Genome-Wide Identification and Expression Analysis of TCP Transcription Factors Responding to Multiple Stresses in Arachis hypogaea L.
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of TCP Family Members in Peanut
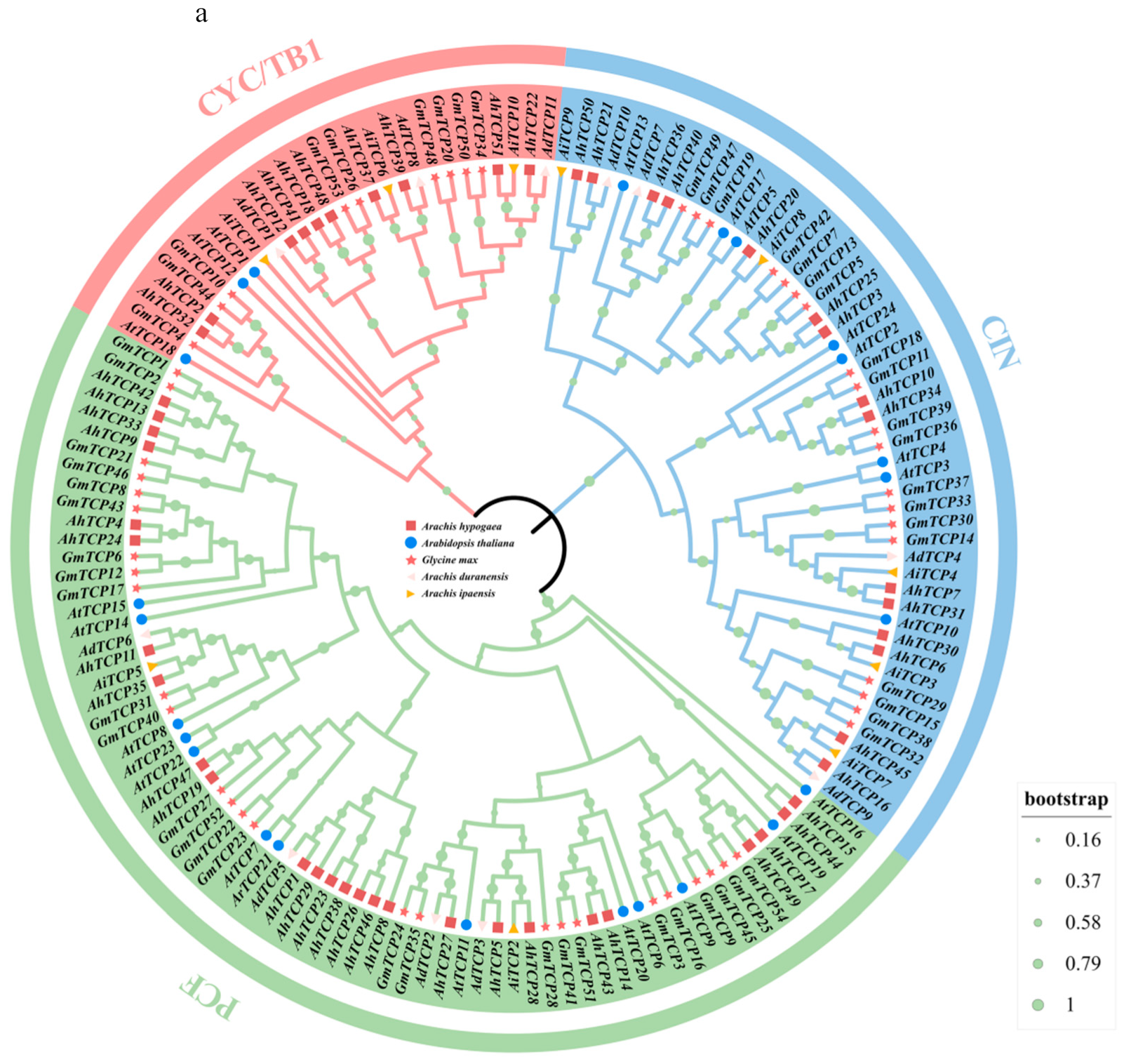
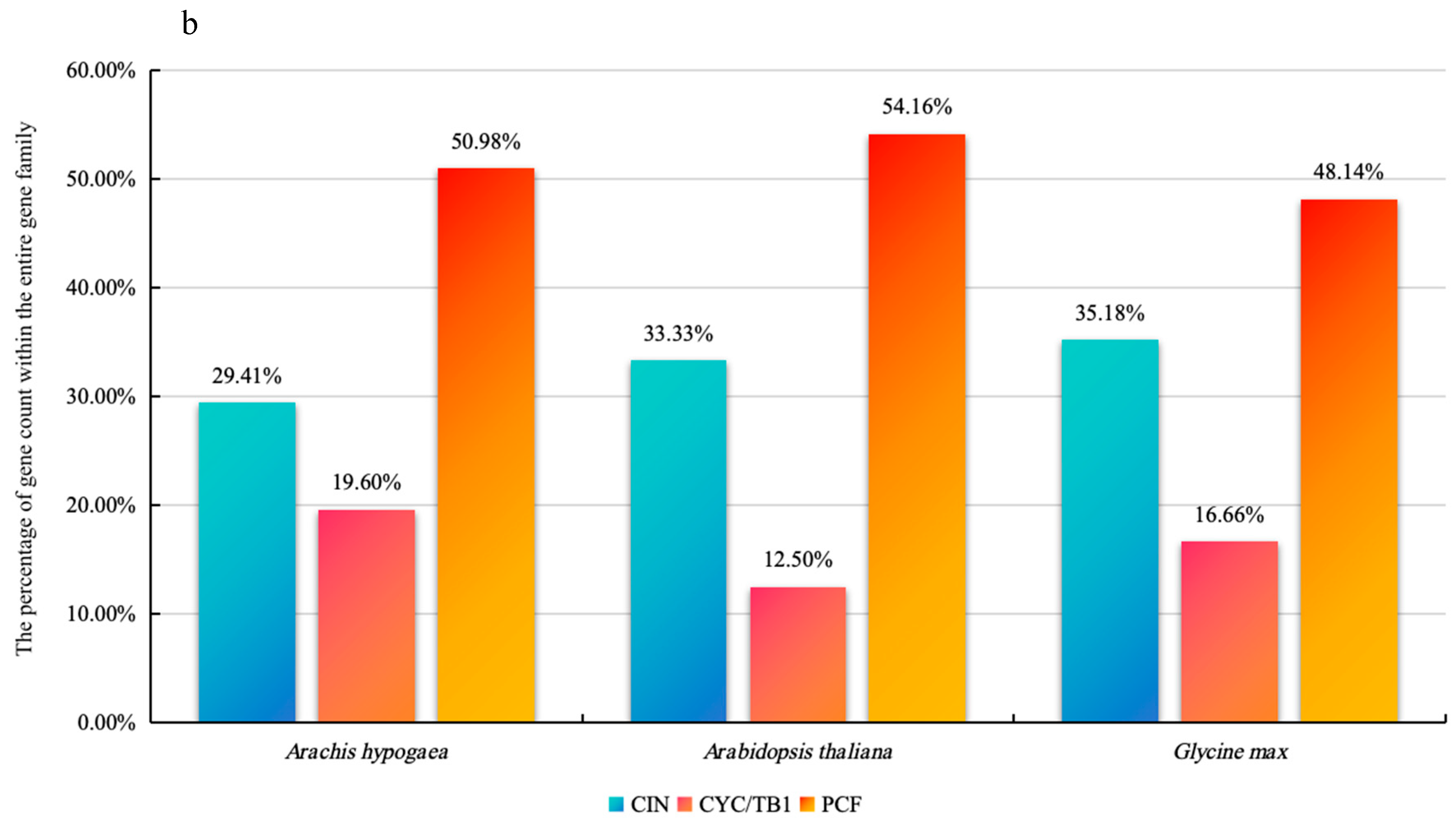
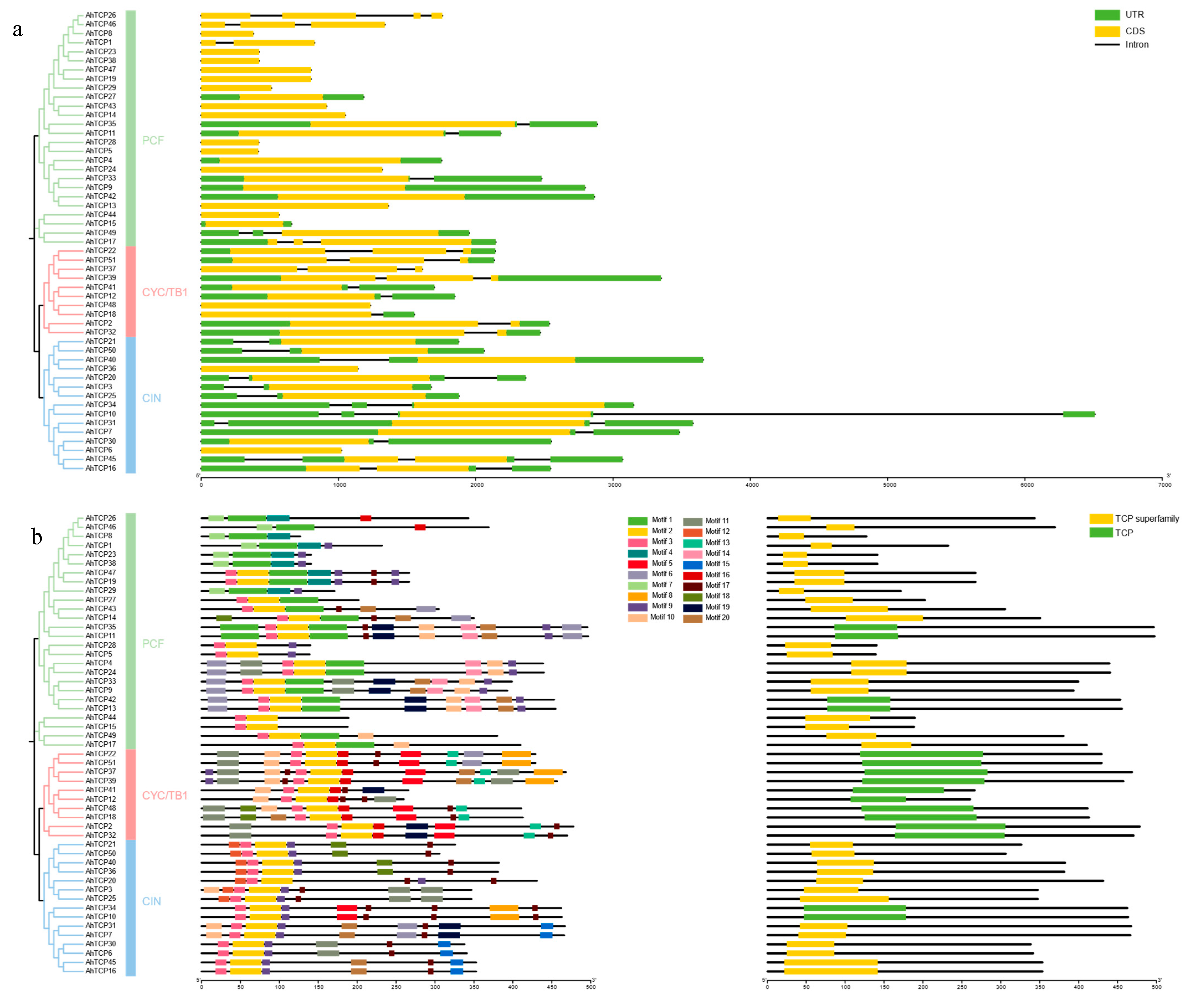
2.2. Phylogenetic Analysis and Classification of AhTCP Genes
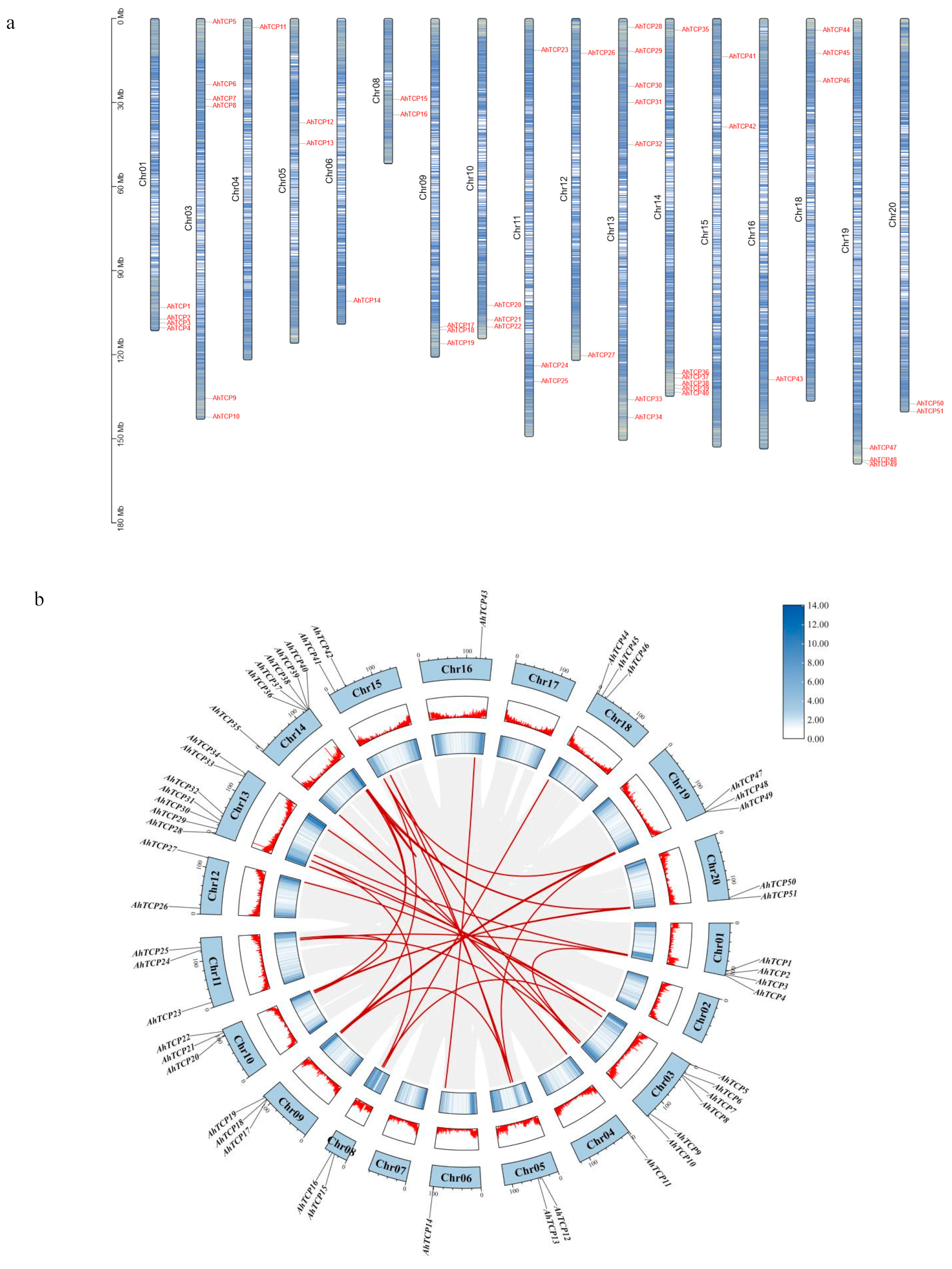
2.3. Chromosome Localization and Collinearity Analysis of AhTCP Genes
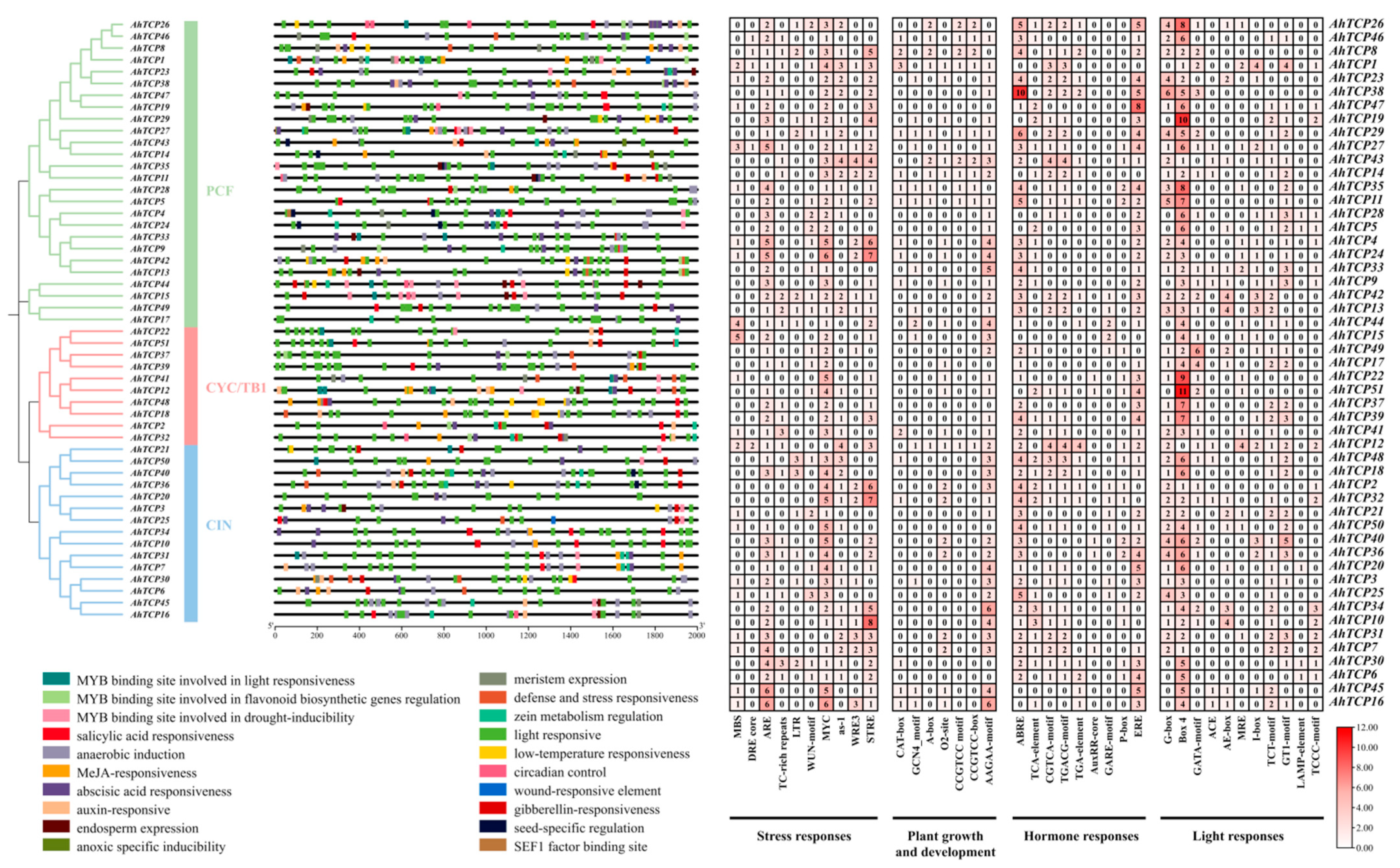
2.4. Cis-Acting Element Analysis of AhTCP Genes
2.5. The Conserved Domain, Motif, and Gene Structure Analysis
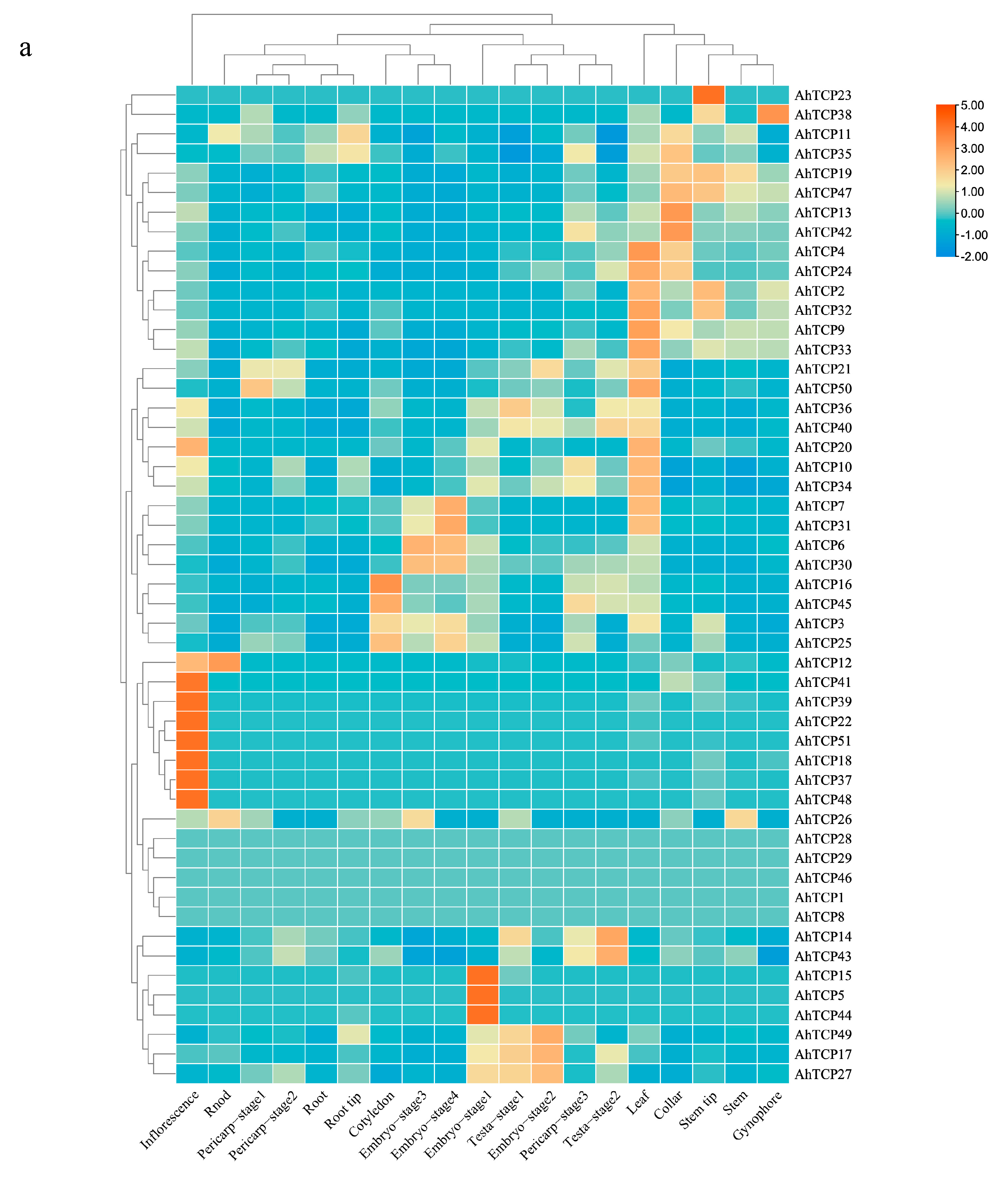
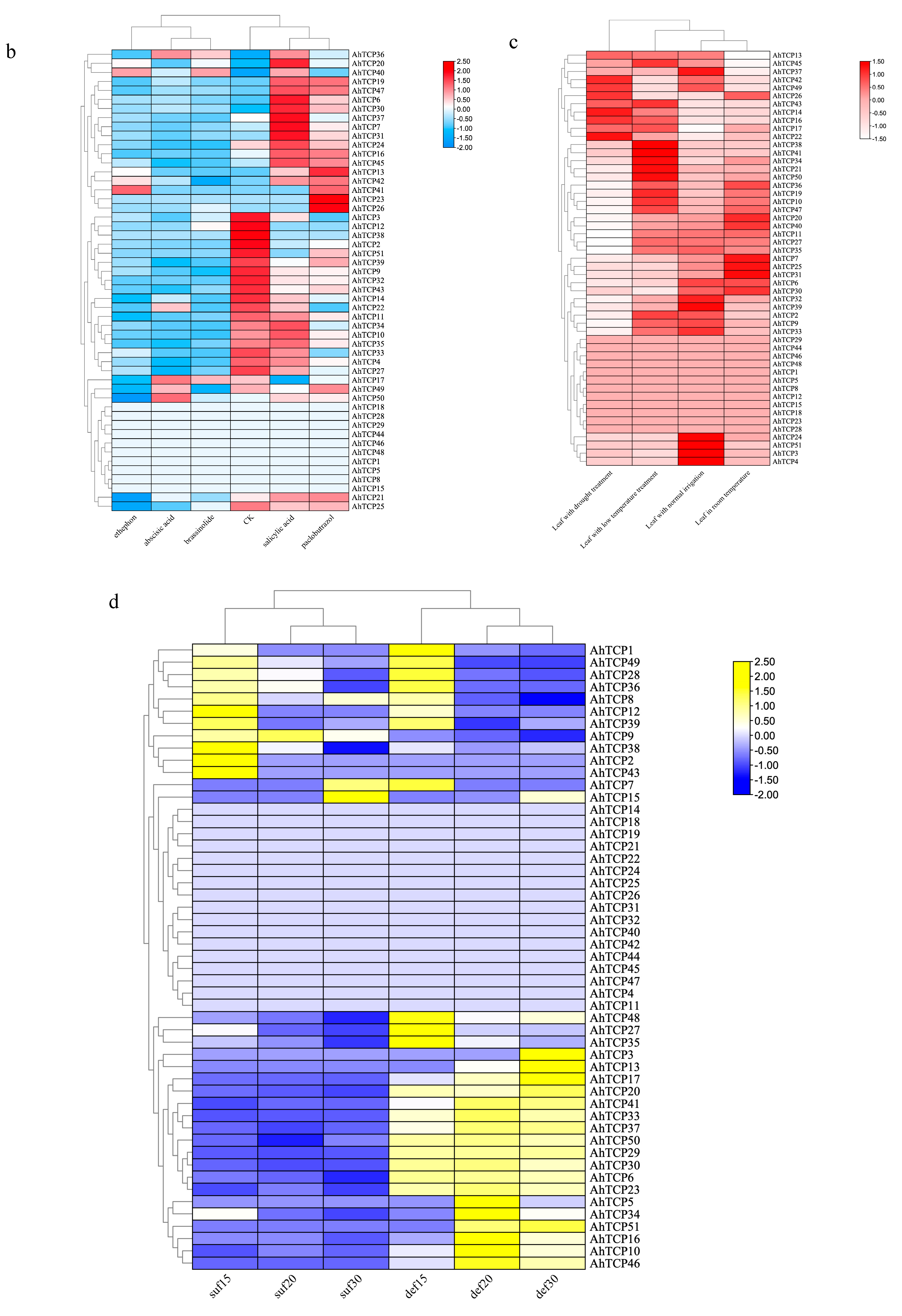
2.6. In Silico Expression Analysis of Peanut TCP Gene Family
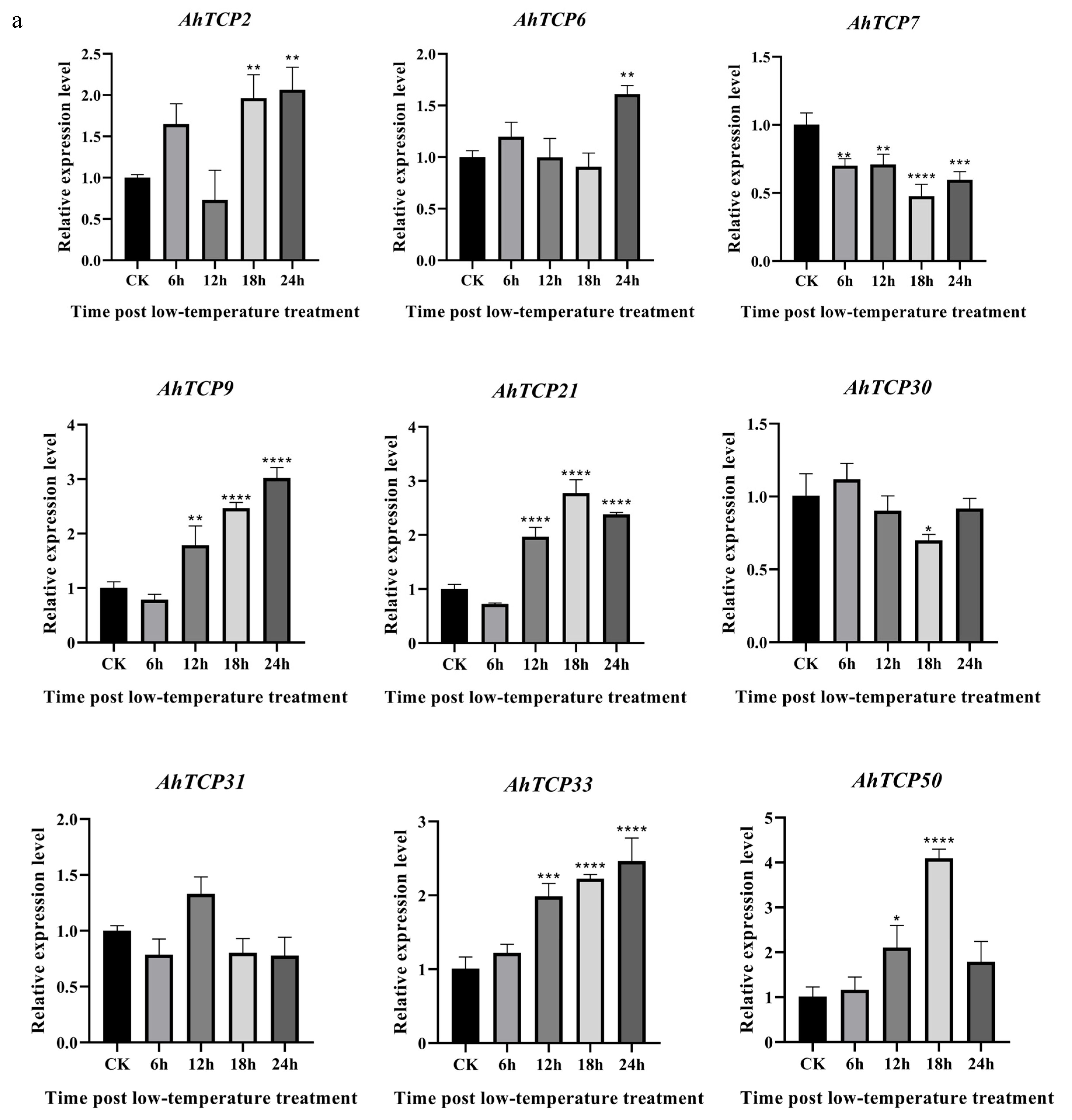
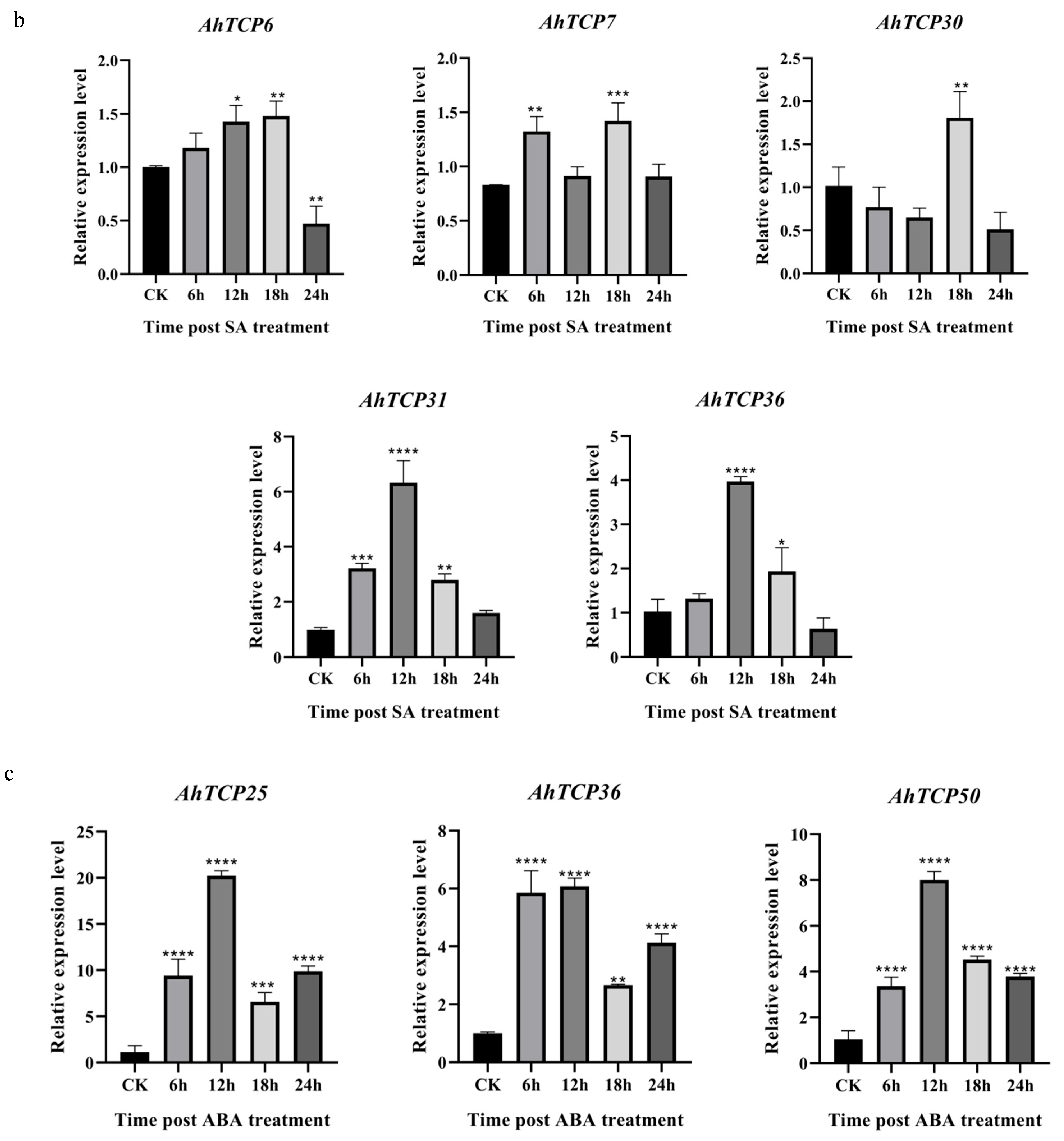
2.7. Expression of Selected AhTCP Genes Under Cold, ABA, and SA Treatments
3. Discussion
4. Materials and Methods
4.1. Identification and Sequence Analysis of Putative AhTCPs
4.2. Chromosomal Location
4.3. Collinearity Analysis of Peanut TCP Gene
4.4. Gene Structure, Conserved Motif, and Phylogenetic Analysis of AhTCP Proteins
4.5. Cis-Acting Element Analysis of AhTCP Genes
4.6. Expression Analysis of the AhTCPs in Different Tissues, Hormone Conditions, and Diverse Stresses
4.7. Stress Treatments and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vadde, B.V.L.; Challa, K.R.; Nath, U. The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J. 2017, 93, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, S.; Xu, L.; Chu, S.; Yan, X.; Lin, L.; Wen, J.; Zheng, B.; Chen, S.; Li, Q. Transcription factor PagMYB31 positively regulates cambium activity and negatively regulates xylem development in poplar. Plant Cell. 2024, 36, 1806–1828. [Google Scholar] [CrossRef]
- Yadav, A.; Sharma, A.; Kumar, A.; Yadav, R.; Misra, J.P.; Kumar, P.; Singh, D.; Kumar, R. Expression analysis of DREB2A transcriptional factor under drought stresses in rice (Oryza sativa L.). Res. J. Biotechnol. 2016, 11, 1–8. [Google Scholar]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Davière, J.-M.; Wild, M.; Regnault, T.; Baumberger, N.; Eisler, H.; Genschik, P.; Achard, P. Class I TCP-DELLA Interactions in Inflorescence Shoot Apex Determine Plant Height. Curr. Biol. 2014, 24, 1923–1928. [Google Scholar] [CrossRef] [PubMed]
- Schommer, C.; Palatnik, J.; Aggarwal, P.; Chetelat, A.; Cubas, P.; Farmer, E.; Nath, U.; Weigel, D. Control of jasmonate bio-synthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, 1991–2001. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, P.; Tyagi, A. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015, 5, 9998. [Google Scholar] [CrossRef]
- Wang, S.; Sun, X.; Hoshino, Y.; Yu, Y.; Jia, B.; Sun, Z.; Sun, M.; Duan, X.; Zhu, Y. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in Rice (Oryza sativa L.). PLoS ONE 2014, 9, e91357. [Google Scholar] [CrossRef]
- Aggarwal, P.; Das Gupta, M.; Joseph, A.; Chatterjee, N.; Srinivasan, N.; Nath, U. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 2010, 22, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Li, P.; Zhao, X.; Tan, M.; Ahmad, M.Z.; Xu, Y.; Tadege, M.; Zhao, J. CsTCPs regulate shoot tip development and catechin biosynthesis in tea plant (Camellia sinensis). Hortic. Res. 2021, 8, 1539–1559. [Google Scholar] [CrossRef] [PubMed]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Palatnik, J.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.; Weigel, D. Control of leaf morphogenesis by mi-croRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Efroni, I.; Blum, E.; Goldshmidt, A.; Eshed, Y. A Protracted and Dynamic Maturation Schedule Underlies Arabidopsis Leaf Development. Plant Cell 2008, 20, 2293–2306. [Google Scholar] [CrossRef]
- Nath, U.; Crawford, B.C.W.; Carpenter, R.; Coen, E. Genetic Control of Surface Curvature. Science 2003, 299, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.P.; Cabral, L.M.; De Veylder, L.; Tanurdzic, M.; Engler, J.d.A.; Geelen, D.; Inzé, D.; Martienssen, R.A.; Ferreira, P.C.G.; Hemerly, A.S. ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 2008, 27, 2746–2756. [Google Scholar] [CrossRef]
- Danisman, S.; van der Wal, F.; Dhondt, S.; Waites, R.; de Folter, S.; Bimbo, A.; van Dijk, A.D.; Muino, J.M.; Cutri, L.; Dornelas, M.C.; et al. Arabidopsis Class I and Class II TCP Transcription Factors Regulate Jasmonic Acid Metabolism and Leaf Development Antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial Control of Gene Expression by miR319-Regulated TCP Transcription Factors in Leaf Development. Plant Physiol. 2017, 176, 1694–1708. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, H.; Zhang, C.; Li, X.; Lu, Y.; Qi, D.; Cui, Y.; Hu, L.; Wang, Z.; Liang, Z.; et al. TCP7 functions redundantly with several Class I TCPs and regulates endoreplication in Arabidopsis. J. Integr. Plant Biol. 2019, 61, 1151–1170. [Google Scholar] [CrossRef]
- Levin, K.; Boden, S. A new branch of understanding for barley inflorescence development. J. Exp. Bot. 2020, 71, 6869–6871. [Google Scholar] [CrossRef] [PubMed]
- Leandro, E.; Pablo, A.; Diana, E.; Federico, D.; Daniel, H. Class I and Class II TCP transcription factors modulate SOC1-dependent flowering at multiple levels. Mol. Plant 2017, 10, 1571–1574. [Google Scholar]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011, 67, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Francesca, R.; Amelia, F.; Lucia, C.; Miguel, A.; David, A.; Simona, M. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 2015, 8, 482–485. [Google Scholar]
- Xu, F.; Dong, H.; Guo, W.; Le, L.; Jing, Y.; Fletcher, J.; Sun, J.; Pu, L. The trxG protein ULT1 regulates Arabidopsis organ size by interacting with TCP14/15 to antagonize the LIM peptidase DA1 for H3K4me3 on target genes. Plant Commun. 2024, 5, 255–274. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Ran, L.; Hu, J.; Ye, X.; Xu, D.; Li, J.; Su, H.; Wang, X.; Ren, S.; Luo, K. miR319a/TCP module and DELLA protein regulate trichome initiation synergistically and improve insect defenses in Populus tomentosa. New Phytol. 2020, 227, 867–883. [Google Scholar] [CrossRef]
- Camoirano, A.; Arce, A.L.; Ariel, F.D.; Alem, A.L.; Gonzalez, D.H.; Viola, I.L. Class I TCP transcription factors regulate trichome branching and cuticle development in Arabidopsis. J. Exp. Bot. 2020, 71, 5438–5453. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP Transcription Factors Control the Morphology of Shoot Lateral Organs via Negative Regulation of the Expression of Boundary-Specific Genes in Arabidopsis. Plant Cell 2006, 19, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 Acts as an Integrator of Branching Signals within Axillary Buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shang, X.; Zhou, N.; Han, Z.; Zhang, C.; Ma, Y.; Fang, W. The effective role of CsTCP5 and CsTCP18 transcription factors from Camellia sinensis (L.) O. Kuntze under drought and low-temperature. Beverage Plant Res. 2023, 3, 268–279. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, Z.; Su, C.; Cui, J.; Meng, J.; Luan, Y. Genome-wide analyses of miRNAs in mycorrhizal plants in response to late blight and elucidation of the role of miR319c in tomato resistance. Hortic. Plant J. 2023, 10, 1371–1382. [Google Scholar] [CrossRef]
- Cao, J.-F.; Zhao, B.; Huang, C.-C.; Chen, Z.-W.; Zhao, T.; Liu, H.-R.; Hu, G.-J.; Shangguan, X.-X.; Shan, C.-M.; Wang, L.-J.; et al. The miR319-Targeted GhTCP4 Promotes the Transition from Cell Elongation to Wall Thickening in Cotton Fiber. Mol. Plant 2020, 13, 1063–1077. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, F.; Wang, Q.; Wang, K.; Jones, D.; Zhang, B. Comprehensive analysis of TCP transcription factors and their ex-pression during cotton (Gossypium arboreum) fiber early development. Sci. Rep. 2016, 6, 21535. [Google Scholar]
- Sun, T. Unexpected Role of a TCP Transcription Factor in Seed Oil Biosynthesis. Plant Physiol. 2020, 184, 550–551. [Google Scholar] [CrossRef]
- Yang, X.; Yan, J.; Zhang, Z.; Lin, T.; Xin, T.; Wang, B.; Wang, S.; Zhao, J.; Zhang, Z.; Lucas, W.J.; et al. Regulation of plant architecture by a new histone acetyltransferase targeting gene bodies. Nat. Plants 2020, 6, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, X.; Xu, M.; Lin, X.; Lin, T.; Qi, J.; Shao, G.; Tian, N.; Yang, Q.; Zhang, Z.; et al. A Rare SNP Identified a TCP Transcription Factor Essential for Tendril Development in Cucumber. Mol. Plant 2015, 8, 1795–1808. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, L.; McSteen, P.; Doebley, J.; Hake, S. Expression Patterns and Mutant Phenotype of teosinte branched1 Correlate with Growth Suppression in Maize and Teosinte. Genetics 2002, 162, 1927–1935. [Google Scholar] [CrossRef]
- Yang, X.; Wang, Y.; Liu, T.; Liu, Q.; Liu, J.; Lu, T.; Yang, R.; Guo, F.; Wang, Y. CYCLOIDEA-like genes control floral symmetry, floral orientation, and nectar guide patterning. Plant Cell 2023, 35, 2799–2820. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhai, Z.; Li, Y.; Geng, S.; Song, G.; Guan, J.; Jia, M.; Wang, F.; Sun, G.; Feng, N.; et al. Genome-wide identification and characterization of LIM gene family in grapevine (Vitis vinifera L.) and their expression analysis at early bud developmental stages. Plant Mol. Biol. Rep. 2024, 42, 1282. [Google Scholar]
- Chai, W.; Jiang, P.; Huang, G.; Jiang, H.; Li, X. Identification and expression profiling analysis of TCP family genes in-volved in growth and development in maize. Physiol. Mol. Biol. Plants 2017, 23, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, Q.; Sun, R.; Xie, F.; Jones, D.; Zhang, B. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 2014, 4, 6645. [Google Scholar] [CrossRef] [PubMed]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Ma, H.; Wang, J.; Zhang, D. Genome-Wide Comparative Analysis and Expression Pattern of TCP Gene Families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 2007, 49, 885–897. [Google Scholar] [CrossRef]
- Zhang, M.; Qin, S.; Yan, J.; Li, L.; Xu, M.; Liu, Y.; Zhang, W. Genome-wide identification and analysis of TCP family genes in Medicago sativa reveal their critical roles in Na+/K+ homeostasis. BMC Plant Biol. 2023, 23, 301. [Google Scholar] [CrossRef]
- Liu, D.-K.; Zhang, C.; Zhao, X.; Ke, S.; Li, Y.; Zhang, D.; Zheng, Q.; Li, M.-H.; Lan, S.; Liu, Z.-J. Genome-wide analysis of the TCP gene family and their expression pattern in Cymbidium goeringii. Front. Plant Sci. 2022, 13, 1068969. [Google Scholar] [CrossRef]
- Shang, X.; Han, Z.; Zhang, D.; Wang, Y.; Qin, H.; Zou, Z.; Zhou, L.; Zhu, X.; Fang, W.; Ma, Y. Genome-Wide Analysis of the TCP Gene Family and Their Expression Pattern Analysis in Tea Plant (Camellia sinensis). Front. Plant Sci. 2022, 13, 840350. [Google Scholar] [CrossRef]
- Zheng, A.; Sun, F.; Cheng, T.; Wang, Y.; Xie, K.; Zhang, C.; Xi, Y. Genome-wide identification of members of the TCP gene family in switchgrass (Panicum virgatum L.) and analysis of their expression. Gene 2019, 702, 89–98. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Z.; Zhao, K.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Genome-wide identification, characterization and expression analysis of the TCP gene family in Prunus mume. Front. Plant Sci. 2016, 7, 1301. [Google Scholar] [CrossRef]
- Sun, X.; Wang, E.; Yu, L.; Liu, S.; Liu, T.; Qin, J.; Jiang, P.; He, S.; Cai, X.; Jing, S.; et al. TCP transcription factor StAST1 represses potato tuberization by regulating tuberigen complex activity. Plant Physiol. 2024, 195, 1347–1364. [Google Scholar] [CrossRef]
- Hu, G.; Zhang, D.; Luo, D.; Sun, W.; Zhou, R.; Hong, Z.; Munir, S.; Ye, Z.; Yang, C.; Zhang, J.; et al. SlTCP24 and SlTCP29 synergistically regulate compound leaf development through interacting with SlAS2 and activating transcription of SlCKX2 in tomato. New Phytol. 2023, 240, 1275–1291. [Google Scholar] [CrossRef]
- Lan, J.; Zhang, J.; Yuan, R.; Yu, H.; An, F.; Sun, L.; Chen, H.; Zhou, Y.; Qian, W.; He, H.; et al. TCP transcription factors suppress cotyledon trichomes by impeding a cell differentiation-regulating complex. Plant Physiol. 2021, 186, 434–451. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, Q.; Fu, H.; Chen, K.; Zhao, S.; Zhang, C.; Cai, T.; Wang, L.; Lu, W.; Dang, H.; et al. Identification of Key Gene Networks and Deciphering Transcriptional Regulators Associated with Peanut Embryo Abortion Mediated by Calcium Deficiency. Front. Plant Sci. 2022, 13, 814015. [Google Scholar] [CrossRef] [PubMed]
- Viola, I.L.; Alem, A.L.; Jure, R.M.; Gonzalez, D.H. Physiological Roles and Mechanisms of Action of Class I TCP Transcription Factors. Int. J. Mol. Sci. 2023, 24, 5437. [Google Scholar] [CrossRef] [PubMed]
- Danisman, S. TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 2016, 7, 1930. [Google Scholar] [CrossRef] [PubMed]
- Manassero, N.G.U.; Viola, I.L.; Welchen, E.; Gonzalez, D.H. TCP transcription factors: Architectures of plant form. Biomol. Concepts 2013, 4, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Danisman, S.; van Dijk, A.D.J.; Bimbo, A.; van der Wal, F.; Hennig, L.; de Folter, S.; Angenent, G.C.; Immink, R.G.H. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 2013, 64, 5673–5685. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yuan, M.; Zheng, B.-Q.; Zhou, L.; Wang, Y. Genome-wide identification and characterization of TCP gene family in Dendrobium nobile and their role in perianth development. Front. Plant Sci. 2024, 15, 1352119. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lv, T.; Shen, Y.; Liu, T.; Liu, M.; Hu, J.; Liu, S.; Jiang, Y.; Zhang, M.; Zhao, M.; et al. Genome-wide identification and integrated analysis of TCP genes controlling ginsenoside biosynthesis in Panax ginseng. BMC Plant Biol. 2024, 24, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhai, Z.; Li, Y.; Geng, S.; Song, G.; Guan, J.; Jia, M.; Wang, F.; Sun, G.; Feng, N.; et al. Genome-Wide Identification and Expression Profiling of the TCP Family Genes in Spike and Grain Development of Wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 1282. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yin, X.; Feng, T.; Kang, Z.; Zhang, X.; Dong, J.; Liang, Z. Genome-wide identification and expression analysis of the TCP genes in Senna tora reveal the regulatory mechanism of their response to MeJA. Ind. Crop. Prod. 2022, 187, 115511. [Google Scholar] [CrossRef]
- Ma, X.; Ma, J.; Fan, D.; Li, C.; Jiang, Y.; Luo, K. Genome-wide identification of TCP family transcription factors from Populus euphratica and their involvement in leaf shape regulation. Sci. Rep. 2016, 6, 32795. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Duan, S.; Xu, C.; Wang, Y.; Zhang, X.; Xu, X.; Chen, L.; Han, Z.; Wu, T. Pan-genome analysis of 13 Malus accessions reveals structural and sequence variations associated with fruit traits. Nat. Commun. 2023, 14, 7377. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.T.; Fink, G.R.; Bartel, D.P. Excised linear introns regulate growth in yeast. Nature 2019, 565, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lyu, H.; Zhu, K.; Van de Peer, Y.; Cheng, Z. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qin, J.; Tian, X.; Xu, S.; Wang, Y.; Li, H.; Wang, X.; Peng, H.; Yao, Y.; Hu, Z.; et al. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2017, 16, 714–726. [Google Scholar] [CrossRef]
- Parenteau, J.; Maignon, L.; Berthoumieux, M.; Catala, M.; Gagnon, V.; Elela, S.A. Introns are mediators of cell response to starvation. Nature 2019, 565, 612–617. [Google Scholar] [CrossRef]
- Pan, J.; Ju, Z.; Ma, X.; Duan, L.; Jia, Z. Genome-wide characterization of TCP family and their potential roles in abiotic stress resistance of oat (Avena sativa L.). Front. Plant Sci. 2024, 15, 1382790. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, W.; Xie, X.; Wang, Z.; Guan, P.; Peng, H.; Jiao, Y.; Ni, Z.; Sun, Q.; Guo, W. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the Triticeae Tribe as a pilot practice in the plant pange-nomic Era. Mol. Plant 2020, 13, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Lei, N.; Yu, X.; Li, S.; Zeng, C.; Zou, L.; Liao, W.; Peng, M. Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress. Sci. Rep. 2017, 7, 10016. [Google Scholar] [CrossRef]
- Tian, C.; Zhai, L.; Zhu, W.; Qi, X.; Yu, Z.; Wang, H.; Chen, F.; Wang, L.; Chen, S. Characterization of the TCP Gene Family in Chrysanthemum nankingense and the Role of CnTCP4 in Cold Tolerance. Plants 2022, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Gustus, C. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics 1995, 141, 333–346. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Wan, H.; Tang, J.; Ni, Z. The sea-island cotton GbTCP4 transcription factor positively regulates drought and salt stress responses. Plant Sci. 2022, 322, 111329. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhong, C.; Zhu, C.; Zhang, J.; Zhu, L.; Wu, L.; Jin, Q. Variability of leaf photosynthetic characteristics in rice and its relationship with resistance to water stress under different nitrogen nutrition regimes. Physiol. Plant 2019, 167, 613–627. [Google Scholar]
- Chen, H.; Zhang, C.; Cai, T.; Deng, Y.; Zhou, S.; Zheng, Y.; Ma, S.; Tang, R.; Varshney, R.; Zhuang, W. Identification of low Ca2+ stress-induced embryo apoptosis response genes in Arachis hypogaea by SSH-associated library lift (SSHaLL). Plant Biotechnol. J. 2016, 14, 682–698. [Google Scholar] [PubMed]
- Jain, M.; Pathak, B.P.; Harmon, A.C.; Tillman, B.L.; Gallo, M. Calcium dependent protein kinase (CDPK) expression during fruit development in cultivated peanut (Arachis hypogaea) under Ca2+-sufficient and -deficient growth regimens. J. Plant Physiol. 2011, 168, 2272–2277. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains iden-tifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar]
- Bailey, T.; Johnson, J.; Grant, C.; Noble, W. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sharif, Y.; Chen, H.; Deng, Y.; Ali, N.; Khan, S.; Zhang, C.; Xie, W.; Chen, K.; Cai, T.; Yang, Q.; et al. Cloning and functional characterization of a peri-carp abundant expression promoter (AhGLP17-1P) from peanut (Arachis hypogaea L.). Front. Genet. 2022, 12, 821281. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Swift, M.L. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]

| Seq_1 | Seq_2 | Ka | Ks | Ka/Ks | Ks/2r (MYA) |
|---|---|---|---|---|---|
| AhTCP4 | AhTCP9 | 0.352897198 | 2.2657562 | 0.1557525 | 139,517,004.99 |
| AhTCP4 | AhTCP24 | 0 | 0.0265001 | 0.0000000 | 1,631,781.68 |
| AhTCP2 | AhTCP32 | 0.014846228 | 0.0383025 | 0.3876048 | 2,358,527.80 |
| AhTCP6 | AhTCP16 | 0.285208236 | 1.9666324 | 0.1450237 | 121,098,056.02 |
| AhTCP9 | AhTCP24 | 0.334979524 | 2.1683105 | 0.1544887 | 133,516,659.16 |
| AhTCP5 | AhTCP27 | 0.489917965 | NaN | NaN | NaN |
| AhTCP10 | AhTCP34 | 0.003780726 | 0.0298712 | 0.1265676 | 1,839,359.90 |
| AhTCP6 | AhTCP30 | 0.009008151 | 0.0377587 | 0.2385718 | 2,325,040.73 |
| AhTCP5 | AhTCP28 | 0.016047684 | 0.0108207 | 1.4830477 | 666,302.14 |
| AhTCP9 | AhTCP42 | 0.262350405 | 1.8892023 | 0.1388683 | 116,330,190.81 |
| AhTCP11 | AhTCP35 | 0.009853829 | 0.0494365 | 0.1993230 | 3,044,119.23 |
| AhTCP12 | AhTCP18 | 0.404993521 | 1.1462073 | 0.3533336 | 70,579,265.77 |
| AhTCP13 | AhTCP42 | 0.005808823 | 0.0586459 | 0.0990491 | 3,611,199.17 |
| AhTCP12 | AhTCP41 | 0.003335655 | 0.0361334 | 0.0923149 | 2,224,964.91 |
| AhTCP12 | AhTCP48 | 0.390690818 | 1.0952004 | 0.3567300 | 67,438,450.47 |
| AhTCP14 | AhTCP43 | 0.004258351 | 0.0297806 | 0.1429908 | 1,833,780.00 |
| AhTCP16 | AhTCP30 | 0.265356041 | 2.2656185 | 0.1171230 | 139,508,527.38 |
| AhTCP15 | AhTCP44 | 0.011964361 | 0.0485017 | 0.2466790 | 2,986,560.63 |
| AhTCP18 | AhTCP41 | 0.518842675 | 1.5176869 | 0.3418641 | 93,453,627.81 |
| AhTCP17 | AhTCP49 | 0.004927137 | 0.0184398 | 0.2672016 | 1,135,454.02 |
| AhTCP18 | AhTCP48 | 0.002091869 | 0.0270387 | 0.0773659 | 1,664,941.75 |
| AhTCP19 | AhTCP47 | 0.006796026 | 0.0295969 | 0.2296192 | 1,822,471.38 |
| AhTCP20 | AhTCP25 | 0.399074138 | 2.0537667 | 0.1943133 | 126,463,464.47 |
| AhTCP21 | AhTCP36 | 0.31837927 | 1.0005152 | 0.3182153 | 61,608,078.22 |
| AhTCP21 | AhTCP40 | 0.328553957 | 1.0062286 | 0.3265202 | 61,959,888.76 |
| AhTCP21 | AhTCP50 | 0.031473149 | 0.0397444 | 0.7918890 | 2,447,315.12 |
| AhTCP22 | AhTCP51 | 0.008109557 | 0.0587456 | 0.1380453 | 3,617,340.68 |
| AhTCP36 | AhTCP40 | 0.02250689 | 0.0348983 | 0.6449280 | 2,148,909.87 |
| AhTCP37 | AhTCP39 | 0.005602267 | 0.0242796 | 0.2307399 | 1,495,047.83 |
| AhTCP36 | AhTCP50 | 0.288229 | 0.8752547 | 0.3293087 | 53,894,994.64 |
| AhTCP40 | AhTCP50 | 0.309598196 | 1.0016415 | 0.3090908 | 61,677,433.92 |
| AhTCP41 | AhTCP48 | 0.416790346 | 1.1703064 | 0.3561378 | 72,063,199.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Niu, S.; Lin, J.; Yang, H.; Zhou, X.; Wang, S.; Liu, X.; Yang, Q.; Zhang, C.; Zhuang, Y.; et al. Genome-Wide Identification and Expression Analysis of TCP Transcription Factors Responding to Multiple Stresses in Arachis hypogaea L. Int. J. Mol. Sci. 2025, 26, 1069. https://doi.org/10.3390/ijms26031069
Zhu Y, Niu S, Lin J, Yang H, Zhou X, Wang S, Liu X, Yang Q, Zhang C, Zhuang Y, et al. Genome-Wide Identification and Expression Analysis of TCP Transcription Factors Responding to Multiple Stresses in Arachis hypogaea L. International Journal of Molecular Sciences. 2025; 26(3):1069. https://doi.org/10.3390/ijms26031069
Chicago/Turabian StyleZhu, Yanting, Sijie Niu, Jingyi Lin, Hua Yang, Xun Zhou, Siwei Wang, Xiaoyan Liu, Qiang Yang, Chong Zhang, Yuhui Zhuang, and et al. 2025. "Genome-Wide Identification and Expression Analysis of TCP Transcription Factors Responding to Multiple Stresses in Arachis hypogaea L." International Journal of Molecular Sciences 26, no. 3: 1069. https://doi.org/10.3390/ijms26031069
APA StyleZhu, Y., Niu, S., Lin, J., Yang, H., Zhou, X., Wang, S., Liu, X., Yang, Q., Zhang, C., Zhuang, Y., Cai, T., Zhuang, W., & Chen, H. (2025). Genome-Wide Identification and Expression Analysis of TCP Transcription Factors Responding to Multiple Stresses in Arachis hypogaea L. International Journal of Molecular Sciences, 26(3), 1069. https://doi.org/10.3390/ijms26031069







