Rapid Simultaneous Detection of the Clinically Relevant Carbapenemase Resistance Genes blaKPC, blaOXA48, blaVIM and blaNDM with the Newly Developed Ready-to-Use qPCR CarbaScan LyoBead
Abstract
:1. Introduction
2. Results
2.1. Evaluation of qPCR Master Mixes
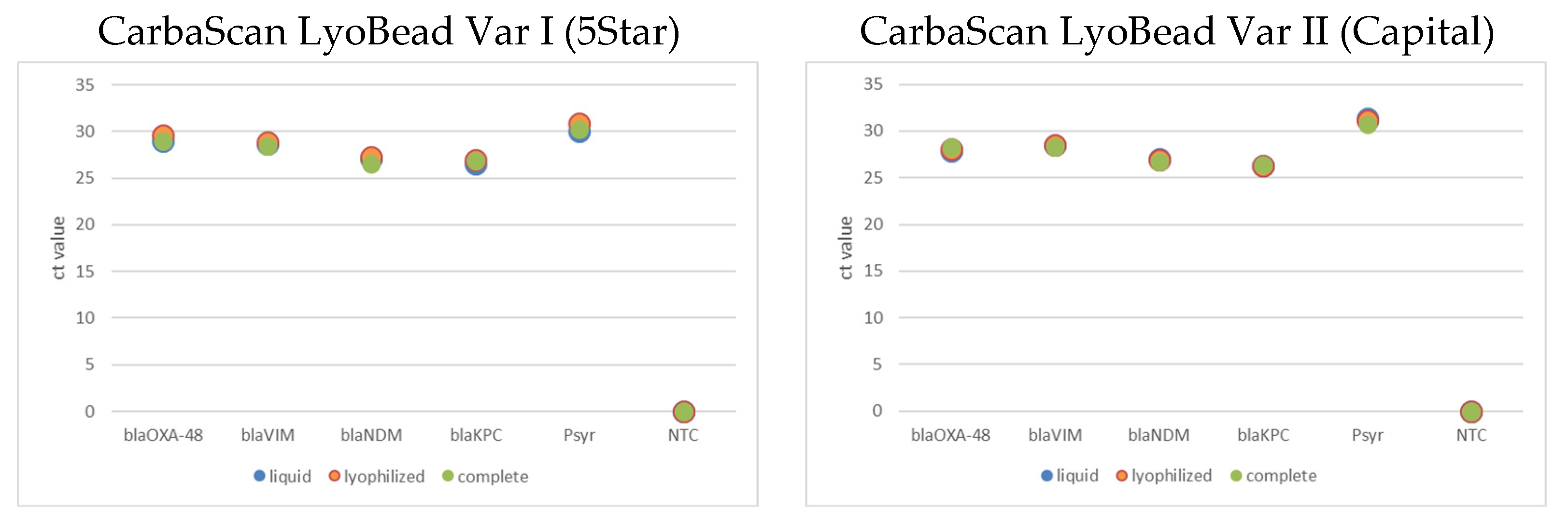
2.2. Lyophilized qPCR and Complete qPCR LyoBead CarbaScan Var I and II
3. Discussion
4. Materials and Methods
4.1. Bioinformatics of Oligonucleotides
4.2. Bacterial Strains and Growth Conditions
4.3. Nucleic Acid Preparation and Nanopore Sequencing
4.4. Genomic DNA Dilution
4.5. qPCR Assays for LyoBead Development and Evaluation
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| blaVIM | |||||
|---|---|---|---|---|---|
| Species | Strain ID | Allelic Variant | Culture Age on Plate | Var I | Var II |
| Pseudomonas aeruginosa | 280207 | blaVIM-2 | 5 days | not detected | not detected |
| 1 day | 15.45 | 18.79 | |||
| Pseudomonas aeruginosa | 280228 | blaVIM-6 | 5 days | not detected | not detected |
| 1 day | 14.48 | 16.39 | |||
| Liquid | Lyophilized | Lyophilized Complete (with Primer and Probes) | |||
|---|---|---|---|---|---|
| Add to mix | LyoBead | LyoBead | LyoBead | ||
| Mastermix 2x (HotStart, YourTaq, 5Star) | Mastermix 4x (Capital) | Resuspension buffer 2x (HotStart, YourTaq, 5Star) | Resuspension buffer 4x (Capital) | Resuspension buffer 4x (Capital, 5Star) | |
| Primer | Primer | ||||
| Probes | Probes | ||||
| template | template | template | |||
| water | water | water | |||
| Marker | Liquid vs. Lyophilized p-Value | Liquid vs. Complete p-Value | Lyophilized vs. Complete p-Value | |
|---|---|---|---|---|
| CarbaScan LyoBead Var I | blaOXA-48 | 0.1162 | 0.0041 | 0.2308 |
| blaVIM | 0.1750 | 0.0060 | 0.0201 | |
| blaNDM | 0.3553 | 0.0149 | 0.0011 | |
| blaKPC | 0.7620 | 0.0105 | 0.1029 | |
| Psyr | 0.0466 | 0.0088 | 0.0011 | |
| CarbaScan LyoBead Var II | blaOXA-48 | 0.1163 | 0.0041 | 0.2309 |
| blaVIM | 0.1750 | 0.0060 | 0.0202 | |
| blaNDM | 0.3554 | 0.0149 | 0.0011 | |
| blaKPC | 0.7620 | 0.0105 | 0.1029 | |
| Psyr | 0.2040 | 0.0691 | 0.1485 |
References
- Kim, M.; Park, J.; Kang, M.; Yang, J.; Park, W. Gain and loss of antibiotic resistant genes in multidrug resistant bacteria: One Health perspective. J. Microbiol. 2021, 59, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Xie, L. Distribution and antimicrobial resistance analysis of gram-negative bacilli isolated from a tertiary hospital in Central China: A 10-year retrospective study from 2012 to 2021. Front. Microbiol. 2023, 14, 1297528. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.L.; Lu, M.C.; Shao, P.L.; Lu, P.L.; Chen, Y.H.; Cheng, S.H.; Ko, W.C.; Lin, C.Y.; Wu, T.S.; Yen, M.Y.; et al. Nationwide surveillance of antimicrobial resistance among clinically important Gram-negative bacteria, with an emphasis on carbapenems and colistin: Results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) in 2018. Int. J. Antimicrob. Agents 2019, 54, 318–328. [Google Scholar] [CrossRef]
- Han, R.; Shi, Q.; Wu, S.; Yin, D.; Peng, M.; Dong, D.; Zheng, Y.; Guo, Y.; Zhang, R.; Hu, F.; et al. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated From Adult and Children Patients in China. Front. Cell. Infect. Microbiol. 2020, 10, 314. [Google Scholar] [CrossRef]
- Hansen, G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021, 10, 75–92. [Google Scholar] [CrossRef]
- Peirano, G.; Chen, L.; Nobrega, D.; Finn, T.; Kreiswirth, B.; DeVinney, R.; Pitout, J.D.D. Genomic Epidemiology of Global Carbapenemase-Producing Escherichia coli, 2015–2017. Emerg. Infect. Dis. J. 2022, 28, 924. [Google Scholar] [CrossRef]
- Caliskan-Aydogan, O.; Alocilja, E.C. A Review of Carbapenem Resistance in Enterobacterales and Its Detection Techniques. Microorganisms 2023, 11, 1491. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Acman, M.; Wang, R.; van Dorp, L.; Shaw, L.P.; Wang, Q.; Luhmann, N.; Yin, Y.; Sun, S.; Chen, H.; Wang, H.; et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat. Commun. 2022, 13, 1131. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Nordmann, P. Functional characterization of Tn 4401, a Tn 3-based transposon involved in bla KPC gene mobilization. Antimicrob. Agents Chemother. 2011, 55, 5370–5373. [Google Scholar] [CrossRef] [PubMed]
- Noel, H.R.; Petrey, J.R.; Palmer, L.D. Mobile genetic elements in Acinetobacter antibiotic-resistance acquisition and dissemination. Ann. N. Y. Acad. Sci. 2022, 1518, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Welch, T.J.; Fricke, W.F.; McDermott, P.F.; White, D.G.; Rosso, M.-L.; Rasko, D.A.; Mammel, M.K.; Eppinger, M.; Rosovitz, M.; Wagner, D. Multiple antimicrobial resistance in plague: An emerging public health risk. PLoS ONE 2007, 2, e309. [Google Scholar] [CrossRef] [PubMed]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Witte, W. Medical consequences of antibiotic use in agriculture. Science 1998, 279, 996–997. [Google Scholar] [CrossRef]
- Chukwudile, B.; Pan, D.; Silva, L.; Gogoi, M.; Al-Oraibi, A.; Bird, P.; George, N.; Thompson, H.A.; Baggaley, R.F.; Hargreaves, S.; et al. Antimicrobial resistance among migrants in Europe: A systematic review and meta-analysis–update from 2017 to 2023. eClinicalMedicine 2024, 75, 102801. [Google Scholar] [CrossRef]
- Ljungquist, O.; Nazarchuk, O.; Kahlmeter, G.; Andrews, V.; Koithan, T.; Wasserstrom, L.; Dmytriiev, D.; Fomina, N.; Bebyk, V.; Matuschek, E.; et al. Highly multidrug-resistant Gram-negative bacterial infections in war victims in Ukraine, 2022. Lancet Infect. Dis. 2023, 23, 784–786. [Google Scholar] [CrossRef]
- Pallett, S.J.C.; Boyd, S.E.; O’Shea, M.K.; Martin, J.; Jenkins, D.R.; Hutley, E.J. The contribution of human conflict to the development of antimicrobial resistance. Commun. Med. 2023, 3, 153. [Google Scholar] [CrossRef]
- Abou Fayad, A.; Rizk, A.; El Sayed, S.; Kaddoura, M.; Jawad, N.K.; Al-Attar, A.; Dewachi, O.; Nguyen, V.K.; Sater, Z.A. Antimicrobial resistance and the Iraq wars: Armed conflict as an underinvestigated pathway with growing significance. BMJ Glob. Health 2023, 7, e010863. [Google Scholar] [CrossRef] [PubMed]
- Geddes, L. The Devastating Impact of War on Antimicrobial Resistance. Available online: https://www.gavi.org/vaccineswork/devastating-impact-war-antimicrobial-resistance (accessed on 19 September 2024).
- Bialvaei, A.Z.; Kafil, H.S.; Asgharzadeh, M.; Yousef Memar, M.; Yousefi, M. Current methods for the identification of carbapenemases. J. Chemother. 2016, 28, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Simner, P.J. Phenotypic Detection of Carbapenemase-Producing Organisms from Clinical Isolates. J. Clin. Microbiol. 2018, 56, e01140-18. [Google Scholar] [CrossRef]
- Yoshioka, N.; Hagiya, H.; Deguchi, M.; Hamaguchi, S.; Kagita, M.; Nishi, I.; Akeda, Y.; Tomono, K. Multiplex Real-Time PCR Assay for Six Major Carbapenemase Genes. Pathogens 2021, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.; Petersen, M.B.; Jørgensen, A.L.; Paulmann, D.; Wang, M. Rapid real-time PCR for the detection of IMP, NDM, VIM, KPC and OXA-48 carbapenemase genes in isolates and spiked stool samples. Diagn. Microbiol. Infect. Dis. 2018, 92, 8–12. [Google Scholar] [CrossRef]
- Weiß, D.; Engelmann, I.; Braun, S.D.; Monecke, S.; Ehricht, R. A multiplex real-time PCR for the direct, fast, economic and simultaneous detection of the carbapenemase genes blaKPC, blaNDM, blaVIM and blaOXA-48. J. Microbiol. Methods 2017, 142, 20–26. [Google Scholar] [CrossRef]
- Tamma, P.D.; Goodman, K.E.; Harris, A.D.; Tekle, T.; Roberts, A.; Taiwo, A.; Simner, P.J. Comparing the Outcomes of Patients With Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin. Infect. Dis. 2017, 64, 257–264. [Google Scholar] [CrossRef]
- Akova, M.; Daikos, G.L.; Tzouvelekis, L.; Carmeli, Y. Interventional strategies and current clinical experience with carbapenemase-producing Gram-negative bacteria. Clin. Microbiol. Infect. 2012, 18, 439–448. [Google Scholar] [CrossRef]
- Mathers, A.J.; Cox, H.L.; Kitchel, B.; Bonatti, H.; Brassinga, A.K.C.; Carroll, J.; Scheld, W.M.; Hazen, K.C.; Sifri, C.D. Molecular dissection of an outbreak of carbapenem-resistant Enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2011, 2, e00204-11. [Google Scholar] [CrossRef]
- Goren, M.G.; Carmeli, Y.; Schwaber, M.J.; Chmelnitsky, I.; Schechner, V.; Navon-Venezia, S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg. Infect. Dis. 2010, 16, 1014. [Google Scholar] [CrossRef]
- Tijet, N.; Boyd, D.; Patel, S.N.; Mulvey, M.R.; Melano, R.G. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4578–4580. [Google Scholar] [CrossRef] [PubMed]
- Amjad, A.; Mirza, I.; Abbasi, S.; Farwa, U.; Malik, N.; Zia, F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran. J. Microbiol. 2011, 3, 189–193. [Google Scholar] [PubMed]
- van der Zwaluw, K.; de Haan, A.; Pluister, G.N.; Bootsma, H.J.; de Neeling, A.J.; Schouls, L.M. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE 2015, 10, e0123690. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, K.; Voets, G.M.; Scharringa, J.; Voskuil, S.; Fluit, A.C.; Rottier, W.C.; Leverstein-Van Hall, M.A.; Cohen Stuart, J.W. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin. Microbiol. Infect. 2014, 20, 345–349. [Google Scholar] [CrossRef]
- Biagi, M.; Wu, T.; Lee, M.; Patel, S.; Butler, D.; Wenzler, E. Searching for the Optimal Treatment for Metallo- and Serine-β-Lactamase Producing Enterobacteriaceae: Aztreonam in Combination with Ceftazidime-avibactam or Meropenem-vaborbactam. Antimicrob. Agents Chemother. 2019, 63, e01426-19. [Google Scholar] [CrossRef]
- Lazar, D.S.; Nica, M.; Dascalu, A.; Oprisan, C.; Albu, O.; Codreanu, D.R.; Kosa, A.G.; Popescu, C.P.; Florescu, S.A. Carbapenem-Resistant NDM and OXA-48-like Producing K. pneumoniae: From Menacing Superbug to a Mundane Bacteria; A Retrospective Study in a Romanian Tertiary Hospital. Antibiotics 2024, 13, 435. [Google Scholar] [CrossRef]
- Probst, K.; Nurjadi, D.; Heeg, K.; Frede, A.M.; Dalpke, A.H.; Boutin, S. Molecular Detection of Carbapenemases in Enterobacterales: A Comparison of Real-Time Multiplex PCR and Whole-Genome Sequencing. Antibiotics 2021, 10, 726. [Google Scholar] [CrossRef]
- Subirats, J.; Royo, E.; Balcázar, J.L.; Borrego, C.M. Real-time PCR assays for the detection and quantification of carbapenemase genes (blaKPC, blaNDM, and blaOXA-48) in environmental samples. Environ. Sci. Pollut. Res. 2017, 24, 6710–6714. [Google Scholar] [CrossRef]
- Peubez, I.; Chaudet, N.; Mignon, C.; Hild, G.; Husson, S.; Courtois, V.; De Luca, K.; Speck, D.; Sodoyer, R. Antibiotic-free selection in E. coli: New considerations for optimal design and improved production. Microb. Cell Factories 2010, 9, 65. [Google Scholar] [CrossRef]
- Hägg, P.; de Pohl, J.W.; Abdulkarim, F.; Isaksson, L.A. A host/plasmid system that is not dependent on antibiotics and antibiotic resistance genes for stable plasmid maintenance in Escherichia coli. J. Biotechnol. 2004, 111, 17–30. [Google Scholar] [CrossRef]
- Fang, L.; Shen, Y.; Chen, R.; Li, C.; Liu, R.; Jia, Y.; Qi, S.; Guo, X. The characterization of an IncN-IncR fusion plasmid co-harboring blaTEM–40, blaKPC–2, and blaIMP–4 derived from ST1393 Klebsiella pneumoniae. Sci. Rep. 2024, 14, 26723. [Google Scholar] [CrossRef] [PubMed]
- Gama, J.A.; Kloos, J.; Johnsen, P.J.; Samuelsen, Ø. Host dependent maintenance of a blaNDM-1-encoding plasmid in clinical Escherichia coli isolates. Sci. Rep. 2020, 10, 9332. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-L.; Ko, W.-C.; Hsueh, P.-R. Geographic patterns of global isolates of carbapenem-resistant Klebsiella pneumoniae and the activity of ceftazidime/avibactam, meropenem/vaborbactam, and comparators against these isolates: Results from the Antimicrobial Testing Leadership and Surveillance (ATLAS) program, 2020. Int. J. Antimicrob. Agents 2022, 60, 106679. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, X.; Dong, N.; Wang, Z.; Li, R. Global distribution and genomic characteristics of carbapenemase-producing Escherichia coli among humans, 2005–2023. Drug Resist. Updates 2024, 72, 101031. [Google Scholar] [CrossRef]
- Braun, S.D.; Völksch, B.; Nüske, J.; Spiteller, D. 3-Methylarginine from Pseudomonas syringae pv. syringae 22d/93 suppresses the bacterial blight caused by its close relative Pseudomonas syringae pv. glycinea. Chembiochem 2008, 9, 1913–1920. [Google Scholar] [CrossRef]
- Collatz, M.; Braun, S.D.; Monecke, S.; Ehricht, R. ConsensusPrime—A Bioinformatic Pipeline for Ideal Consensus Primer Design. BioMedInformatics 2022, 2, 637–642. [Google Scholar] [CrossRef]
- Nanopore, O. Guppy Basecaller. Available online: https://nanoporetech.com/document/Guppy-protocol (accessed on 1 March 2021).
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Polevikov, E. Flye Assembler. Available online: https://github.com/mikolmogorov/Flye?tab=License-1-ov-file (accessed on 1 March 2021).
- Sovic, I. Racon. Available online: https://github.com/isovic/racon?tab=MIT-1-ov-file (accessed on 1 March 2021).
- Nanopore, O. Medaka. Available online: https://github.com/nanoporetech/medaka?tab=License-1-ov-file (accessed on 1 March 2021).
- Seemann, T. Abricate. Available online: https://github.com/tseemann/abricate (accessed on 1 March 2021).



| Version | TP 1 | FP 2 | TN 3 | FN 4 | Specificity (c. i) 5 | Sensitivity (c. i.) 5 | Accuracy (c. i.) 5 |
|---|---|---|---|---|---|---|---|
| qPCR CarbaScan LyoBead Var I | 32 | 1 | 86 | 1 | 98.9% (0.938–1.000) | 93.9% (0.798–0.993) | 97.5% (0.929–0.995) |
| qPCR CarbaScan LyoBead Var II | 33 | 0 | 87 | 0 | 100.0% (0.958–1.000) | 100.0% (0.894–1.000) | 100.0% (0.970–1.000) |
| #. | Oligo Name | Oligo Sequence | 5′ Label | 3′ Quencher | Tm 1 | Accession Number |
|---|---|---|---|---|---|---|
| 1 | blaKPC_fwd | CTGTATCGCCGTCTAGTTCTG | 61.9 | EU447304.1 [15:896] | ||
| 2 | blaKPC_rev | AGTTTAGCGAATGGTTCCG | 62.1 | |||
| 3 | blaKPC_probe | TGTCTTGTCTCTCATGGCCGCTGG | FAM | BHQ-1 | 75.4 | |
| 4 | blaNDM_fwd | GCATTAGCCGCTGCATT | 63.1 | FN396876.1 [2407:3219] | ||
| 5 | blaNDM_rev | GATCGCCAAACCGTTGG | 65.7 | |||
| 6 | blaNDM_probe | ACGATTGGCCAGCAAATGGAAACTGG | ROX | BHQ-2 | 76.1 | |
| 7 | blaOXA48/181_fwd | TTCCCAATAGCTTGATCGC | 63.1 | Consensus (OXA-48-group) | ||
| 8 | blaOXA48/181_rev | CCATCCCACTTAAAGACTTGG | 62.6 | |||
| 9 | blaOXA48/181_probe | TCGATTTGGGCGTGGTTAAGGATGAAC | HEX | BHQ-1 | 74.8 | |
| 10 | blaVIM_fwd | TGGCAACGTACGCATCACC | 68.5 | Consensus | ||
| 11 | blaVIM_rev | CGCAGCACCGGGATAGAA | 67.7 | |||
| 12 | blaVIM_probe | TCTCTAGAAGGACTCTCATCGAGCGGG | Cy5 | BHQ-3 | 73.0 | |
| 13 | ipc_psyr_fw | GGTTTGGTAGACGGTTCGA | 63.0 | CP000075.1 [21609082160931] | ||
| 14 | ipc_psyr_rv | GACCGAGAAAGACGTAAGCA | 62.4 | |||
| 15 | ipc_psyr_probe | TGGGTCAGGTTGCCCATTGACAGA | TAMRA | BHQ-2 | 75.5 |
| Organism | Strain ID 1 | Resistance Genes (Location) 2 |
|---|---|---|
| Acinetobacter baumannii | 95932 | |
| Acinetobacter baumannii | 215784 | blaVIM-2 (p) |
| Acinetobacter baumannii | 240611 | blaNDM-1 (c) |
| Acinetobacter baumannii | 240737 | blaNDM-2 (c) |
| Acinetobacter baumannii | 301751 | blaNDM-1 (c) |
| Citrobacter freundii | 240619 | blaOXA-48 (p), blaVIM-1 (p) |
| Citrobacter freundii | 242274 | blaNDM-1 (p) |
| Citrobacter freundii | 279615 | blaVIM-1 (p) |
| Enterobacter cloacae | 97966 | blaOXA-163 (p) |
| Escherichia coli | 97947 | blaNDM-4 (p) |
| Escherichia coli | 98219 | blaNDM-5 (p) |
| Escherichia coli | 240608 | blaOXA-48 (c) |
| Escherichia coli | 240615 | blaKPC-2 (p) |
| Escherichia coli | 240776 | blaNDM-7 (p) |
| Escherichia coli | 240780 | blaVIM-4 (p) |
| Escherichia coli | 319495 | |
| Klebsiella pneumoniae | 79748 | blaKPC-2 (p), blaVIM-1 (p) |
| Klebsiella pneumoniae | 95473 | blaOXA-48 (p) |
| Klebsiella pneumoniae | 95506 | blaOXA-181 (c) |
| Klebsiella pneumoniae | 95515 | blaOXA-232 (p) |
| Klebsiella pneumoniae | 95571 | blaOXA-48 (p) |
| Klebsiella pneumoniae | 95573 | blaOXA-48 (p) |
| Klebsiella pneumoniae | 215756 | blaKPC-2 (p) |
| Klebsiella pneumoniae | 219840 | blaKPC-2 (p), blaVIM-1 (p) |
| Klebsiella pneumoniae | 223971 | blaKPC-3 (p) |
| Klebsiella pneumoniae | 238631 | blaKPC-2 (p) |
| Klebsiella pneumoniae | 239644 | blaOXA-48-like (p) |
| Klebsiella pneumoniae | 240799 | blaNDM-1 (p), blaOXA-232 (p) |
| Klebsiella pneumoniae | 242816 | blaKPC-2 (p), blaVIM-2 (p) |
| Klebsiella pneumoniae | 245295 | blaKPC-3 (p) |
| Klebsiella pneumoniae | 272567 | blaVIM-1 (p) |
| Klebsiella pneumoniae | 274401 | blaKPC-2 (p) |
| Klebsiella pneumoniae | 280220 | blaOXA-232 (p) |
| Klebsiella pneumoniae | 280236 | |
| Pseudomonas aeruginosa | 279584 | blaVIM-4 (c) |
| Pseudomonas aeruginosa | 280207 | blaVIM-6 (c) |
| Pseudomonas aeruginosa | 280228 | blaVIM-2 (c) |
| Pseudomonas syringae (IPC) | 305664 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reinicke, M.; Diezel, C.; Teimoori, S.; Haase, B.; Monecke, S.; Ehricht, R.; Braun, S.D. Rapid Simultaneous Detection of the Clinically Relevant Carbapenemase Resistance Genes blaKPC, blaOXA48, blaVIM and blaNDM with the Newly Developed Ready-to-Use qPCR CarbaScan LyoBead. Int. J. Mol. Sci. 2025, 26, 1218. https://doi.org/10.3390/ijms26031218
Reinicke M, Diezel C, Teimoori S, Haase B, Monecke S, Ehricht R, Braun SD. Rapid Simultaneous Detection of the Clinically Relevant Carbapenemase Resistance Genes blaKPC, blaOXA48, blaVIM and blaNDM with the Newly Developed Ready-to-Use qPCR CarbaScan LyoBead. International Journal of Molecular Sciences. 2025; 26(3):1218. https://doi.org/10.3390/ijms26031218
Chicago/Turabian StyleReinicke, Martin, Celia Diezel, Salma Teimoori, Bernd Haase, Stefan Monecke, Ralf Ehricht, and Sascha D. Braun. 2025. "Rapid Simultaneous Detection of the Clinically Relevant Carbapenemase Resistance Genes blaKPC, blaOXA48, blaVIM and blaNDM with the Newly Developed Ready-to-Use qPCR CarbaScan LyoBead" International Journal of Molecular Sciences 26, no. 3: 1218. https://doi.org/10.3390/ijms26031218
APA StyleReinicke, M., Diezel, C., Teimoori, S., Haase, B., Monecke, S., Ehricht, R., & Braun, S. D. (2025). Rapid Simultaneous Detection of the Clinically Relevant Carbapenemase Resistance Genes blaKPC, blaOXA48, blaVIM and blaNDM with the Newly Developed Ready-to-Use qPCR CarbaScan LyoBead. International Journal of Molecular Sciences, 26(3), 1218. https://doi.org/10.3390/ijms26031218






