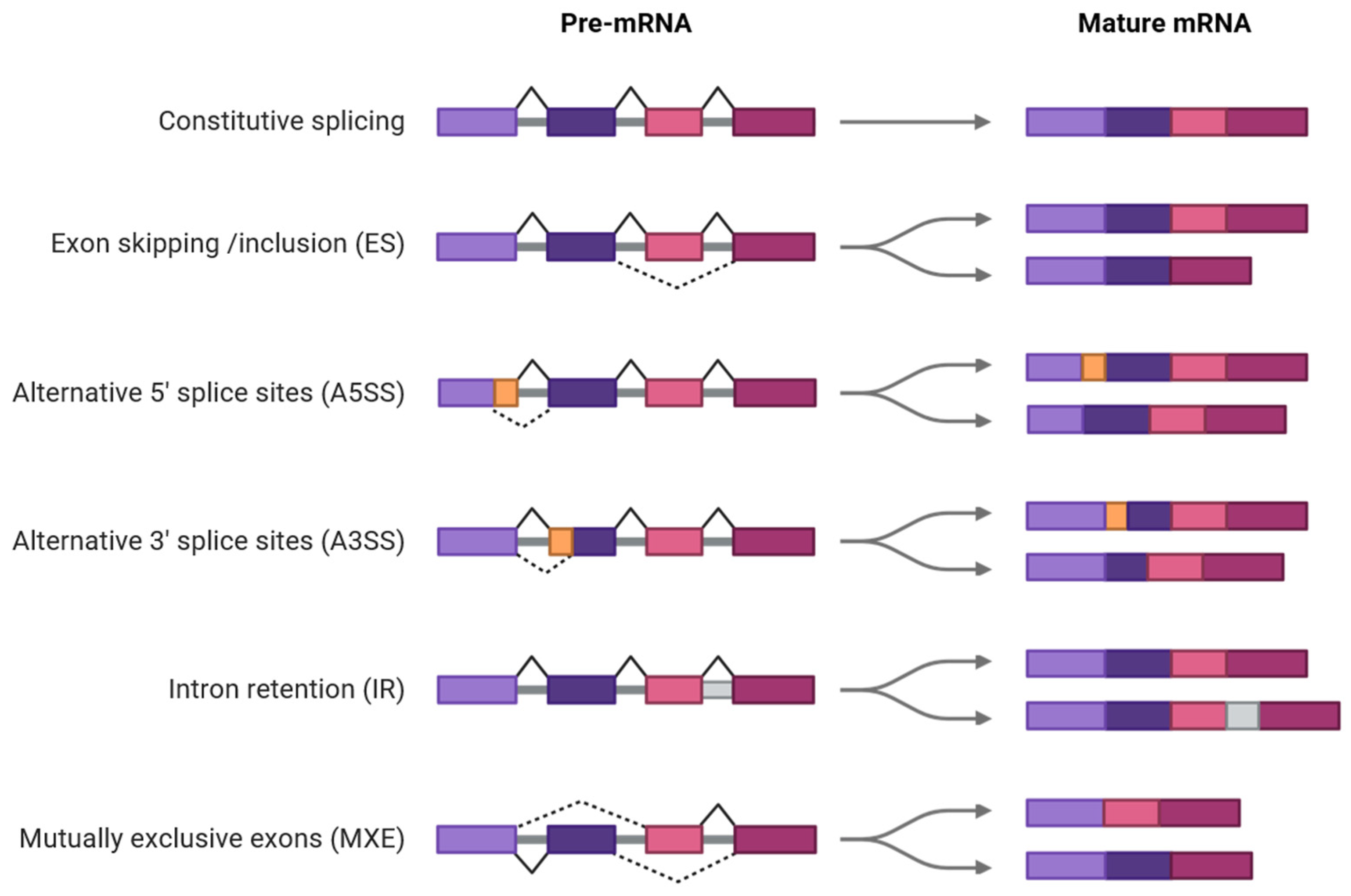
2. Prevalence and Variability of AS Across Plant Species
3. The Role of AS in Plant Growth and Development
4. Abiotic Stress-Induced Alternative Splicing Variants
4.1. Drought Stress
4.2. Salinity Stress
4.3. Temperature Stress
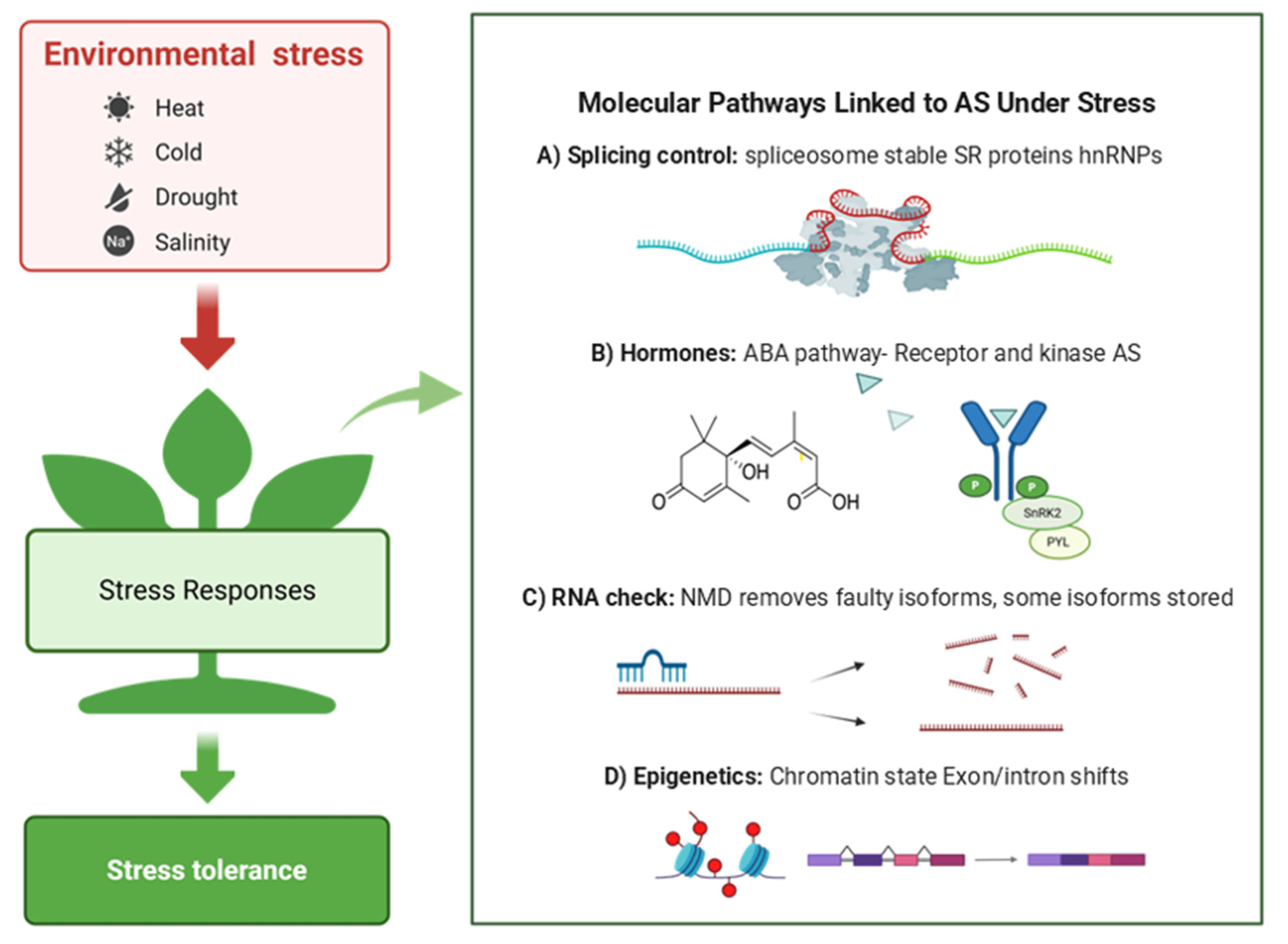
4.4. Molecular Pathways Linked to AS Under Stress
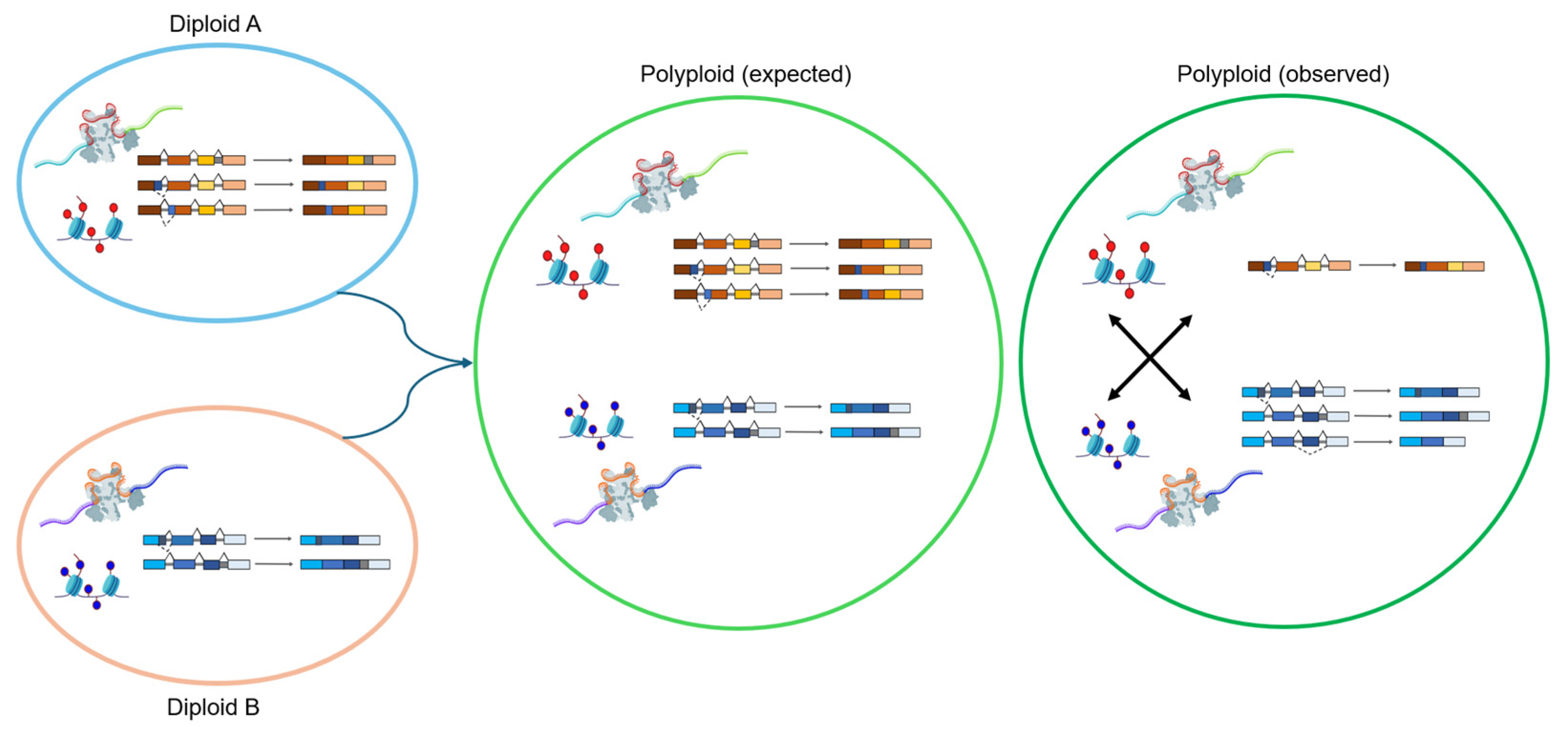
5. Alternative Splicing in Polyploids
5.1. Brassica napus
5.2. Gossypium hirsutum
5.3. Triticum aestivum
5.4. Other Polyploid Species
6. Future Directions and Implications
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef]
- Khaipho-Burch, M.; Cooper, M.; Crossa, J.; de Leon, N.; Holland, J.; Lewis, R.; McCouch, S.; Murray, S.C.; Rabbi, I.; Ronald, P. Genetic modification can improve crop yields—But stop overselling it. Nature 2023, 621, 470–473. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Dinneny, J.R. Developmental responses to water and salinity in root systems. Annu. Rev. Cell Dev. Biol. 2019, 35, 239–257. [Google Scholar] [CrossRef]
- He, W.; Zhang, X.; Lv, P.; Wang, W.; Wang, J.; He, Y.; Song, Z.; Cai, D. Full-length transcriptome reconstruction reveals genetic differences in hybrids of Oryza sativa and Oryza punctata with different ploidy and genome compositions. BMC Plant Biol. 2022, 22, 131. [Google Scholar] [CrossRef]
- Alhabsi, A.; Ling, Y.; Crespi, M.; Reddy, A.S.; Mahfouz, M. Alternative splicing dynamics in plant adaptive responses to stress. Annu. Rev. Plant Biol. 2025, 76, 687–717. [Google Scholar] [CrossRef] [PubMed]
- Dikaya, V.; El Arbi, N.; Rojas-Murcia, N.; Nardeli, S.M.; Goretti, D.; Schmid, M. Insights into the role of alternative splicing in plant temperature response. J. Exp. Bot. 2021, 72, 7384–7403. [Google Scholar] [CrossRef] [PubMed]
- Godinho, D.P.; Yanez, R.J.; Duque, P. Pathogen-responsive alternative splicing in plant immunity. Trends Plant Sci. 2025, 30, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-X.; Guo, Q.-H.; Xu, W.-B.; Liu, P.; Yan, K. Rapid regulation of alternative splicing in response to environmental stresses. Front. Plant Sci. 2022, 13, 832177. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.; Xu, X.; Zhou, W.; Wu, L. Alternative splicing: An efficient regulatory approach towards plant developmental plasticity. Wiley Interdiscip. Rev. RNA 2023, 14, e1758. [Google Scholar] [CrossRef]
- Punzo, P.; Grillo, S.; Batelli, G. Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 2020, 48, 2117–2126. [Google Scholar] [CrossRef]
- Rosenkranz, R.R.; Ullrich, S.; Löchli, K.; Simm, S.; Fragkostefanakis, S. Relevance and regulation of alternative splicing in plant heat stress response: Current understanding and future directions. Front. Plant Sci. 2022, 13, 911277. [Google Scholar] [CrossRef]
- Singh, A.; Roychoudhury, A. Gene regulation at transcriptional and post-transcriptional levels to combat salt stress in plants. Physiol. Plant. 2021, 173, 1556–1572. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Mo, R.; Cui, D.; Cheng, W.; Wang, H.; Qin, J.; Liu, Z. Alternative splicing in the regulatory circuit of plant temperature response. Int. J. Mol. Sci. 2023, 24, 3878. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed]
- Sybilska, E.; Daszkowska-Golec, A. A complex signaling trio in seed germination: Auxin-JA-ABA. Trends Plant Sci. 2023, 28, 873–875. [Google Scholar] [CrossRef]
- Liu, G.; Wang, H.; Gao, H.; Yu, S.; Liu, C.; Wang, Y.; Sun, Y.; Zhang, D. Alternative Splicing of Functional Genes in Plant Growth, Development, and Stress Responses. Int. J. Mol. Sci. 2025, 26, 5864. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Petrillo, E.; Dubrovina, A.S.; Kiselev, K.V. Regulation of alternative splicing in plant stress responses. Front. Plant Sci. 2023, 13, 1120961. [Google Scholar] [CrossRef]
- Liu, Y.; Do, S.; Huynh, H.; Li, J.-X.; Liu, Y.-G.; Du, Z.-Y.; Chen, M.-X. Importance of pre-mRNA splicing and its study tools in plants. Adv. Biotechnol. 2024, 2, 4. [Google Scholar] [CrossRef]
- Szakonyi, D.; Duque, P. Alternative splicing as a regulator of early plant development. Front. Plant Sci. 2018, 9, 1174. [Google Scholar] [CrossRef]
- Marasco, L.E.; Kornblihtt, A.R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 2023, 24, 242–254. [Google Scholar] [CrossRef]
- Du, Y.; Cao, L.; Wang, S.; Guo, L.; Tan, L.; Liu, H.; Feng, Y.; Wu, W. Differences in alternative splicing and their potential underlying factors between animals and plants. J. Adv. Res. 2024, 64, 83–98. [Google Scholar] [CrossRef]
- de la Fuente, R.; Díaz-Villanueva, W.; Arnau, V.; Moya, A. Evolutionary trends of alternative splicing. eLife 2024, 13, RP94802. [Google Scholar] [CrossRef]
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative splicing and protein diversity: Plants versus animals. Front. Plant Sci. 2019, 10, 708. [Google Scholar] [CrossRef]
- McCue, K.; Burge, C.B. An interpretable model of pre-mRNA splicing for animal and plant genes. Sci. Adv. 2024, 10, eadn1547. [Google Scholar] [CrossRef]
- Wright, C.J.; Smith, C.W.; Jiggins, C.D. Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet. 2022, 23, 697–710. [Google Scholar] [CrossRef]
- Kim, S.; Kim, T.-H. Alternative splicing for improving abiotic stress tolerance and agronomic traits in crop plants. J. Plant Biol. 2020, 63, 409–420. [Google Scholar] [CrossRef]
- Islam, M.M.; Deepo, D.M.; Nasif, S.O.; Siddique, A.B.; Hassan, O.; Siddique, A.B.; Paul, N.C. Cytogenetics and consequences of polyploidization on different biotic-abiotic stress tolerance and the potential mechanisms involved. Plants 2022, 11, 2684. [Google Scholar] [CrossRef]
- Yu, H.; Bi, X.; Li, Z.; Fu, X.; Li, Y.; Li, Y.; Yang, Y.; Liu, D.; Li, G.; Dong, W. Transcriptomic Analysis of Alternative Splicing Events during Different Fruit Ripening Stages of Coffea arabica L. Genes 2024, 15, 459. [Google Scholar] [CrossRef]
- Tossi, V.E.; Martínez Tosar, L.J.; Laino, L.E.; Iannicelli, J.; Regalado, J.J.; Escandón, A.S.; Baroli, I.; Causin, H.F.; Pitta-Álvarez, S.I. Impact of polyploidy on plant tolerance to abiotic and biotic stresses. Front. Plant Sci. 2022, 13, 869423. [Google Scholar] [CrossRef]
- Gao, P.; Quilichini, T.D.; Zhai, C.; Qin, L.; Nilsen, K.T.; Li, Q.; Sharpe, A.G.; Kochian, L.V.; Zou, J.; Reddy, A.S. Alternative splicing dynamics and evolutionary divergence during embryogenesis in wheat species. Plant Biotechnol. J. 2021, 19, 1624–1643. [Google Scholar] [CrossRef]
- de Jong, G.W.; Adams, K.L. Subgenome-dominant expression and alternative splicing in response to Sclerotinia infection in polyploid Brassica napus and progenitors. Plant J. 2023, 114, 142–158. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Quiroz, L.F.; Reddy, A.S.; Spillane, C.; Ortiz, R. Alternative splicing variation: Accessing and exploiting in crop improvement programs. Int. J. Mol. Sci. 2023, 24, 15205. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Ashman, T.-L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26, Erratum in Plant Cell 2021, 33, 2899. [Google Scholar] [CrossRef]
- Marquardt, S.; Petrillo, E.; Manavella, P.A. Cotranscriptional RNA processing and modification in plants. Plant Cell 2023, 35, 1654–1670. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Rodríguez, F.S.; Aballay, F.E.; Cartagena, C.M.; Servi, L.; Petrillo, E. Alternative splicing in plants: Current knowledge and future directions for assessing the biological relevance of splice variants. J. Exp. Bot. 2023, 74, 2251–2272. [Google Scholar] [CrossRef]
- Martín, G.; Márquez, Y.; Mantica, F.; Duque, P.; Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 2021, 22, 35. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, W.; Zhu, D.; Zhang, B.; Xu, Q.; Shi, C.; He, H.; Dai, X.; Li, Y.; He, W. Population-level exploration of alternative splicing and its unique role in controlling agronomic traits of rice. Plant Cell 2024, 36, 4372–4387. [Google Scholar] [CrossRef]
- Zhou, Y.; Lu, Q.; Zhang, J.; Zhang, S.; Weng, J.; Di, H.; Zhang, L.; Li, X.; Liang, Y.; Dong, L. Genome-wide profiling of alternative splicing and gene fusion during Rice black-streaked dwarf virus stress in maize (Zea mays L.). Genes 2022, 13, 456. [Google Scholar] [CrossRef]
- Zhu, R.; Yue, C.; Wu, S.; Wu, M.; Xu, Z.; Liu, X.; Wang, R.; Wang, M. Alternative Splicing of BnABF4L Mediates Response to Abiotic Stresses in Rapeseed (Brassica napus L.). Biotechnol. Biofuels Bioprod. 2025, 18, 51. [Google Scholar] [CrossRef]
- Bao, Y.; Yin, X.; Song, X.; Liu, L.; Liu, Y.; Meng, S. Genome-wide identification of novel alternative splicing events and related isoforms linked to cotton domestication and fiber domestication. Gene 2025, 963, 149601. [Google Scholar] [CrossRef]
- Alyahya, N.; Taybi, T. Transcriptome-wide characterization of alternative splicing regulation in Najran wheat (Triticum aestivum) under salt stress. Curr. Plant Biol. 2024, 38, 100334. [Google Scholar] [CrossRef]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef]
- Mei, W.; Boatwright, L.; Feng, G.; Schnable, J.C.; Barbazuk, W.B. Evolutionarily conserved alternative splicing across monocots. Genetics 2017, 207, 465–480. [Google Scholar] [CrossRef]
- Mei, W.; Liu, S.; Schnable, J.C.; Yeh, C.-T.; Springer, N.M.; Schnable, P.S.; Barbazuk, W.B. A comprehensive analysis of alternative splicing in paleopolyploid maize. Front. Plant Sci. 2017, 8, 694. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; Gong, W. Genome-wide identification and comparative analysis of alternative splicing across four legume species. Planta 2019, 249, 1133–1142. [Google Scholar] [CrossRef]
- Misra, C.S.; Sousa, A.G.; Barros, P.M.; Kermanov, A.; Becker, J.D. Cell-type-specific alternative splicing in the Arabidopsis germline. Plant Physiol. 2023, 192, 85–101. [Google Scholar] [CrossRef]
- Abulfaraj, A.A.; Alshareef, S.A. Concordant gene expression and alternative splicing regulation under abiotic stresses in Arabidopsis. Genes 2024, 15, 675. [Google Scholar] [CrossRef]
- An, Y.; Wu, J.; Chen, Y.; Shize, L. Comprehensive analysis of alternative splicing in Rosa roxburghii Tratt revealed its role in flavonoid synthesis. Front. Plant Sci. 2025, 16, 1627126. [Google Scholar] [CrossRef]
- Yu, S.; Wan, J.; Xu, T.; Zhang, J.; Cao, L.; Liu, J.; Liu, H.; Ren, X.; Yang, Z. A gene expression atlas of Nicotiana tabacum across various tissues at transcript resolution. Front. Plant Sci. 2025, 16, 1500654. [Google Scholar] [CrossRef]
- Jiang, G.; Zhang, D.; Li, Z.; Liang, H.; Deng, R.; Su, X.; Jiang, Y.; Duan, X. Alternative splicing of MaMYB16L regulates starch degradation in banana fruit during ripening. J. Integr. Plant Biol. 2021, 63, 1341–1352. [Google Scholar] [CrossRef]
- Yuan, H.; Yu, H.; Huang, T.; Shen, X.; Xia, J.; Pang, F.; Wang, J.; Zhao, M. The complexity of the Fragaria x ananassa (octoploid) transcriptome by single-molecule long-read sequencing. Hortic. Res. 2019, 6, 46, Erratum in Hortic. Res. 2019, 6, 63. [Google Scholar] [CrossRef]
- Chen, Q.; Lin, X.; Tang, W.; Deng, Q.; Wang, Y.; Lin, Y.; He, W.; Zhang, Y.; Li, M.; Luo, Y.; et al. Transcriptomic Complexity in Strawberry Fruit Development and Maturation Revealed by Nanopore Sequencing. Front. Plant Sci. 2022, 13, 872054. [Google Scholar] [CrossRef]
- Deng, Q.; Lu, H.; Liu, D.; Huang, Y.; Feng, J.; Wei, D.; Wang, Z.; Tang, Q. Modulation of flowering by an alternatively spliced AGL18-1 transcript in Brassica juncea. Crop J. 2025, 13, 456–467. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Wei, X.; Datta, T.; Wei, F.; Xie, Z. Polyploidization: A biological force that enhances stress resistance. Int. J. Mol. Sci. 2024, 25, 1957. [Google Scholar] [CrossRef]
- Scarrow, M.; Wang, Y.; Sun, G. Molecular regulatory mechanisms underlying the adaptability of polyploid plants. Biol. Rev. 2021, 96, 394–407. [Google Scholar] [CrossRef]
- Li, M.; Hu, M.; Xiao, Y.; Wu, X.; Wang, J. The activation of gene expression and alternative splicing in the formation and evolution of allopolyploid Brassica napus. Hortic. Res. 2022, 9, uhab075. [Google Scholar] [CrossRef]
- Park, S.; Ahn, E.; Zhang, D.; Meinhardt, L.W. Comparative genome-wide characterization and evolutionary insights into the AP2/ERF gene family in three Coffea species (C. canephora, C. eugenioides, and C. arabica). BMC Genom. 2025, 26, 653. [Google Scholar]
- Lam, P.Y.; Wang, L.; Lo, C.; Zhu, F.-Y. Alternative splicing and its roles in plant metabolism. Int. J. Mol. Sci. 2022, 23, 7355. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, G.; Guo, C.; He, Y.; Li, Z.; Ning, G.; Shi, X.; Bao, M. The FLOWERING LOCUS T orthologous gene of Platanus acerifolia is expressed as alternatively spliced forms with distinct spatial and temporal patterns. Plant Biol. 2011, 13, 809–820. [Google Scholar] [CrossRef]
- Zhang, J.-Z.; Li, Z.-M.; Mei, L.; Yao, J.-L.; Hu, C.-G. PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuation in expression level. Planta 2009, 229, 847–859. [Google Scholar] [CrossRef]
- Ye, L.X.; Wu, Y.M.; Zhang, J.X.; Zhang, J.X.; Zhou, H.; Zeng, R.F.; Zheng, W.X.; Qiu, M.Q.; Zhou, J.J.; Xie, Z.Z. A bZIP transcription factor (CiFD) regulates drought- and low-temperature-induced flowering by alternative splicing in citrus. J. Integr. Plant Biol. 2023, 65, 674–691. [Google Scholar] [CrossRef]
- Capovilla, G.; Symeonidi, E.; Wu, R.; Schmid, M. Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5117–5127. [Google Scholar] [CrossRef]
- Jin, S.; Kim, S.Y.; Susila, H.; Nasim, Z.; Youn, G.; Ahn, J.H. FLOWERING LOCUS M isoforms differentially affect the subcellular localization and stability of SHORT VEGETATIVE PHASE to regulate temperature-responsive flowering in Arabidopsis. Mol. Plant 2022, 15, 1696–1709. [Google Scholar] [CrossRef]
- Ma, H.; Pei, J.; Zhuo, J.; Tang, Q.; Hou, D.; Lin, X. The CONSTANS-LIKE gene PeCOL13 regulates flowering through intron-retained alternative splicing in Phyllostachys edulis. Int. J. Biol. Macromol. 2024, 274, 133393. [Google Scholar] [CrossRef]
- Gil, K.E.; Park, M.J.; Lee, H.J.; Park, Y.J.; Han, S.H.; Kwon, Y.J.; Seo, P.J.; Jung, J.H.; Park, C.M. Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 2017, 89, 128–140. [Google Scholar] [CrossRef]
- Zhang, D.; Li, M.; Aslam, M.M.; Huang, M.; Chen, M.-X.; Liu, Y.-G.; Zhang, J. The role of Arabidopsis Splicing Factor 30 in floral transition and the implications of its alternative splicing. Plant Physiol. 2025, 198, kiaf335. [Google Scholar] [CrossRef]
- Hong, Y.; Yao, J.; Shi, H.; Chen, Y.; Zhu, J.-K.; Wang, Z. The Arabidopsis spliceosomal protein SmEb modulates ABA responses by maintaining proper alternative splicing of HAB1. Stress Biol. 2021, 1, 4. [Google Scholar] [CrossRef]
- Sybilska, E.; Collin, A.; Sadat Haddadi, B.; Mur, L.A.; Beckmann, M.; Guo, W.; Simpson, C.G.; Daszkowska-Golec, A. The cap-binding complex modulates ABA-responsive transcript splicing during germination in barley (Hordeum vulgare). Sci. Rep. 2024, 14, 18278. [Google Scholar] [CrossRef]
- Kashkan, I.; Hrtyan, M.; Retzer, K.; Humpolíčková, J.; Jayasree, A.; Filepová, R.; Vondráková, Z.; Simon, S.; Rombaut, D.; Jacobs, T.B. Mutually opposing activity of PIN7 splicing isoforms is required for auxin-mediated tropic responses in Arabidopsis thaliana. New Phytol. 2022, 233, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Miao, P.; Qin, W.; Hu, W.; Wei, Z.; Ding, W.; Zhang, H.; Wang, Z. A novel single nucleotide mutation of TFL1 alters the plant architecture of Gossypium arboreum through changing the pre-mRNA splicing. Plant Cell Rep. 2024, 43, 26. [Google Scholar] [CrossRef]
- Zheng, J.; Wen, S.; Yu, Z.; Luo, K.; Rong, J.; Ding, M. Alternative splicing during fiber development in G. hirsutum. Int. J. Mol. Sci. 2023, 24, 11812. [Google Scholar] [CrossRef]
- Wang, M.; Wang, P.; Liang, F.; Ye, Z.; Li, J.; Shen, C.; Pei, L.; Wang, F.; Hu, J.; Tu, L. A global survey of alternative splicing in allopolyploid cotton: Landscape, complexity and regulation. New Phytol. 2018, 217, 163–178. [Google Scholar] [CrossRef]
- Lightfoot, D.J.; Malone, K.M.; Timmis, J.N.; Orford, S.J. Evidence for alternative splicing of MADS-box transcripts in developing cotton fibre cells. Mol. Genet. Genom. 2008, 279, 75–85. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, X.; Wu, Y.n.; Wang, R.; Zhang, Y.; Shi, F.; Zhao, H.; Yu, P.; Wang, Y.; Chen, M. TaPP2C-a5 fine-tunes wheat seed dormancy and germination with a Triticeae-specific, alternatively spliced transcript. J. Adv. Res. 2025; in press. [Google Scholar] [CrossRef]
- Zhang, Y.; Miao, H.; Xiao, Y.; Wang, C.; Zhang, J.; Shi, X.; Xie, S.; Wang, C.; Li, T.; Deng, P. An intron-located single nucleotide variation of TaGS5-3D is related to wheat grain size through accumulating intron retention transcripts. Theor. Appl. Genet. 2023, 136, 193. [Google Scholar] [CrossRef]
- Wu, B.; Zhang, X.; Hu, K.; Zheng, H.; Zhang, S.; Liu, X.; Ma, M.; Zhao, H. Two alternative splicing variants of a wheat gene TaNAK1, TaNAK1. 1 and TaNAK1. 2, differentially regulate flowering time and plant architecture leading to differences in seed yield of transgenic Arabidopsis. Front. Plant Sci. 2022, 13, 1014176. [Google Scholar] [CrossRef]
- Song, T.; Yu, Y.; Zhang, M.; Zhou, H.; Zhang, S.; Yu, M.; Zhou, J.; Cheng, J.; Xiang, J.; Yang, S. A wheat TaTOE1-B1 transcript TaTOE1-B1-3 can delay the flowering time of transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 12645. [Google Scholar] [CrossRef] [PubMed]
- Verta, J.-P.; Jacobs, A. The role of alternative splicing in adaptation and evolution. Trends Ecol. Evol. 2022, 37, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Li, W.; Peng, Y.; Cao, Y.; Qu, J.; Sun, F.; Yang, Q.; Lu, Y.; Zhang, X.; Zheng, L.; et al. ZmPP2C26 Alternative Splicing Variants Negatively Regulate Drought Tolerance in Maize. Front. Plant Sci. 2022, 13, 851531. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, J.; Pu, X.; Lv, H.; Liu, Y.; Ma, H.; Wu, F.; Wang, Q.; Feng, X.; Liu, T.; et al. Maize DNA Methylation in Response to Drought Stress Is Involved in Target Gene Expression and Alternative Splicing. Int. J. Mol. Sci. 2021, 22, 8285. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Hwang, H.; Bang, G.; Ha, J.; Park, Y.; Kim, J.Y. Understanding the molecular mechanisms of drought tolerance in wild soybean (Glycine soja) through multi-omics-based alternative splicing predictions. Environ. Exp. Bot. 2024, 225, 105872. [Google Scholar] [CrossRef]
- Cheng, J.; Wu, T.; Zhou, Y.; Al-Saud, N.B.S.; Cheng, B.; Admas, T.; Zhang, W.; Pan, R. The alternative splicing of HvLHCA4.2 enhances drought tolerance in barley by regulating ROS scavenging and stomatal closure. Int. J. Biol. Macromol. 2025, 307 Pt 4, 142384. [Google Scholar] [CrossRef] [PubMed]
- Collin, A.; Matkowski, H.; Sybilska, E.; Biantari, A.; Krol, O.; Daszkowska-Golec, A. ABA-induced alternative splicing drives transcriptomic reprogramming for drought tolerance in barley. BMC Plant Biol. 2025, 25, 445. [Google Scholar] [CrossRef]
- Yang, H.; Li, P.; Jin, G.; Gui, D.; Liu, L.; Zhang, C. Temporal regulation of alternative splicing events in rice memory under drought stress. Plant Divers. 2022, 44, 116–125. [Google Scholar] [CrossRef]
- Ning, M.; Li, Q.; Wang, Y.; Li, Q.; Tao, Y.; Zhang, F.; Hu, F.; Huang, L. Alternative splicing drives the functional diversification of a bHLH transcription factor in the control of growth and drought tolerance in rice. Sci. Bull. 2025, 70, 153–156. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.-H.; Guo, Q.-H.; Liu, P.; Li, Y.; Wu, C.-A.; Yang, G.-D.; Huang, J.-G.; Zhang, S.-Z.; Zheng, C.-C. Salt responsive alternative splicing of a RING finger E3 ligase modulates the salt stress tolerance by fine-tuning the balance of COP9 signalosome subunit 5A. PLoS Genet. 2021, 17, e1009898. [Google Scholar] [CrossRef]
- Lan, Y.; Zhang, K.; He, T.; Wang, H.; Jiang, C.; Yan, H.; Xiang, Y. Systematic analysis of the serine/arginine-rich protein splicing factors (SRs) and focus on salt tolerance of PtSC27 in Populus trichocarpa. Plant Physiol. Biochem. 2022, 173, 97–109. [Google Scholar] [CrossRef]
- Jian, G.; Mo, Y.; Hu, Y.; Huang, Y.; Ren, L.; Zhang, Y.; Hu, H.; Zhou, S.; Liu, G.; Guo, J. Variety-specific transcriptional and alternative splicing regulations modulate salt tolerance in rice from early stage of stress. Rice 2022, 15, 56. [Google Scholar] [CrossRef]
- Yu, H.; Du, Q.; Campbell, M.; Yu, B.; Walia, H.; Zhang, C. Genome-wide discovery of natural variation in pre-mRNA splicing and prioritising causal alternative splicing to salt stress response in rice. New Phytol. 2021, 230, 1273–1287. [Google Scholar] [CrossRef]
- Long, L.; Zhao, J.-R.; Guo, D.-D.; Ma, X.-N.; Xu, F.-C.; Yang, W.-W.; Gao, W. Identification of NHXs in Gossypium species and the positive role of GhNHX1 in salt tolerance. BMC Plant Biol. 2020, 20, 147. [Google Scholar] [CrossRef]
- Mangena, P. Cell mutagenic autopolyploidy enhances salinity stress tolerance in leguminous crops. Cells 2023, 12, 2082. [Google Scholar] [CrossRef]
- Song, X.; Zhang, M.; Wang, T.T.; Duan, Y.Y.; Ren, J.; Gao, H.; Fan, Y.J.; Xia, Q.M.; Cao, H.X.; Xie, K.D. Polyploidization leads to salt stress resilience via ethylene signaling in citrus plants. New Phytol. 2025, 246, 176–191. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, Y.; Yao, J.; Huang, H.; Qian, B.; Liu, X.; Chen, Y.; Pang, J.; Zhan, X.; Zhu, J.K. Modulation of plant development and chilling stress responses by alternative splicing events under control of the spliceosome protein SmEb in Arabidopsis. Plant Cell Environ. 2022, 45, 2762–2779. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Song, Y.; Wu, S.; Peng, Y.; Ming, Y.; Li, Z.; Zhang, X.; Song, W.; Su, Z.; Gong, Z. Regulation of alternative splicing by CBF-mediated protein condensation in plant response to cold stress. Nat. Plants 2025, 11, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lv, W.; Shao, L.; Fu, Y.; Liu, H.; Yang, C.; Chen, A.; Xie, X.; Wang, Z.; Li, C. PacBio and Illumina RNA sequencing identify alternative splicing events in response to cold stress in two poplar species. Front. Plant Sci. 2021, 12, 737004. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Zhang, C.; Yang, L.; Liu, Y.; Wang, G.; Li, S.; Dirk, L.M.; Downie, A.B.; Zhao, T. Two alternatively spliced variants of ZmHSF12 regulate the balance of plant growth and heat tolerance in maize and Arabidopsis. Plant J. 2025, 123, e70372. [Google Scholar] [CrossRef]
- Ma, J.; Li, S.; Wang, T.; Tao, Z.; Huang, S.; Lin, N.; Zhao, Y.; Wang, C.; Li, P. Cooperative condensation of RNA-DIRECTED DNA METHYLATION 16 splicing isoforms enhances heat tolerance in Arabidopsis. Nat. Commun. 2025, 16, 433. [Google Scholar] [CrossRef]
- Jo, S.H.; Park, H.J.; Jung, H.; Lee, G.S.; Moon, J.H.; Kim, H.-S.; Lee, H.-J.; Jung, C.; Cho, H.S. PROTEIN PHOSPHATASE 2A B′ η drives spliceosome subunit dephosphorylation to mediate alternative splicing following heat stress. Plant Cell 2025, 37, koaf117. [Google Scholar] [CrossRef]
- Cecchini, N.M.; Torres, J.R.; López, I.L.; Cobo, S.; Nota, F.; Alvarez, M.E. Alternative splicing of an exitron determines the subnuclear localization of the Arabidopsis DNA glycosylase MBD4L under heat stress. Plant J. 2022, 110, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, Z.; Qu, L.; Hu, Y.; Lu, D. Transcriptomic and alternative splicing analyses provide insights into the roles of exogenous salicylic acid ameliorating waxy maize seedling growth under heat stress. BMC Plant Biol. 2022, 22, 432. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Vosseberg, J.; Schinkel, M.; Gremmen, S.; Snel, B. The spread of the first introns in proto-eukaryotic paralogs. Commun. Biol. 2022, 5, 476. [Google Scholar] [CrossRef]
- Kováčová, T.; Souček, P.; Hujová, P.; Freiberger, T.; Grodecká, L. Splicing enhancers at intron–exon borders participate in acceptor splice sites recognition. Int. J. Mol. Sci. 2020, 21, 6553. [Google Scholar] [CrossRef]
- Ling, Y.; Mahfouz, M.M.; Zhou, S. Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 2021, 26, 1153–1170. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Cumbie, J.S.; Dharmawardhana, P.; Jaiswal, P.; Chang, J.H.; Palusa, S.G.; Reddy, A.; Megraw, M.; Mockler, T.C. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant 2015, 8, 207–227. [Google Scholar] [CrossRef]
- Sybilska, E.; Daszkowska-Golec, A. Alternative splicing in ABA signaling during seed germination. Front. Plant Sci. 2023, 14, 1144990. [Google Scholar] [CrossRef]
- Raxwal, V.K.; Riha, K. The biological functions of nonsense-mediated mRNA decay in plants: RNA quality control and beyond. Biochem. Soc. Trans. 2023, 51, 31–39. [Google Scholar] [CrossRef]
- Ganie, S.A.; Reddy, A.S. Stress-induced changes in alternative splicing landscape in rice: Functional significance of splice isoforms in stress tolerance. Biology 2021, 10, 309. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Yao, S.; Liang, F.; Gill, R.A.; Huang, J.; Cheng, X.; Liu, Y.; Tong, C.; Liu, S. A global survey of the transcriptome of allopolyploid Brassica napus based on single-molecule long-read isoform sequencing and Illumina-based RNA sequencing data. Plant J. 2020, 103, 843–857. [Google Scholar] [CrossRef]
- Zhou, R.; Moshgabadi, N.; Adams, K.L. Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc. Natl. Acad. Sci. USA 2011, 108, 16122–16127. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, M.; Wang, J. Characteristics of duplicated gene expression and DNA methylation regulation in different tissues of allopolyploid Brassica napus. BMC Plant Biol. 2024, 24, 518. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.E. The Effects of Abiotic Stress on Isoform Composition in Polyploid Brassica napus. MS. Thesis, University of British Columbia, Vancouver, BC, Canada, 2021. [Google Scholar]
- Ma, J.-Q.; Xu, W.; Xu, F.; Lin, A.; Sun, W.; Jiang, H.-H.; Lu, K.; Li, J.-N.; Wei, L.-J. Differential alternative splicing genes and isoform regulation networks of rapeseed (Brassica napus L.) infected with Sclerotinia sclerotiorum. Genes 2020, 11, 784. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.-Q.; Wei, L.-J.; Lin, A.; Zhang, C.; Sun, W.; Yang, B.; Lu, K.; Li, J.-N. The alternative splicing landscape of Brassica napus infected with Leptosphaeria maculans. Genes 2019, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Li, W.; Wang, S.; Zhang, X.; Coules, A.; Ding, G.; Xu, F.; Ren, J.; Lu, C.; Shi, L. Differential alternative splicing genes in response to boron deficiency in Brassica napus. Genes 2019, 10, 224. [Google Scholar] [CrossRef]
- Yang, L.; Yang, L.; Zhao, C.; Liu, J.; Tong, C.; Zhang, Y.; Cheng, X.; Jiang, H.; Shen, J.; Xie, M. Differential alternative splicing genes and isoform co-expression networks of Brassica napus under multiple abiotic stresses. Front. Plant Sci. 2022, 13, 1009998. [Google Scholar] [CrossRef]
- Lee, J.S.; Adams, K.L. Global insights into duplicated gene expression and alternative splicing in polyploid Brassica napus under heat, cold, and drought stress. Plant Genome 2020, 13, e20057. [Google Scholar] [CrossRef]
- Lin, A.; Ma, J.; Xu, F.; Xu, W.; Jiang, H.; Zhang, H.; Qu, C.; Wei, L.; Li, J. Differences in alternative splicing between yellow and black-seeded rapeseed. Plants 2020, 9, 977. [Google Scholar] [CrossRef]
- Liu, L.; Wu, D.; Gu, Y.; Liu, F.; Liu, B.; Mao, F.; Yi, X.; Tang, T.; Zhao, X. Comprehensive profiling of alternative splicing landscape during secondary dormancy in oilseed rape (Brassica napus L.). Mol. Breed. 2022, 42, 44. [Google Scholar] [CrossRef]
- Senchina, D.S.; Alvarez, I.; Cronn, R.C.; Liu, B.; Rong, J.; Noyes, R.D.; Paterson, A.H.; Wing, R.A.; Wilkins, T.A.; Wendel, J.F. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 2003, 20, 633–643. [Google Scholar] [CrossRef]
- Wendel, J.F.; Cronn, R.C. Polyploidy and the evolutionary history of cotton. Adv. Agron. 2003, 87, 139–186. [Google Scholar]
- Wang, M.; Tu, L.; Lin, M.; Lin, Z.; Wang, P.; Yang, Q.; Ye, Z.; Shen, C.; Li, J.; Zhang, L. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 2017, 49, 579–587. [Google Scholar] [CrossRef]
- Viot, C.R.; Wendel, J.F. Evolution of the cotton genus Gossypium and its domestication in the americas: A review. Crit. Rev. Plant Sci. 2023, 42, 1–33. [Google Scholar] [CrossRef]
- Min, X.J. A survey of alternative splicing in allotetraploid cotton (Gossypium hirsutum L.). Comput. Mol. Biol. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, B.; Zheng, H.; Hu, Y.; Lu, G.; Yang, C.; Zhang, L. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 2015, 5, 14139. [Google Scholar] [CrossRef]
- Yuan, D.; Tang, Z.; Wang, M.; Gao, W.; Tu, L.; Jin, X.; Chen, L.; He, Y.; Zhang, L.; Zhu, L. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 2015, 5, 17662. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Peng, Z.; He, S.; Gong, W.; Xu, F.; Pan, Z.; Jia, Y.; Geng, X.; Du, X. Integration of proteomic and transcriptomic profiles reveals multiple levels of genetic regulation of salt tolerance in cotton. BMC Plant Biol. 2018, 18, 128. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Zhang, C.; Zhu, W.; Yan, G.; Chen, X.; Qiu, P.; Ruzimurod, B.; Ye, W.; Qaraevna, B.Z.; Yin, Z. Molecular mechanism that underlies cotton response to salt and drought stress revealed by complementary transcriptomic and iTRAQ analyses. Environ. Exp. Bot. 2023, 209, 105288. [Google Scholar] [CrossRef]
- Chen, D.-Y.; Chen, Q.-Y.; Wang, D.-D.; Mu, Y.-P.; Wang, M.-Y.; Huang, J.-R.; Mao, Y.-B. Differential transcription and alternative splicing in cotton underly specialized defense responses against pests. Front. Plant Sci. 2020, 11, 573131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, X.; Song, Y.; Sun, Z.; Zhang, J.; Wu, H.; Yang, Y.; Wang, Z.; He, D. Comparative transcriptome analysis reveals genes associated with the gossypol synthesis and gland morphogenesis in Gossypium hirsutum. Genes 2022, 13, 1452. [Google Scholar] [CrossRef] [PubMed]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; International Wheat Genome Sequencing Consortium; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B.; Mayer, K.F. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Cheng, L.; Bao, Z.; Kong, Q.; Lassois, L.; Stein, N.; Huang, S.; Zhou, Q. Genome analyses and breeding of polyploid crops. Nat. Plants 2025, 11, 1714–1728. [Google Scholar] [CrossRef]
- Li, J.; Gao, X.; Chen, X.; Fan, Z.; Zhang, Y.; Wang, Z.; Shi, J.; Wang, C.; Zhang, H.; Wang, L. Comparative transcriptome responses of leaf and root tissues to salt stress in wheat strains with different salinity tolerances. Front. Genet. 2023, 14, 1015599. [Google Scholar] [CrossRef]
- Ma, Z.; Li, M.; Zhang, H.; Zhao, B.; Liu, Z.; Duan, S.; Meng, X.; Li, G.; Guo, X. Alternative splicing of TaHsfA2-7 is involved in the improvement of thermotolerance in wheat. Int. J. Mol. Sci. 2023, 24, 1014. [Google Scholar] [CrossRef]
- Wen, J.; Qin, Z.; Sun, L.; Zhang, Y.; Wang, D.; Peng, H.; Yao, Y.; Hu, Z.; Ni, Z.; Sun, Q. Alternative splicing of TaHSFA6e modulates heat shock protein–mediated translational regulation in response to heat stress in wheat. New Phytol. 2023, 239, 2235–2247. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Yu, K.; Han, L.; Li, X.; Wang, H.; Liu, Y.; Zhang, Y. Global profiling of alternative splicing landscape responsive to salt stress in wheat (Triticum aestivum L.). Plant Growth Regul. 2020, 92, 107–116. [Google Scholar] [CrossRef]
- Liu, Z.; Qin, J.; Tian, X.; Xu, S.; Wang, Y.; Li, H.; Wang, X.; Peng, H.; Yao, Y.; Hu, Z. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2018, 16, 714–726. [Google Scholar] [CrossRef]
- Bai, J.; Wang, Y.; Liu, Z.; Guo, H.; Zhang, F.; Guo, L.; Yuan, S.; Duan, W.; Li, Y.; Tan, Z. Global survey of alternative splicing and gene modules associated with fertility regulation in a thermosensitive genic male sterile wheat. J. Exp. Bot. 2022, 73, 2157–2174. [Google Scholar] [CrossRef]
- Zhang, Z.; Xun, H.; Lv, R.; Gou, X.; Ma, X.; Li, J.; Zhao, J.; Li, N.; Gong, L.; Liu, B. Effects of homoeologous exchange on gene expression and alternative splicing in a newly formed allotetraploid wheat. Plant J. 2022, 111, 1267–1282. [Google Scholar] [CrossRef]
- Yu, K.; Feng, M.; Yang, G.; Sun, L.; Qin, Z.; Cao, J.; Wen, J.; Li, H.; Zhou, Y.; Chen, X. Changes in alternative splicing in response to domestication and polyploidization in wheat. Plant Physiol. 2020, 184, 1955–1968. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.-A.; Peng, D.-L. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef]
- Kagale, S.; Nixon, J.; Khedikar, Y.; Pasha, A.; Provart, N.J.; Clarke, W.E.; Bollina, V.; Robinson, S.J.; Coutu, C.; Hegedus, D.D. The developmental transcriptome atlas of the biofuel crop Camelina sativa. Plant J. 2016, 88, 879–894. [Google Scholar] [CrossRef]
- Gong, W.; Song, Q.; Ji, K.; Gong, S.; Wang, L.; Chen, L.; Zhang, J.; Yuan, D. Full-length transcriptome from Camellia oleifera seed provides insight into the transcript variants involved in oil biosynthesis. J. Agric. Food Chem. 2020, 68, 14670–14683. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, H.; Zhuang, Y.; Chen, K.; Zhang, C.; Cai, T.; Yang, Q.; Fu, H.; Chen, X.; Chitkineni, A. Multiple strategies, including 6mA methylation, affecting plant alternative splicing in allopolyploid peanut. Plant Biotechnol. J. 2024, 22, 1681–1702. [Google Scholar] [CrossRef] [PubMed]
- Kashkan, I.; Timofeyenko, K.; Růžička, K. How alternative splicing changes the properties of plant proteins. Quant. Plant Biol. 2022, 3, e14. [Google Scholar] [CrossRef] [PubMed]
- Reixachs-Solé, M.; Eyras, E. Uncovering the impacts of alternative splicing on the proteome with current omics techniques. Wiley Interdiscip. Rev. RNA 2022, 13, e1707. [Google Scholar] [CrossRef]
- Kjer-Hansen, P.; Weatheritt, R.J. The function of alternative splicing in the proteome: Rewiring protein interactomes to put old functions into new contexts. Nat. Struct. Mol. Biol. 2023, 30, 1844–1856. [Google Scholar] [CrossRef]
- Manuel, J.M.; Guilloy, N.; Khatir, I.; Roucou, X.; Laurent, B. Re-evaluating the impact of alternative RNA splicing on proteomic diversity. Front. Genet. 2023, 14, 1089053. [Google Scholar] [CrossRef]
- Petrillo, E.; Kalyna, M.; Mandadi, K.K.; Tu, S.-L.; Simpson, C.G. Alternative splicing regulation in plants. Front. Plant Sci. 2020, 11, 913. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Shi, X.; Ning, N.; Wu, H.; Mei, J.; Gu, X.; Ruan, H.; Zhang, M.; Li, Z.; Ma, S. The Exserohilum turcicum effector EtEC81 reprograms alternative splicing in maize and activates immunity. Cell Rep. 2025, 44, 115501. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatima, F.; Yoo, M.-J. The Role of Alternative Splicing in Polyploids in Response to Abiotic Stress. Int. J. Mol. Sci. 2025, 26, 10146. https://doi.org/10.3390/ijms262010146
Fatima F, Yoo M-J. The Role of Alternative Splicing in Polyploids in Response to Abiotic Stress. International Journal of Molecular Sciences. 2025; 26(20):10146. https://doi.org/10.3390/ijms262010146
Chicago/Turabian StyleFatima, Faiza, and Mi-Jeong Yoo. 2025. "The Role of Alternative Splicing in Polyploids in Response to Abiotic Stress" International Journal of Molecular Sciences 26, no. 20: 10146. https://doi.org/10.3390/ijms262010146
APA StyleFatima, F., & Yoo, M.-J. (2025). The Role of Alternative Splicing in Polyploids in Response to Abiotic Stress. International Journal of Molecular Sciences, 26(20), 10146. https://doi.org/10.3390/ijms262010146






