Multimer Detection System: A Universal Assay System for Differentiating Protein Oligomers from Monomers
Abstract
1. Introduction
2. Oligomer Formation and Fibrillization
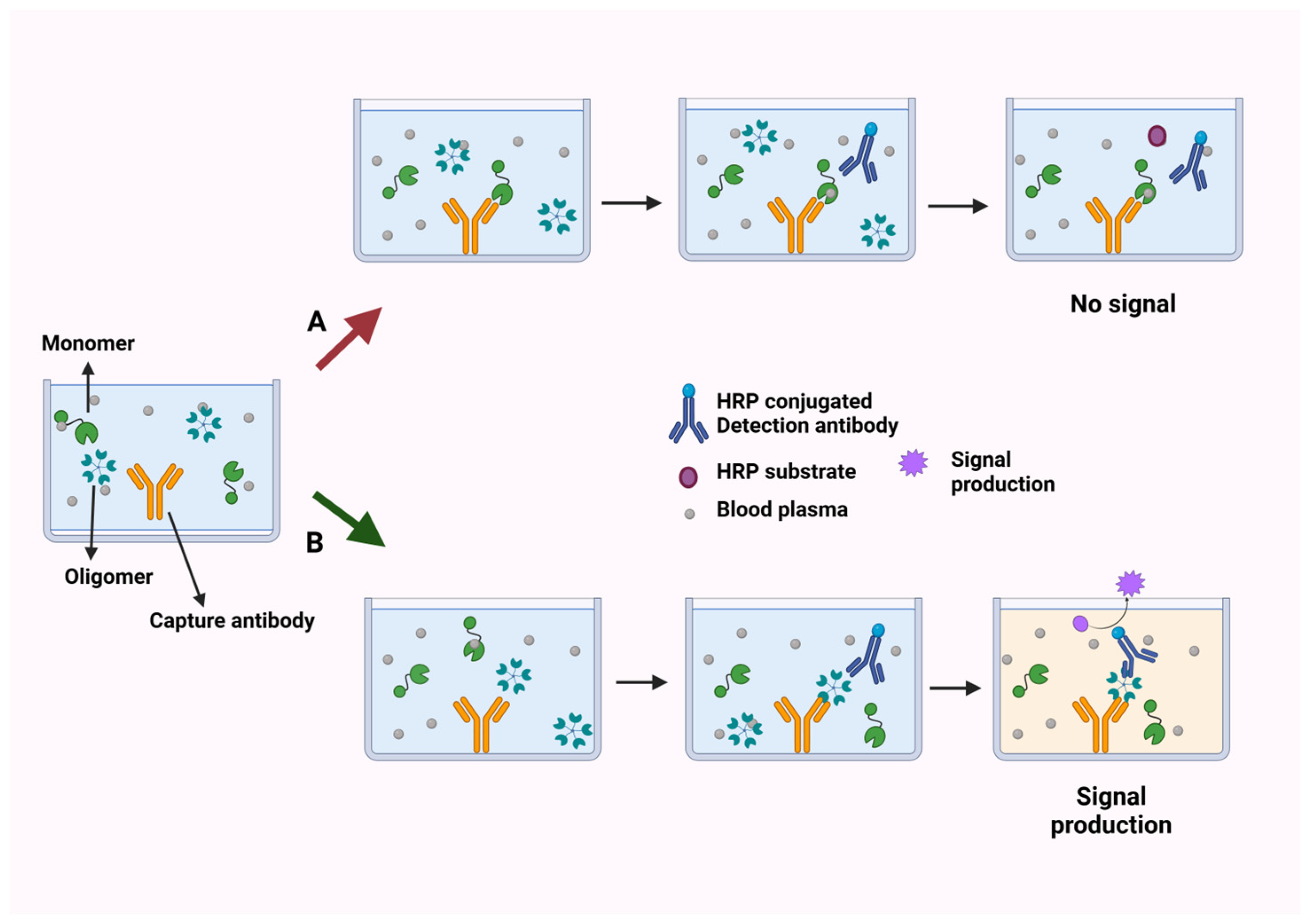
3. Multimer Detection System
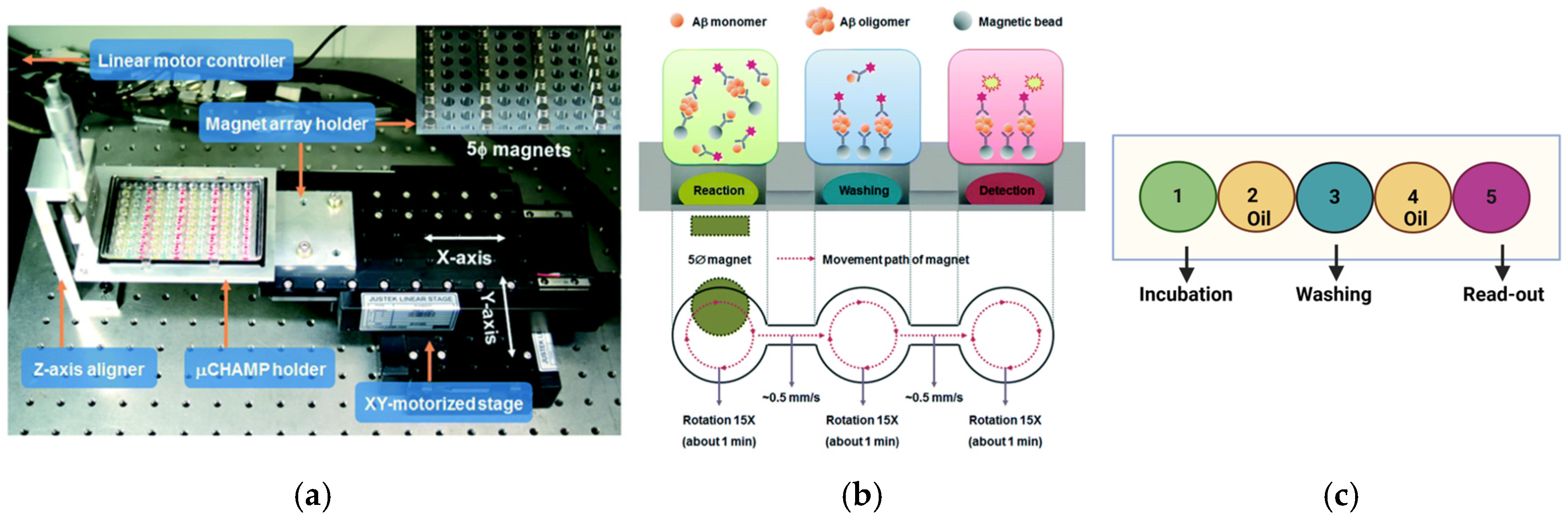
4. Expansion of MDS
5. Applications of MDS in NDs
5.1. Prion Disease
5.2. Alzheimer’s Disease (AD)
5.3. Frontotemporal Dementia (FTD)
5.4. Synucleinopathies and Tauopathies
5.5. Huntington’s Disease
6. Future Applications: Hybrid Oligomers
7. Gaps and Limitations
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dai, M.; Masahiro, K. Oligomerization of Proteins and Neurodegenerative Diseases. In Oligomerization of Chemical and Biological Compounds; Claire, L., Ed.; IntechOpen: Rijeka, Croatia, 2014; Chapter 9. [Google Scholar]
- Carrell, R.W.; Lomas, D.A. Conformational disease. Lancet 1997, 350, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Donald, J.E.; Kulp, D.W.; DeGrado, W.F. Salt bridges: Geometrically specific, designable interactions. Proteins Struct. Funct. Bioinform. 2011, 79, 898–915. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Imperiali, B. Protein oligomerization: How and why. Bioorg. Med. Chem. 2005, 13, 5013–5020. [Google Scholar] [CrossRef]
- Thibaudeau, T.A.; Anderson, R.T.; Smith, D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar] [CrossRef]
- Mirdha, L.; Chakraborty, H. Fluorescence-based techniques for the detection of the oligomeric status of proteins: Implication in amyloidogenic diseases. Eur. Biophys. J. 2021, 50, 671–685. [Google Scholar] [CrossRef]
- McKenzie, D.M.; Wirth, D.; Pogorelov, T.V.; Hristova, K. Utility of FRET in studies of membrane protein oligomerization: The concept of the effective dissociation constant. Biophys. J. 2023, 122, 4113–4120. [Google Scholar] [CrossRef]
- Shen, H.; Fu, C.; Zhang, J.; Feng, B.; Yu, S. Protocol for determining protein dynamics using FT-IR spectroscopy. STAR Protoc. 2023, 4, 102587. [Google Scholar] [CrossRef]
- Cortivo, G.D.; Marino, V.; Iacobucci, C.; Vallone, R.; Arlt, C.; Rehkamp, A.; Sinz, A.; Dell’Orco, D. Oligomeric state, hydrodynamic properties and target recognition of human Calcium and Integrin Binding protein 2 (CIB2). Sci. Rep. 2019, 9, 15058. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, A.; Tang, X.; Qin, J. Electrochemical sensitive detection of amyloid-β oligomer harnessing cellular prion protein on AuNPs embedded poly (pyrrole-3-carboxylic acid) matrix. Mater. Today Adv. 2022, 14, 100250. [Google Scholar] [CrossRef]
- Rushworth, J.V.; Ahmed, A.; Griffiths, H.H.; Pollock, N.M.; Hooper, N.M.; Millner, P.A. A label-free electrical impedimetric biosensor for the specific detection of Alzheimer’s amyloid-beta oligomers. Biosens. Bioelectron. 2014, 56, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Eaton, P.; Quaresma, P.; Soares, C.; Neves, C.; de Almeida, M.P.; Pereira, E.; West, P. A direct comparison of experimental methods to measure dimensions of synthetic nanoparticles. Ultramicroscopy 2017, 182, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Ying, C.; Li, J.; Mayer, M. Simultaneous Determination of the Size and Shape of Single α-Synuclein Oligomers in Solution. ACS Nano 2023, 17, 12325–12335. [Google Scholar] [CrossRef]
- Correcirc, D.H.A.; Ramos, C.H.I. The use of circular dichroism spectroscopy to study protein folding, form and function. Afr. J. Biochem. Res. 2009, 3, 164–173. [Google Scholar]
- Singh, S.; DeMarco, M.L. In Vitro Conversion Assays Diagnostic for Neurodegenerative Proteinopathies. J. Appl. Lab. Med. 2020, 5, 142–157. [Google Scholar] [CrossRef]
- Paciotti, S.; Bellomo, G.; Gatticchi, L.; Parnetti, L. Are We Ready for Detecting α-Synuclein Prone to Aggregation in Patients? The Case of “Protein-Misfolding Cyclic Amplification” and “Real-Time Quaking-Induced Conversion” as Diagnostic Tools. Front. Neurol. 2018, 9, 415. [Google Scholar] [CrossRef]
- Peden, A.H.; McGuire, L.; Appleford, N.E.J.; Mallinson, G.; Wilham, J.M.; Orrú, C.D.; Caughey, B.; Ironside, J.W.; Knight, R.S.G.; Will, R.G.; et al. Sensitive and specific detection of sporadic Creutzfeldt-Jakob disease brain prion protein using real-time quaking-induced conversion. J. Gen. Virol. 2012, 93 Pt 2, 438–449. [Google Scholar] [CrossRef]
- Srivastava, A.; Alam, P.; Caughey, B. RT-QuIC and Related Assays for Detecting and Quantifying Prion-like Pathological Seeds of α-Synuclein. Biomolecules 2022, 12, 576. [Google Scholar] [CrossRef]
- Rajasekhar, K.; Narayanaswamy, N.; Murugan, N.A.; Viccaro, K.; Lee, H.-G.; Shah, K.; Govindaraju, T. Aβ plaque-selective NIR fluorescence probe to differentiate Alzheimer’s disease from tauopathies. Biosens. Bioelectron. 2017, 98, 54–61. [Google Scholar] [CrossRef]
- Liu, L.; Chang, Y.; Yu, J.; Jiang, M.; Xia, N. Two-in-one polydopamine nanospheres for fluorescent determination of beta-amyloid oligomers and inhibition of beta-amyloid aggregation. Sens. Actuators B Chem. 2017, 251, 359–365. [Google Scholar] [CrossRef]
- Ren, W.; Xu, M.; Liang, S.H.; Xiang, H.; Tang, L.; Zhang, M.; Ding, D.; Li, X.; Zhang, H.; Hu, Y. Discovery of a novel fluorescent probe for the sensitive detection of β-amyloid deposits. Biosens. Bioelectron. 2016, 75, 136–141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, L.; Xia, N.; Zhang, J.; Mao, W.; Wu, Y.; Ge, X. A graphene oxide-based fluorescent platform for selective detection of amyloid-β oligomers. Anal. Methods 2015, 7, 8727–8732. [Google Scholar] [CrossRef]
- Yi, X.; Feng, C.; Hu, S.; Li, H.; Wang, J. Surface plasmon resonance biosensors for simultaneous monitoring of amyloid-beta oligomers and fibrils and screening of select modulators. Analyst 2016, 141, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Mordechai, S.; Shufan, E.; Porat Katz, B.S.; Salman, A. Early diagnosis of Alzheimer’s disease using infrared spectroscopy of isolated blood samples followed by multivariate analyses. Analyst 2017, 142, 1276–1284. [Google Scholar] [CrossRef]
- Zhang, A.; Portugal Barron, D.; Chen, E.W.; Guo, Z. A protein aggregation platform that distinguishes oligomers from amyloid fibrils. Analyst 2023, 148, 2283–2294. [Google Scholar] [CrossRef]
- Khan, A.N.; Khan, R.H. Protein misfolding and related human diseases: A comprehensive review of toxicity, proteins involved, and current therapeutic strategies. Int. J. Biol. Macromol. 2022, 223, 143–160. [Google Scholar] [CrossRef]
- Limbocker, R.; Cremades, N.; Cascella, R.; Tessier, P.M.; Vendruscolo, M.; Chiti, F. Characterization of Pairs of Toxic and Nontoxic Misfolded Protein Oligomers Elucidates the Structural Determinants of Oligomer Toxicity in Protein Misfolding Diseases. Acc. Chem. Res. 2023, 56, 1395–1405. [Google Scholar] [CrossRef]
- Wells, C.; Brennan, S.E.; Keon, M.; Ooi, L. The role of amyloid oligomers in neurodegenerative pathologies. Int. J. Biol. Macromol. 2021, 181, 582–604. [Google Scholar] [CrossRef]
- Verma, M.; Vats, A.; Taneja, V. Toxic species in amyloid disorders: Oligomers or mature fibrils. Ann. Indian Acad. Neurol. 2015, 18, 138–145. [Google Scholar]
- Lee, S.J.C.; Nam, E.; Lee, H.J.; Savelieff, M.G.; Lim, M.H. Towards an understanding of amyloid-β oligomers: Characterization, toxicity mechanisms, and inhibitors. Chem. Soc. Rev. 2017, 46, 310–323. [Google Scholar] [CrossRef]
- Harte, N.P.; Klyubin, I.; McCarthy, E.K.; Min, S.; Garrahy, S.A.; Xie, Y.; Davey, G.P.; Boland, J.J.; Rowan, M.J.; Mok, K.H. Amyloid Oligomers and Mature Fibrils Prepared from an Innocuous Protein Cause Diverging Cellular Death Mechanisms. J. Biol. Chem. 2015, 290, 28343–28352. [Google Scholar] [CrossRef] [PubMed]
- Winner, B.; Jappelli, R.; Maji, S.K.; Desplats, P.A.; Boyer, L.; Aigner, S.; Hetzer, C.; Loher, T.J.; Vilar, M.; Campioni, S.; et al. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA 2011, 108, 4194–4199. [Google Scholar] [CrossRef] [PubMed]
- Dear, A.J.; Michaels, T.C.T.; Meisl, G.; Klenerman, D.; Wu, S.; Perrett, S.; Linse, S.; Dobson, C.M.; Knowles, T.P.J. Kinetic diversity of amyloid oligomers. Proc. Natl. Acad. Sci. USA 2020, 117, 12087–12094. [Google Scholar] [CrossRef] [PubMed]
- Šarić, A.; Chebaro, Y.C.; Knowles, T.P.; Frenkel, D. Crucial role of nonspecific interactions in amyloid nucleation. Proc. Natl. Acad. Sci. USA 2014, 111, 17869–17874. [Google Scholar] [CrossRef]
- Cheon, M.; Chang, I.; Mohanty, S.; Luheshi, L.M.; Dobson, C.M.; Vendruscolo, M.; Favrin, G. Structural Reorganisation and Potential Toxicity of Oligomeric Species Formed during the Assembly of Amyloid Fibrils. PLoS Comput. Biol. 2007, 3, e173. [Google Scholar] [CrossRef]
- Ahmed, M.; Davis, J.; Aucoin, D.; Sato, T.; Ahuja, S.; Aimoto, S.; Elliott, J.I.; Van Nostrand, W.E.; Smith, S.O. Structural conversion of neurotoxic amyloid-β1–42 oligomers to fibrils. Nat. Struct. Mol. Biol. 2010, 17, 561–567. [Google Scholar] [CrossRef]
- Thirumalai, D.; Reddy, G.; Straub, J.E. Role of water in protein aggregation and amyloid polymorphism. Acc. Chem. Res. 2011, 45, 83–92. [Google Scholar] [CrossRef]
- Muschol, M.; Hoyer, W. Amyloid oligomers as on-pathway precursors or off-pathway competitors of fibrils. Front. Mol. Biosci. 2023, 10, 1120416. [Google Scholar] [CrossRef]
- An, S.S.A.; Lim, K.T.; Oh, H.J.; Lee, B.S.; Zukic, E.; Ju, Y.R.; Yokoyama, T.; Kim, S.Y.; Welker, E. Differentiating blood samples from scrapie-infected and non-infected hamsters by detecting disease-associated prion proteins using Multimer Detection System. Biochem. Biophys. Res. Commun. 2010, 392, 505–509. [Google Scholar] [CrossRef]
- Van Giau, V.; An, S.S.A. Epitope Mapping Immunoassay Analysis of the Interaction between β-Amyloid and Fibrinogen. Int. J. Mol. Sci. 2019, 20, 496. [Google Scholar] [CrossRef]
- Lim, K.; Kim, S.Y.; Lee, B.; Segarra, C.; Kang, S.; Ju, Y.; Schmerr, M.J.; Coste, J.; Kim, S.Y.; Yokoyama, T. Magnetic microparticle-based multimer detection system for the detection of prion oligomers in sheep. Int. J. Nanomed. 2015, 10, 241–250. [Google Scholar]
- An, S.S.A.; Lee, B.-S.; Yu, J.S.; Lim, K.; Kim, G.J.; Lee, R.; Kim, S.; Kang, S.; Park, Y.H.; Wang, M.J. Dynamic changes of oligomeric amyloid β levels in plasma induced by spiked synthetic Aβ 42. Alzheimer’s Res. Ther. 2017, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Park, M.C.; Kim, M.; Lim, G.T.; Kang, S.M.; An, S.S.A.; Kim, T.S.; Kang, J.Y. Droplet-based magnetic bead immunoassay using microchannel-connected multiwell plates (μCHAMPs) for the detection of amyloid beta oligomers. Lab On A Chip 2016, 16, 2245–2253. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Kim, M.; Kang, S.M.; Lim, K.T.; Kim, T.S.; Kang, J.Y. Magnetic bead droplet immunoassay of oligomer amyloid β for the diagnosis of Alzheimer′s disease using micro-pillars to enhance the stability of the oil–water interface. Biosens. Bioelectron. 2015, 67, 724–732. [Google Scholar] [CrossRef]
- Properzi, F.; Pocchiari, M. Identification of misfolded proteins in body fluids for the diagnosis of prion diseases. Int. J. Cell Biol. 2013, 2013, 839329. [Google Scholar] [CrossRef]
- Chesebro, B. Introduction to the transmissible spongiform encephalopathies or prion diseases. Br. Med. Bull. 2003, 66, 1–20. [Google Scholar] [CrossRef]
- Bélondrade, M.; Nicot, S.; Mayran, C.; Bruyere-Ostells, L.; Almela, F.; Di Bari, M.A.; Levavasseur, E.; Watts, J.C.; Fournier-Wirth, C.; Lehmann, S.; et al. Sensitive protein misfolding cyclic amplification of sporadic Creutzfeldt–Jakob disease prions is strongly seed and substrate dependent. Sci. Rep. 2021, 11, 4058. [Google Scholar] [CrossRef]
- Vascellari, S.; Orrù, C.D.; Caughey, B. Real-time quaking-induced conversion assays for prion diseases, synucleinopathies, and tauopathies. Front. Aging Neurosci. 2022, 14, 853050. [Google Scholar] [CrossRef]
- Dong, T.T.; Satoh, K. The Latest Research on RT-QuIC Assays-A Literature Review. Pathogens 2021, 10, 305. [Google Scholar] [CrossRef]
- Sbartai, A.; Jaffrezic-Renault, N.; Bougard, D.; Segarra, C.; Fournier-Wirth, C.; Kang, S.; Lim, K.; An, S.S.A. Magnetic microparticle-based multimer detection system for the electrochemical detection of prion oligomers in sheep using a recyclable BDD electrode. Microchem. J. 2021, 164, 106089. [Google Scholar] [CrossRef]
- Cline, E.N.; Bicca, M.A.; Viola, K.L.; Klein, W.L. The amyloid-β oligomer hypothesis: Beginning of the third decade. J. Alzheimer’s Dis. 2018, 64, S567–S610. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Jamerlan, A.; An, S.S.A.; Hulme, J. Advances in amyloid beta oligomer detection applications in Alzheimer’s disease. TrAC Trends Anal. Chem. 2020, 129, 115919. [Google Scholar] [CrossRef]
- Lee, H.; Ugay, D.; Hong, S.; Kim, Y. Alzheimer’s Disease Diagnosis Using Misfolding Proteins in Blood. Dement. Neurocogn Disord. 2020, 19, 1–18. [Google Scholar] [CrossRef]
- Youn, Y.C.; Lee, B.S.; Kim, G.J.; Ryu, J.S.; Lim, K.; Lee, R.; Suh, J.; Park, Y.H.; Pyun, J.M.; Ryu, N.; et al. Blood Amyloid-β Oligomerization as a Biomarker of Alzheimer’s Disease: A Blinded Validation Study. J. Alzheimer’s Dis. 2020, 75, 493–499. [Google Scholar] [CrossRef]
- Dominguez, J.C.; Ampil, E.; Anlacan, M.; Reyes, A.; Sañosa, M.; Valdez, M.; Ong, E.; Laxamana, L.; Cenina, A.; Dumlao, L. Multimer Detection System-Oligomeric Aβ (MDS-OAβ) for Alzheimer’s Disease: Real-World Experience from the Philippines. Alzheimer’s Dement. 2019, 15, P1025. [Google Scholar] [CrossRef]
- Minn, D.; Kang, S. Plasma oligomeric amyloid-β levels after spiking synthetic amyloid beta according to age. Alzheimer’s Dement. 2023, 19, e075556. [Google Scholar] [CrossRef]
- Wang, M.J.; Yi, S.; Han, J.-Y.; Park, S.Y.; Jang, J.-W.; Chun, I.K.; Kim, S.E.; Lee, B.S.; Kim, G.J.; Yu, J.S. Oligomeric forms of amyloid-β protein in plasma as a potential blood-based biomarker for Alzheimer’s disease. Alzheimer’s Res. Ther. 2017, 9, 98. [Google Scholar] [CrossRef]
- Youn, Y.C.; Kang, S.; Suh, J.; Park, Y.H.; Kang, M.J.; Pyun, J.M.; Choi, S.H.; Jeong, J.H.; Park, K.W.; Lee, H.W.; et al. Blood amyloid-β oligomerization associated with neurodegeneration of Alzheimer’s disease. Alzheimer’s Res. Ther. 2019, 11, 40. [Google Scholar] [CrossRef]
- Fagan, A.M.; Xiong, C.; Jasielec, M.S.; Bateman, R.J.; Goate, A.M.; Benzinger, T.L.; Ghetti, B.; Martins, R.N.; Masters, C.L.; Mayeux, R. Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer’s disease. Sci. Transl. Med. 2014, 6, 226ra30. [Google Scholar] [CrossRef]
- Jack, C.R.; Knopman, D.S.; Jagust, W.J.; Petersen, R.C.; Weiner, M.W.; Aisen, P.S.; Shaw, L.M.; Vemuri, P.; Wiste, H.J.; Weigand, S.D. Tracking pathophysiological processes in Alzheimer’s disease: An updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013, 12, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y. Follow-up Comparisons of Two Plasma Biomarkers of Alzheimer’s Disease, Neurofilament Light Chain, and Oligomeric Aβ: A Pilot Study. Curr. Alzheimer Res. 2023, 20, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Mofrad, R.B.; Scheltens, P.; Kim, S.; Kang, S.; Youn, Y.C.; An, S.S.A.; Tomassen, J.; van Berckel, B.N.M.; Visser, P.J.; van der Flier, W.M.; et al. Plasma amyloid-β oligomerization assay as a pre-screening test for amyloid status. Alzheimers Res. Ther. 2021, 13, 133. [Google Scholar] [CrossRef] [PubMed]
- Pyun, J.-M.; Youn, Y.C.; Park, Y.H.; Kim, S. Integration of amyloid-β oligomerization tendency as a plasma biomarker in Alzheimer’s disease diagnosis. Front. Neurol. 2023, 13, 1028448. [Google Scholar] [CrossRef] [PubMed]
- An SS, L.K.; Oh, H.J. Differential Detection of Multimeric and Monomeric Forms of Multimer-Forming Polypeptides. U.S. Patent No. 8,026,070, 27 September 2011. [Google Scholar]
- Bagyinszky, E.; Kim, M.; Park, Y.H.; An, S.S.A.; Kim, S. PSEN1 His214Asn Mutation in a Korean Patient with Familial EOAD and the Importance of Histidine-Tryptophan Interactions in TM-4 Stability. Int. J. Mol. Sci. 2023, 25, 116. [Google Scholar] [CrossRef]
- Yang, Y.; Bagyinszky, E.; An, S.S.A. Patient with PSEN1 Glu318Gly and Other Possible Disease Risk Mutations, Diagnosed with Early Onset Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 15461. [Google Scholar] [CrossRef]
- Bagyinszky, E.; Kang, M.J.; Van Giau, V.; Shim, K.; Pyun, J.M.; Suh, J.; An, S.S.A.; Kim, S. Novel amyloid precursor protein mutation, Val669Leu (“Seoul APP”), in a Korean patient with early-onset Alzheimer’s disease. Neurobiol. Aging 2019, 84, 236.e1–236.e7. [Google Scholar] [CrossRef]
- Dominguez, J.C.; Yu, J.R.T.; De Guzman, M.F.; Ampil, E.; Guevarra, A.C.; Joson, M.L.; Reandelar, M., Jr.; Martinez, M.S.; Ligsay, A.; Ocampo, F.; et al. Multimer Detection System-Oligomerized Amyloid Beta (MDS-OAβ): A Plasma-Based Biomarker Differentiates Alzheimer’s Disease from Other Etiologies of Dementia. Int. J. Alzheimers Dis. 2022, 2022, 9960832. [Google Scholar] [CrossRef]
- Wang, S.-M.; Kang, D.W.; Um, Y.H.; Kim, S.; Lee, C.U.; Scheltens, P.; Lim, H.K. Plasma oligomer beta-amyloid is associated with disease severity and cerebral amyloid deposition in Alzheimer’s disease spectrum. Alzheimer’s Res. Ther. 2024, 16, 55. [Google Scholar] [CrossRef]
- Youn, Y.C.; Kim, H.R.; Shin, H.-W.; Jeong, H.-B.; Han, S.-W.; Pyun, J.-M.; Ryoo, N.; Park, Y.H.; Kim, S. Prediction of amyloid PET positivity via machine learning algorithms trained with EDTA-based blood amyloid-β oligomerization data. BMC Med. Inform. Decis. Mak. 2022, 22, 286. [Google Scholar] [CrossRef]
- Medeiros, R.; Baglietto-Vargas, D.; LaFerla, F.M. The role of tau in Alzheimer’s disease and related disorders. CNS Neurosci. Ther. 2011, 17, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Gyparaki, M.T.; Arab, A.; Sorokina, E.M.; Santiago-Ruiz, A.N.; Bohrer, C.H.; Xiao, J.; Lakadamyali, M. Tau forms oligomeric complexes on microtubules that are distinct from pathological oligomers in disease. Biophys. J. 2020, 120, 31a. [Google Scholar]
- Gyparaki, M.T.; Arab, A.; Sorokina, E.M.; Santiago-Ruiz, A.N.; Bohrer, C.H.; Xiao, J.; Lakadamyali, M. Tau forms oligomeric complexes on microtubules that are distinct from tau aggregates. Proc. Natl. Acad. Sci. USA 2021, 118, e2021461118. [Google Scholar] [CrossRef] [PubMed]
- Feinstein, H.E.; Benbow, S.J.; Lapointe, N.E.; Patel, N.; Ramachandran, S.; Do, T.D.; Gaylord, M.; Huskey, N.E.; Dressler, N.K.; Korff, M.; et al. Oligomerization of the microtubule-associated protein tau is mediated by its N-terminal sequences: Implications for normal and pathological tau action. J. Neurochem. 2016, 137, 939–954. [Google Scholar] [CrossRef]
- Weickert, S.; Wawrzyniuk, M.; John, L.H.; Rüdiger, S.G.D.; Drescher, M. The mechanism of Hsp90-induced oligomerizaton of Tau. Sci. Adv. 2020, 6, eaax6999. [Google Scholar] [CrossRef]
- Wegmann, S.; Nicholls, S.B.; Takeda, S.; Fan, Z.; Hyman, B.T. Formation, release, and internalization of stable tau oligomers in cells. J. Neurochem. 2016, 139, 1163–1174. [Google Scholar] [CrossRef]
- Kolarova, M.; García-Sierra, F.; Bartos, A.; Ricny, J.; Ripova, D. Structure and pathology of tau protein in Alzheimer disease. Int. J. Alzheimer’s Dis. 2012, 2012, 731526. [Google Scholar] [CrossRef]
- Margittai, M.; Langen, R. Template-assisted filament growth by parallel stacking of tau. Proc. Natl. Acad. Sci. USA 2004, 101, 10278–10283. [Google Scholar] [CrossRef]
- Ferrari, L.; Stucchi, R.; Konstantoulea, A.; Kamp, G.V.D.; Kos, R.; Geerts, W.J.C.; Förster, F.G.; Altelaar, M.; Hoogenraad, C.C.; Rüdiger, S.G.D. Fibril formation rewires interactome of the Alzheimer protein Tau by π-stacking. bioRxiv 2019. [Google Scholar] [CrossRef]
- Mandelkow, E.; von Bergen, M.; Biernat, J.; Mandelkow, E.-M. Structural Principles of Tau and the Paired Helical Filaments of Alzheimer’s Disease. Brain Pathol. 2007, 17, 83–90. [Google Scholar] [CrossRef]
- Falcon, B.; Zhang, W.; Murzin, A.G.; Murshudov, G.N.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Ghetti, B.F.; Scheres, S.H.W.; Goedert, M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 2018, 561, 137–140. [Google Scholar] [CrossRef]
- Ávila, J.; Jiménez, J.S.; Sayas, C.L.; Bolós, M.; Zabala, J.C.; Rivas, G.; Hernández, F. Tau Structures. Front. Aging Neurosci. 2016, 8, 262. [Google Scholar] [CrossRef]
- Oakley, S.; Maina, M.B.; Marshall, K.E.; Al-Hilaly, Y.K.; Harrington, C.R.; Wischik, C.M.; Serpell, L.C. Tau Filament Self-Assembly and Structure: Tau as a Therapeutic Target. Front. Neurol. 2020, 11, 590754. [Google Scholar] [CrossRef]
- Campese, N.; Palermo, G.; Del Gamba, C.; Beatino, M.F.; Galgani, A.; Belli, E.; Del Prete, E.; Della Vecchia, A.; Vergallo, A.; Siciliano, G.; et al. Progress regarding the context-of-use of tau as biomarker of Alzheimer’s disease and other neurodegenerative diseases. Expert Rev. Proteom. 2021, 18, 27–48. [Google Scholar] [CrossRef]
- An, S.S.A.; Hulme, J. Plasma amyloid-beta oligomer and phosphorylated tau: Diagnostic tools for progressive Alzheimer’s disease. Neural Regen. Res. 2023, 18, 2391–2392. [Google Scholar] [CrossRef]
- Holper, S.; Watson, R.; Yassi, N. Tau as a Biomarker of Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 7307. [Google Scholar] [CrossRef]
- Hill, E.; Wall, M.J.; Moffat, K.G.; Karikari, T.K. Understanding the Pathophysiological Actions of Tau Oligomers: A Critical Review of Current Electrophysiological Approaches. Front. Mol. Neurosci. 2020, 13, 155. [Google Scholar] [CrossRef]
- Blömeke, L.; Pils, M.; Kraemer-Schulien, V.; Dybala, A.; Schaffrath, A.; Kulawik, A.; Rehn, F.; Cousin, A.; Nischwitz, V.; Willbold, J.; et al. Quantitative detection of α-Synuclein and Tau oligomers and other aggregates by digital single particle counting. NPJ Park. Dis. 2022, 8, 68. [Google Scholar] [CrossRef]
- Kurz, A.; Kurz, C.; Ellis, K.A.; Lautenschlager, N.T. What is frontotemporal dementia? Maturitas 2014, 79, 216–219. [Google Scholar] [CrossRef]
- Dacka, M.; Porzak, M.; Bochyński, K.; Białogłowski, K.; Dąbrowska, P.; Żuber, M.; Ciuba, K.; Molenda, K.; Borodziuk, F.; Borodziuk, B. Frontotemporal Dementia: A Clinical Review. J. Educ. Health Sport 2024, 59, 235–246. [Google Scholar] [CrossRef]
- Mohandas, E.; Rajmohan, V. Frontotemporal dementia: An updated overview. Indian. J. Psychiatry 2009, 51, S65–S69. [Google Scholar]
- Cardarelli, R.; Kertesz, A.; Knebl, J.A. Frontotemporal dementia: A review for primary care physicians. Am. Fam. Physician 2010, 82, 1372–1377. [Google Scholar]
- Swift, I.J.; Sogorb-Esteve, A.; Heller, C.; Synofzik, M.; Otto, M.; Graff, C.; Galimberti, D.; Todd, E.G.; Heslegrave, A.J.; van der Ende, E.L.; et al. Fluid biomarkers in frontotemporal dementia: Past, present and future. J. Neurol. Neurosurg. Psychiatry 2020, 92, 204–215. [Google Scholar] [CrossRef]
- van der Ende, E.L.; van Swieten, J.C. Fluid Biomarkers of Frontotemporal Lobar Degeneration. Adv. Exp. Med. Biol. 2021, 1281, 123–139. [Google Scholar]
- Ooi, S.; Patel, S.K.; Eratne, D.; Kyndt, C.; Reidy, N.; Lewis, C.; Lee, S.; Darby, D.; Brodtmann, A. Plasma Neurofilament Light Chain and Clinical Diagnosis in Frontotemporal Dementia Syndromes. J. Alzheimer’s Dis. 2022, 89, 1221–1231. [Google Scholar] [CrossRef]
- Landqvist Waldö, M.; Frizell Santillo, A.; Passant, U.; Zetterberg, H.; Rosengren, L.E.; Nilsson, C.; Englund, E. Cerebrospinal fluid neurofilament light chain protein levels in subtypes of frontotemporal dementia. BMC Neurol. 2013, 13, 54. [Google Scholar] [CrossRef]
- Verde, F.; Otto, M.; Silani, V. Neurofilament Light Chain as Biomarker for Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Front. Neurosci. 2021, 15, 679199. [Google Scholar] [CrossRef]
- Rohrer, J.D.; Geser, F.; Zhou, J.; Gennatas, E.D.; Sidhu, M.; Trojanowski, J.Q.; DeArmond, S.J.; Miller, B.L.; Seeley, W.W. TDP-43 subtypes are associated with distinct atrophy patterns in frontotemporal dementia. Neurology 2010, 75, 2204–2211. [Google Scholar] [CrossRef]
- Davidson, Y.S.; Robinson, A.C.; Flood, L.; Rollinson, S.; Benson, B.C.; Asi, Y.T.; Richardson, A.M.T.; Jones, M.S.; Snowden, J.S.; Pickering-Brown, S.; et al. Heterogeneous ribonuclear protein E2 (hnRNP E2) is associated with TDP-43-immunoreactive neurites in Semantic Dementia but not with other TDP-43 pathological subtypes of Frontotemporal Lobar Degeneration. Acta Neuropathol. Commun. 2017, 5, 54. [Google Scholar] [CrossRef]
- Jamerlan, A.M.; Shim, K.H.; Youn, Y.C.; Teunissen, C.; An, S.S.A.; Scheltens, P.; Kim, S. Increased oligomeric TDP-43 in the plasma of Korean frontotemporal dementia patients with semantic dementia. Alzheimer’s Dement. 2023, 19, 4020–4027. [Google Scholar] [CrossRef]
- Leyton, C.E.; Hodges, J.R. Frontotemporal dementias: Recent advances and current controversies. Ann. Indian Acad. Neurol. 2010, 13 (Suppl. S2), S74–S80. [Google Scholar]
- Corriveau-Lecavalier, N.; Botha, H.; Graff-Radford, J.; Switzer, A.R.; Przybelski, S.A.; Wiste, H.J.; Murray, M.E.; Reichard, R.R.; Dickson, D.W.; Nguyen, A.T.; et al. Clinical criteria for a limbic-predominant amnestic neurodegenerative syndrome. Brain Commun. 2024, 6, fcae183. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Overk, C.R.; Oueslati, A.; Masliah, E. The many faces of α-synuclein: From structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013, 14, 38–48. [Google Scholar] [CrossRef]
- Bengoa-Vergniory, N.; Roberts, R.F.; Wade-Martins, R.; Alegre-Abarrategui, J. Alpha-synuclein oligomers: A new hope. Acta Neuropathol. 2017, 134, 819–838. [Google Scholar] [CrossRef]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.L.; Guerrero-Muñoz, M.J.; Jackson, G.R.; Kayed, R. Preparation and Characterization of Neurotoxic Tau Oligomers. Biochemistry 2010, 49, 10039–10041. [Google Scholar] [CrossRef]
- Sengupta, U.; Guerrero-Munoz, M.J.; Castillo-Carranza, D.L.; Lasagna-Reeves, C.A.; Gerson, J.E.; Paulucci-Holthauzen, A.A.; Krishnamurthy, S.; Farhed, M.; Jackson, G.R.; Kayed, R. Pathological Interface Between Oligomeric Alpha-Synuclein and Tau in Synucleinopathies. Biol. Psychiatry 2015, 78, 672–683. [Google Scholar] [CrossRef]
- Giasson, B.I.; Forman, M.S.; Higuchi, M.; Golbe, L.I.; Graves, C.L.; Kotzbauer, P.T.; Trojanowski, J.Q.; Lee, V.M.-Y. Initiation and Synergistic Fibrillization of Tau and Alpha-Synuclein. Science 2003, 300, 636–640. [Google Scholar] [CrossRef]
- Lemarié, F.L.; Sanders, S.S.; Nguyen, Y.; Martin, D.D.O.; Hayden, M.R. Full-length huntingtin is palmitoylated at multiple sites and post-translationally myristoylated following caspase-cleavage. Front. Physiol. 2023, 14, 1086112. [Google Scholar] [CrossRef]
- Yang, J.; Yang, X. Phase Transition of Huntingtin: Factors and Pathological Relevance. Front. Genet. 2020, 11, 754. [Google Scholar] [CrossRef]
- Babinchak, W.M.; Surewicz, W.K. Liquid-Liquid Phase Separation and its Mechanistic Role in Pathological Protein Aggregation. J. Mol. Biol. 2020, 432, 1910–1925. [Google Scholar] [CrossRef]
- Landles, C.; Milton, R.E.; Ali, N.; Flomen, R.; Flower, M.; Schindler, F.; Gomez-Paredes, C.; Bondulich, M.K.; Osborne, G.F.; Goodwin, D.; et al. Subcellular Localization and Formation Of Huntingtin Aggregates Correlates With Symptom Onset And Progression In A Huntington’s Disease Model. Brain Commun. 2020, 2, fcaa066. [Google Scholar] [CrossRef]
- Barnat, M.; Capizzi, M.; Aparicio, E.; Boluda, S.; Wennagel, D.; Kacher, R.; Kassem, R.; Lenoir, S.; Agasse, F.; Braz, B.Y.; et al. Huntington’s disease alters human neurodevelopment. Science 2020, 369, 787–793. [Google Scholar] [CrossRef]
- Jarosińska, O.D.; Rüdiger, S.G.D. Molecular Strategies to Target Protein Aggregation in Huntington’s Disease. Front. Mol. Biosci. 2021, 8, 769184. [Google Scholar] [CrossRef]
- Barron, J.C.; Hurley, E.P.; Parsons, M.P. Huntingtin and the Synapse. Front. Cell. Neurosci. 2021, 15, 689332. [Google Scholar] [CrossRef]
- Huelsmeier, J.; Walker, E.; Bakthavachalu, B.; Ramaswami, M. Ataxin-2 Disordered Region Promotes Huntingtin Protein Aggregation And Neurodegeneration In Drosophila Models Of Huntington’s Disease. bioRxiv 2021. [Google Scholar] [CrossRef]
- Fox, J.H.; Connor, T.; Stiles, M.; Kama, J.A.; Lu, Z.; Dorsey, K.; Liebermann, G.; Sapp, E.; Cherny, R.A.; Banks, M.; et al. Cysteine Oxidation within N-terminal Mutant Huntingtin Promotes Oligomerization and Delays Clearance of Soluble Protein. J. Biol. Chem. 2011, 286, 18320–18330. [Google Scholar] [CrossRef]
- Kotler, S.A.; Tugarinov, V.; Schmidt, T.; Ceccon, A.; Libich, D.S.; Ghirlando, R.; Schwieters, C.D.; Clore, G.M. Probing initial transient oligomerization events facilitating Huntingtin fibril nucleation at atomic resolution by relaxation-based NMR. Proc. Natl. Acad. Sci. USA 2019, 116, 3562–3571. [Google Scholar] [CrossRef]
- Legleiter, J. Structural flexibility of huntingtin oligomers plays a key role in directly binding lipid membranes. Biophys. J. 2023, 122, 351a–352a. [Google Scholar] [CrossRef]
- Kim, M.W.; Chelliah, Y.; Kim, S.W.; Otwinowski, Z.; Bezprozvanny, I. Secondary Structure of Huntingtin Amino-Terminal Region. Structure 2009, 17, 1205–1212. [Google Scholar] [CrossRef]
- Sedighi, F.; Skeens, A.; Adegbuyiro, A.; Bard, J.; Siriwardhana, C.; Donley, E.; Geldenhuys, W.J.; Legleiter, J. Oligomerization enhances huntingtin membrane activity but is suppressed by covalent crosslinking. bioRxiv 2023. [Google Scholar] [CrossRef]
- Legleiter, J.; Mitchell, E.J.; Lotz, G.P.; Sapp, E.; Ng, C.; Difiglia, M.; Thompson, L.M.; Muchowski, P.J. Mutant Huntingtin Fragments Form Oligomers in a Polyglutamine Length-dependent Manner in Vitro and in Vivo. J. Biol. Chem. 2010, 285, 14777–14790. [Google Scholar] [CrossRef]
- Bonfanti, S.; Lionetti, M.C.; Fumagalli, M.R.; Chirasani, V.R.; Tiana, G.; Dokholyan, N.V.; Zapperi, S.; La Porta, C.A.M. Molecular mechanisms of heterogeneous oligomerization of huntingtin proteins. Sci. Rep. 2019, 9, 7615. [Google Scholar] [CrossRef] [PubMed]
- Herrera, F.; Tenreiro, S.; Miller-Fleming, L.; Outeiro, T.F. Visualization of cell-to-cell transmission of mutant huntingtin oligomers. PLoS Curr. 2011, 3, RRN1210. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, M.; Kodali, R.B.; Sahoo, B.; Thakur, A.K.; Mayasundari, A.; Mishra, R.K.; Peterson, C.B.; Wetzel, R. Slow amyloid nucleation via α-helix-rich oligomeric intermediates in short polyglutamine-containing huntingtin fragments. J. Mol. Biol. 2012, 415, 881–899. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, B.; Singer, D.; Kodali, R.B.; Zuchner, T.; Wetzel, R. Aggregation Behavior of Chemically Synthesized, Full-Length Huntingtin Exon1. Biochemistry 2014, 53, 3897–3907. [Google Scholar] [CrossRef]
- Tsigelny, I.F.; Crews, L.A.; Desplats, P.A.; Shaked, G.M.; Sharikov, Y.; Mizuno, H.; Spencer, B.; Rockenstein, E.; Trejo, M.; Platoshyn, O.; et al. Mechanisms of Hybrid Oligomer Formation in the Pathogenesis of Combined Alzheimer’s and Parkinson’s Diseases. PLoS ONE 2008, 3, e3135. [Google Scholar] [CrossRef]
- Guerrero-Munoz, M.J.; Castillo-Carranza, D.L.; Krishnamurthy, S.; Paulucci-Holthauzen, A.; Sengupta, U.; Lasagna-Reeves, C.A.; Ahmad, Y.; Jackson, G.R.; Kayed, R. Amyloid-β oligomers as a template for secondary amyloidosis in Alzheimer’s disease. Neurobiol. Dis. 2014, 71, 14–23. [Google Scholar] [CrossRef]
- Kayed, R.; Dettmer, U.; Lesné, S.E. Soluble endogenous oligomeric α-synuclein species in neurodegenerative diseases: Expression, spreading, and cross-talk. J. Park. Dis. 2020, 10, 791–818. [Google Scholar] [CrossRef]
- Arseni, D.; Nonaka, T.; Jacobsen, M.H.; Murzin, A.G.; Cracco, L.; Peak-Chew, S.Y.; Garringer, H.J.; Kawakami, I.; Suzuki, H.; Onaya, M.; et al. Heteromeric amyloid filaments of ANXA11 and TDP-43 in FTLD-TDP Type C. Nature 2024, 634, 662–668. [Google Scholar] [CrossRef]
- Robinson, J.L.; Suh, E.; Xu, Y.; Hurtig, H.I.; Elman, L.; McMillan, C.; Irwin, D.J.; Porta, S.; Van Deerlin, V.M.; Lee, E.B. Annexin A11 aggregation in FTLD–TDP type C and related neurodegenerative disease proteinopathies. Acta Neuropathol. 2024, 147, 104. [Google Scholar] [CrossRef]
- Freibaum, B.D.; Chitta, R.; High, A.A.; Taylor, P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010, 9, 1104–1120. [Google Scholar] [CrossRef]
- Smith, B.N.; Topp, S.D.; Fallini, C.; Shibata, H.; Chen, H.-J.; Troakes, C.; King, A.; Ticozzi, N.; Kenna, K.P.; Soragia-Gkazi, A.; et al. Mutations in the vesicular trafficking protein annexin A11 are associated with amyotrophic lateral sclerosis. Sci. Transl. Med. 2017, 9, eaaad9157. [Google Scholar] [CrossRef] [PubMed]
- Baloh, R.H. How do the RNA-binding proteins TDP-43 and FUS relate to amyotrophic lateral sclerosis and frontotemporal degeneration, and to each other? Curr. Opin. Neurol. 2012, 25, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S.; Caccamo, A.; Tran, L.; Lambert, M.P.; Glabe, C.G.; Klein, W.L.; LaFerla, F.M. Temporal Profile of Amyloid-β (Aβ) Oligomerization in an in Vivo Model of Alzheimer Disease. J. Biol. Chem. 2006, 281, 1599–1604. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Reed, M.N.; Kotilinek, L.A.; Grant, M.K.O.; Forster, C.L.; Qiang, W.; Shapiro, S.L.; Reichl, J.H.; Chiang, A.C.A.; Jankowsky, J.L.; et al. Quaternary Structure Defines a Large Class of Amyloid-β Oligomers Neutralized by Sequestration. Cell Rep. 2015, 11, 1760–1771. [Google Scholar] [CrossRef] [PubMed]
- Bonnycastle, L.L.C.; Mehroke, J.S.; Rashed, M.; Gong, X.; Scott, J.K. Probing the basis of antibody reactivity with a panel of constrained peptide libraries displayed by filamentous phage. J. Mol. Biol. 1996, 258, 747–762. [Google Scholar] [CrossRef]
- Anders, R.F. Multiple cross-reactivities amongst antigens of Plasmodium falciparum impair the development of protective immunity against malaria. Parasite Immunol. 1986, 8, 529–539. [Google Scholar] [CrossRef]
- Bartels, T.; Choi, J.G.; Selkoe, D.J. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 2011, 477, 107–110. [Google Scholar] [CrossRef]
- Gurry, T.; Ullman, O.; Fisher, C.K.; Perovic, I.; Pochapsky, T.C.; Stultz, C.M. The dynamic structure of α-synuclein multimers. J. Am. Chem. Soc. 2013, 135, 3865–3872. [Google Scholar] [CrossRef]
- Jeong, H.R.; An, S.S. Causative factors for formation of toxic islet amyloid polypeptide oligomer in type 2 diabetes mellitus. Clin. Interv. Aging 2015, 10, 1873–1879. [Google Scholar]
- Kim, S.; An, S.S.A. Role of p53 isoforms and aggregations in cancer. Medicine 2016, 95, e3993. [Google Scholar] [CrossRef]
- Park, G.Y.; Jamerlan, A.; Shim, K.H.; An, S.S.A. Diagnostic and Treatment Approaches Involving Transthyretin in Amyloidogenic Diseases. Int. J. Mol. Sci. 2019, 20, 2982. [Google Scholar] [CrossRef]
- Estaun-Panzano, J.; Arotcarena, M.L.; Bezard, E. Monitoring α-synuclein aggregation. Neurobiol. Dis. 2023, 176, 105966. [Google Scholar] [CrossRef]
| MDS-OAβ | Amyloid PET | MMSE Score | Most Probable Diagnosis |
|---|---|---|---|
| Negative | Abnormal | Normal | Other NDs |
| Normal | Normal | Normal | |
| AD-compatible atrophy | Cognitive impairment | Other NDs | |
| Normal | Cognitive impairment | Other reasons for cognitive impairment | |
| Positive | AD-compatible atrophy | Normal | Preclinical AD |
| Normal | Normal | Preclinical Amyloidopathies | |
| AD-compatible atrophy | Cognitive impairment | AD | |
| Normal | Cognitive impairment | An early stage of AD |
| Protein | Associated Disease | Sample Source Used in MDS | Disease Differentiation Potential | Key Publications |
|---|---|---|---|---|
| Prion | CJD, Scrapie | Scrapie-infected hamster plasma | Detection of prion protein multimers in plasma of scrapie-infected hamsters | An, 2010 [40] |
| Scrapie-infected sheep plasma | MDS detected PrP Sc in plasma samples from scrapie-infected sheep with clinical samples with 100% accuracy. | Lim, 2015 [42] | ||
| A-beta | AD, LATE-NC, DLB | AD plasma | Differentiated AD patients from healthy controls with 78.3% sensitivity and 86.5% specificity | Lee, 2020 [55] |
| AD plasma (heparin-treated) | Differentiated AD patients from normal controls with 100% sensitivity and 92.31% specificity using a cutoff of 0.78 ng/mL | Youn, 2020 [56] | ||
| Abnormal amyloid status in plasma | Identification of individuals with abnormal amyloid status with an AUC of 0.74, which improved to 0.81 when combined with APOE4 and age | Mofrad, 2021 [64] | ||
| AD plasma | Differentiated dementias due to AD versus non-AD etiologies | Dominguez, 2022 [70] | ||
| TDP-43 | AD, LATE, DLB, FTD | FTD plasma (semantic dementia) | The ratio of o-TDP-43 to t-TDP-43 concentrations in semantic dementia | Jamerlan, 2023 [102] |
| A-syn | LATE-NC, PD, DLB | No data yet | ||
| Tau | CJD, AD, LATE-NC, DLB, PNFA, bvFTD | No data yet | ||
| Huntingtin | HD | No data yet | ||
| Hybrid | PNFA, bvFTD | No data yet |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamerlan, A.M.; Shim, K.H.; Sharma, N.; An, S.S.A. Multimer Detection System: A Universal Assay System for Differentiating Protein Oligomers from Monomers. Int. J. Mol. Sci. 2025, 26, 1199. https://doi.org/10.3390/ijms26031199
Jamerlan AM, Shim KH, Sharma N, An SSA. Multimer Detection System: A Universal Assay System for Differentiating Protein Oligomers from Monomers. International Journal of Molecular Sciences. 2025; 26(3):1199. https://doi.org/10.3390/ijms26031199
Chicago/Turabian StyleJamerlan, Angelo Moscoso, Kyu Hwan Shim, Niti Sharma, and Seong Soo A. An. 2025. "Multimer Detection System: A Universal Assay System for Differentiating Protein Oligomers from Monomers" International Journal of Molecular Sciences 26, no. 3: 1199. https://doi.org/10.3390/ijms26031199
APA StyleJamerlan, A. M., Shim, K. H., Sharma, N., & An, S. S. A. (2025). Multimer Detection System: A Universal Assay System for Differentiating Protein Oligomers from Monomers. International Journal of Molecular Sciences, 26(3), 1199. https://doi.org/10.3390/ijms26031199







