Ubiquitin-Proteasome-Mediated Protein Degradation and Disorders of the Central Nervous System
Abstract
:1. Introduction
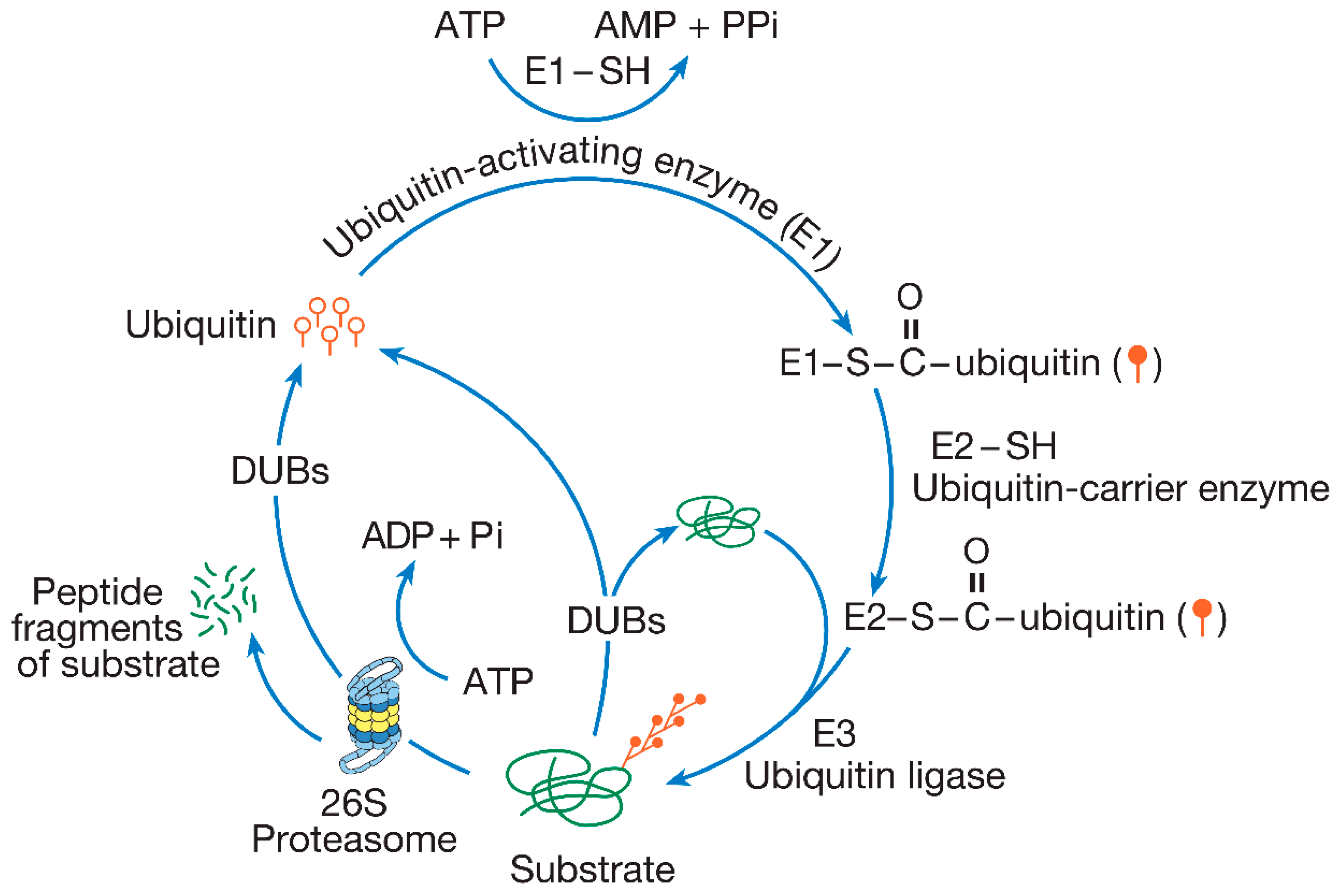
2. The Ubiquitin–Proteasome Pathway
2.1. Ubiquitin Conjugation
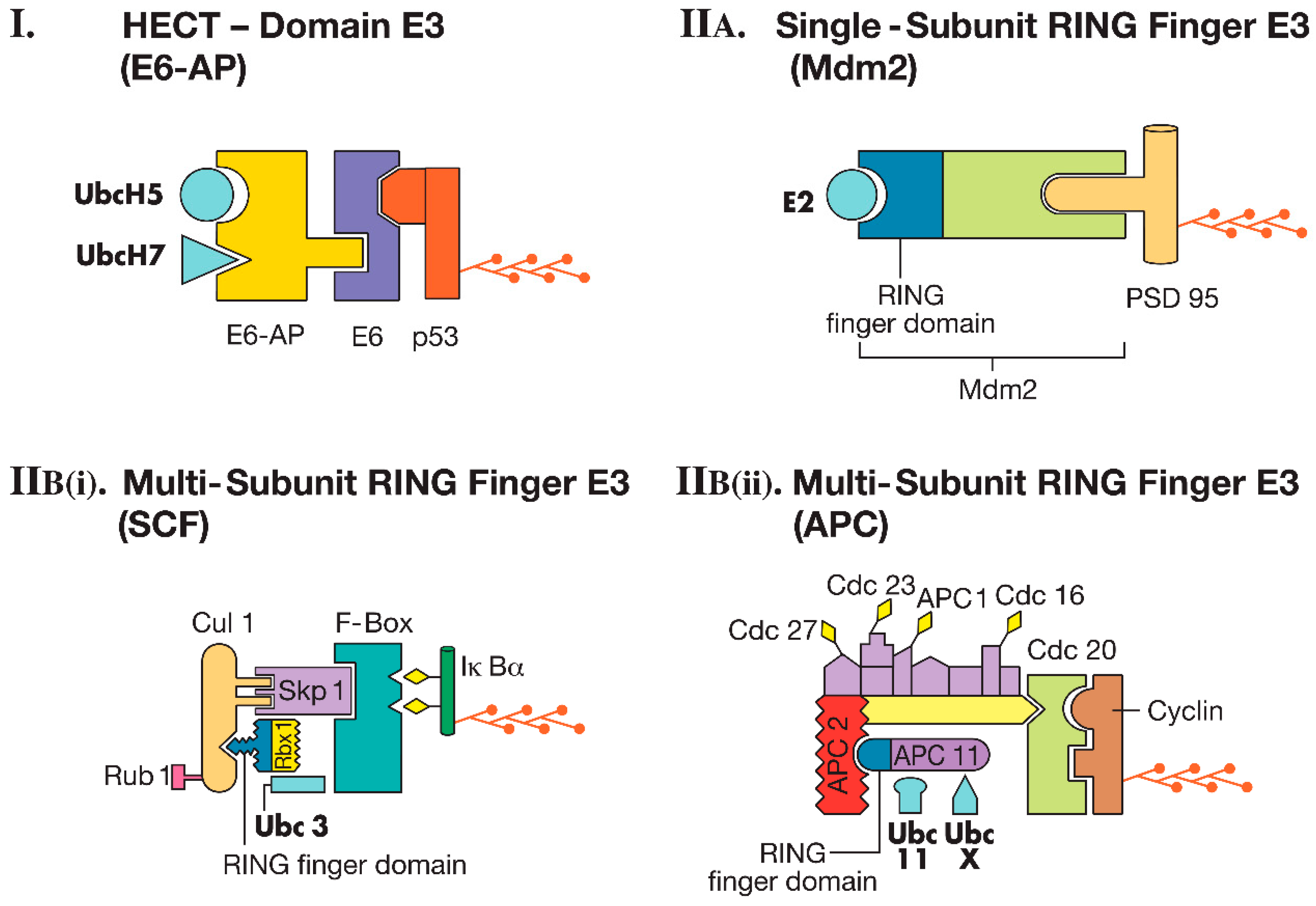
2.2. Ubiquitin-Conjugating Enzymes: E1, E2, and E3
2.2.1. HECT Domain E3s
2.2.2. RING Finger E3s
Single-Subunit RING Finger E3s
Multi-Subunit RING Finger E3s
2.3. The Proteasome
2.3.1. The Catalytic 20S Core
2.3.2. The 19S Regulatory Complex (RC)
2.4. Deubiquitinating Enzymes (DUBs)
3. Association Between the UPP and the Disorders of the Nervous System
3.1. Angelman Syndrome (AS)
3.2. Autism
3.3. Major Depressive Disorder (MDD)
3.4. Schizophrenia
3.5. Amyotrophic Lateral Sclerosis (ALS)
3.6. Huntington’s Disease (HD)
3.7. Parkinson’s Disease (PD)
3.8. Spinocerebellar Ataxia (SCA)
4. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Hegde, A.N. Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Prog. Neurobiol. 2004, 73, 311–357. [Google Scholar] [CrossRef]
- Spano, D.; Catara, G. Targeting the Ubiquitin-Proteasome System and Recent Advances in Cancer Therapy. Cells 2023, 13, 29. [Google Scholar] [CrossRef]
- Hegde, A.N.; Upadhya, S.C. The ubiquitin-proteasome pathway in health and disease of the nervous system. Trends Neurosci. 2007, 30, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Cole, G.M.; Timiras, P.S. Ubiquitin-protein conjugates in Alzheimer’s lesions. Neurosci. Lett. 1987, 79, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Glotzer, M.; Murray, A.W.; Kirschner, M.W. Cyclin is degraded by the ubiquitin pathway. Nature 1991, 349, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Shabbeer, S.; Omer, D.; Berneman, D.; Weitzman, O.; Alpaugh, A.; Pietraszkiewicz, A.; Metsuyanim, S.; Shainskaya, A.; Papa, M.Z.; Yarden, R.I. BRCA1 targets G2/M cell cycle proteins for ubiquitination and proteasomal degradation. Oncogene 2013, 32, 5005–5016. [Google Scholar] [CrossRef] [PubMed]
- Paccosi, E.; Artemi, G.; Filippi, S.; Balzerano, A.; Costanzo, F.; Laghezza-Masci, V.; Proietti, S.; Proietti-De-Santis, L. Cockayne syndrome group A protein localizes at centrosomes during mitosis and regulates Cyclin B1 ubiquitination. Eur. J. Cell Biol. 2023, 102, 151325. [Google Scholar] [CrossRef]
- Kishino, T.; Lalande, M.; Wagstaff, J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 1997, 15, 70–73. [Google Scholar] [CrossRef]
- Matsuura, T.; Sutcliffe, J.S.; Fang, P.; Galjaard, R.J.; Jiang, Y.H.; Benton, C.S.; Rommens, J.M.; Beaudet, A.L. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 1997, 15, 74–77. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Armstrong, D.; Albrecht, U.; Atkins, C.M.; Noebels, J.L.; Eichele, G.; Sweatt, J.D.; Beaudet, A.L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 1998, 21, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Hegde, A.N. The ubiquitin-proteasome pathway and synaptic plasticity. Learn. Mem. 2010, 17, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Reis, N.; Lee, Y.; Glickman, M.H.; Vierstra, R.D. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001, 20, 7096–7107. [Google Scholar] [CrossRef] [PubMed]
- Hengstermann, A.; Linares, L.K.; Ciechanover, A.; Whitaker, N.J.; Scheffner, M. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells. Proc. Natl. Acad. Sci. USA 2001, 98, 1218–1223. [Google Scholar] [CrossRef]
- Deshaies, R.J. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 1999, 15, 435–467. [Google Scholar] [CrossRef] [PubMed]
- Page, A.M.; Hieter, P. The anaphase-promoting complex: New subunits and regulators. Annu. Rev. Biochem. 1999, 68, 583–609. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S.; Cotton, T.R.; Trevelyan, S.J.; Richardson, L.W.; Lee, W.T.; Silke, J.; Lechtenberg, B.C. The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family. Nat. Commun. 2023, 14, 168. [Google Scholar] [CrossRef] [PubMed]
- Riley, B.E.; Lougheed, J.C.; Callaway, K.; Velasquez, M.; Brecht, E.; Nguyen, L.; Shaler, T.; Walker, D.; Yang, Y.; Regnstrom, K.; et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 2013, 4, 1982. [Google Scholar] [CrossRef] [PubMed]
- Pao, K.C.; Wood, N.T.; Knebel, A.; Rafie, K.; Stanley, M.; Mabbitt, P.D.; Sundaramoorthy, R.; Hofmann, K.; van Aalten, D.M.F.; Virdee, S. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 2018, 556, 381–385. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef]
- Williams, C.A. Neurological aspects of the Angelman syndrome. Brain Dev. 2005, 27, 88–94. [Google Scholar] [CrossRef]
- Albrecht, U.; Sutcliffe, J.S.; Cattanach, B.M.; Beechey, C.V.; Armstrong, D.; Eichele, G.; Beaudet, A.L. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat. Genet. 1997, 17, 75–78. [Google Scholar] [CrossRef]
- Rougeulle, C.; Glatt, H.; Lalande, M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat. Genet. 1997, 17, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Talis, A.L.; Howley, P.M. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem. 1999, 274, 18785–18792. [Google Scholar] [CrossRef]
- Kühne, C.; Banks, L. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J. Biol. Chem. 1998, 273, 34302–34309. [Google Scholar] [CrossRef] [PubMed]
- Nuber, U.; Schwarz, S.E.; Scheffner, M. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur. J. Biochem. 1998, 254, 643–649. [Google Scholar] [CrossRef]
- Weeber, E.J.; Jiang, Y.H.; Elgersma, Y.; Varga, A.W.; Carrasquillo, Y.; Brown, S.E.; Christian, J.M.; Mirnikjoo, B.; Silva, A.; Beaudet, A.L.; et al. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J. Neurosci. 2003, 23, 2634–2644. [Google Scholar] [CrossRef] [PubMed]
- van Woerden, G.M.; Harris, K.D.; Hojjati, M.R.; Gustin, R.M.; Qiu, S.; de Avila Freire, R.; Jiang, Y.H.; Elgersma, Y.; Weeber, E.J. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of alphaCaMKII inhibitory phosphorylation. Nat. Neurosci. 2007, 10, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Greer, P.L.; Hanayama, R.; Bloodgood, B.L.; Mardinly, A.R.; Lipton, D.M.; Flavell, S.W.; Kim, T.K.; Griffith, E.C.; Waldon, Z.; Maehr, R.; et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 2010, 140, 704–716. [Google Scholar] [CrossRef]
- Sun, J.; Zhu, G.; Liu, Y.; Standley, S.; Ji, A.; Tunuguntla, R.; Wang, Y.; Claus, C.; Luo, Y.; Baudry, M.; et al. UBE3A Regulates Synaptic Plasticity and Learning and Memory by Controlling SK2 Channel Endocytosis. Cell Rep. 2015, 12, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Liu, Y.; Jia, Y.; Hao, X.; Lin, W.J.; Tran, J.; Lynch, G.; Baudry, M.; Bi, X. UBE3A-mediated p18/LAMTOR1 ubiquitination and degradation regulate mTORC1 activity and synaptic plasticity. eLife 2018, 7, e37993. [Google Scholar] [CrossRef] [PubMed]
- Reiter, L.T.; Seagroves, T.N.; Bowers, M.; Bier, E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum. Mol. Genet. 2006, 15, 2825–2835. [Google Scholar] [CrossRef]
- Silva-Santos, S.; van Woerden, G.M.; Bruinsma, C.F.; Mientjes, E.; Jolfaei, M.A.; Distel, B.; Kushner, S.A.; Elgersma, Y. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J. Clin. Investig. 2015, 125, 2069–2076. [Google Scholar] [CrossRef]
- Crider, A.; Pandya, C.D.; Peter, D.; Ahmed, A.O.; Pillai, A. Ubiquitin-proteasome dependent degradation of GABAAα1 in autism spectrum disorder. Mol. Autism 2014, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Tsai, N.P.; Wilkerson, J.R.; Guo, W.; Maksimova, M.A.; DeMartino, G.N.; Cowan, C.W.; Huber, K.M. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell 2012, 151, 1581–1594. [Google Scholar] [CrossRef] [PubMed]
- Glessner, J.T.; Wang, K.; Cai, G.; Korvatska, O.; Kim, C.E.; Wood, S.; Zhang, H.; Estes, A.; Brune, C.W.; Bradfield, J.P.; et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009, 459, 569–573. [Google Scholar] [CrossRef]
- Soueid, J.; Hamze, Z.; Bedran, J.; Chahrour, M.; Boustany, R.M. A novel autism-associated UBLCP1 mutation impacts proteasome regulation/activity. Transl. Psychiatry 2023, 13, 404. [Google Scholar] [CrossRef] [PubMed]
- Copping, N.A.; Christian, S.G.B.; Ritter, D.J.; Islam, M.S.; Buscher, N.; Zolkowska, D.; Pride, M.C.; Berg, E.L.; LaSalle, J.M.; Ellegood, J.; et al. Neuronal overexpression of Ube3a isoform 2 causes behavioral impairments and neuroanatomical pathology relevant to 15q11.2-q13.3 duplication syndrome. Hum. Mol. Genet. 2017, 26, 3995–4010. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.J.; Berrios, J.; Newbern, J.M.; Snider, W.D.; Philpot, B.D.; Hahn, K.M.; Zylka, M.J. An Autism-Linked Mutation Disables Phosphorylation Control of UBE3A. Cell 2015, 162, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Simon, J.M.; Ptacek, T.S.; Yi, J.J.; Loo, L.; Mao, H.; Wolter, J.M.; McCoy, E.S.; Paranjape, S.R.; Taylor-Blake, B.; et al. Autism-linked UBE3A gain-of-function mutation causes interneuron and behavioral phenotypes when inherited maternally or paternally in mice. Cell Rep. 2023, 42, 112706. [Google Scholar] [CrossRef]
- Zhang, J.; Gambin, T.; Yuan, B.; Szafranski, P.; Rosenfeld, J.A.; Balwi, M.A.; Alswaid, A.; Al-Gazali, L.; Shamsi, A.M.A.; Komara, M.; et al. Haploinsufficiency of the E3 ubiquitin-protein ligase gene TRIP12 causes intellectual disability with or without autism spectrum disorders, speech delay, and dysmorphic features. Hum. Genet. 2017, 136, 377–386. [Google Scholar] [CrossRef]
- Belaish, S.; Israel-Elgali, I.; Shapira, G.; Krieger, I.; Segev, A.; Nitzan, U.; Majer, M.; Bloch, Y.; Weizman, A.; Gurwitz, D.; et al. Genome wide analysis implicates upregulation of proteasome pathway in major depressive disorder. Transl. Psychiatry 2021, 11, 409. [Google Scholar] [CrossRef]
- Minelli, A.; Magri, C.; Barbon, A.; Bonvicini, C.; Segala, M.; Congiu, C.; Bignotti, S.; Milanesi, E.; Trabucchi, L.; Cattane, N.; et al. Proteasome system dysregulation and treatment resistance mechanisms in major depressive disorder. Transl. Psychiatry 2015, 5, e687. [Google Scholar] [CrossRef] [PubMed]
- Mouri, A.; Sasaki, A.; Watanabe, K.; Sogawa, C.; Kitayama, S.; Mamiya, T.; Miyamoto, Y.; Yamada, K.; Noda, Y.; Nabeshima, T. MAGE-D1 regulates expression of depression-like behavior through serotonin transporter ubiquitylation. J. Neurosci. 2012, 32, 4562–4580. [Google Scholar] [CrossRef]
- Fukuo, Y.; Kishi, T.; Kushima, I.; Yoshimura, R.; Okochi, T.; Kitajima, T.; Matsunaga, S.; Kawashima, K.; Umene-Nakano, W.; Naitoh, H.; et al. Possible association between ubiquitin-specific peptidase 46 gene and major depressive disorders in the Japanese population. J. Affect. Disord. 2011, 133, 150–157. [Google Scholar] [CrossRef] [PubMed]
- St Clair, D.; Blackwood, D.; Muir, W.; Carothers, A.; Walker, M.; Spowart, G.; Gosden, C.; Evans, H.J. Association within a family of a balanced autosomal translocation with major mental illness. Lancet 1990, 336, 13–16. [Google Scholar] [CrossRef]
- Leliveld, S.R.; Hendriks, P.; Michel, M.; Sajnani, G.; Bader, V.; Trossbach, S.; Prikulis, I.; Hartmann, R.; Jonas, E.; Willbold, D.; et al. Oligomer assembly of the C-terminal DISC1 domain (640–854) is controlled by self-association motifs and disease-associated polymorphism S704C. Biochemistry 2009, 48, 7746–7755. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, A.; Tomoda, T.; Chang, J.; Takaki, M.; Zhan, C.; Morita, M.; Cascio, M.B.; Elashvili, S.; Koizumi, H.; Takanezawa, Y.; et al. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum. Mol. Genet. 2006, 15, 3313–3323. [Google Scholar] [CrossRef] [PubMed]
- Yalla, K.; Elliott, C.; Day, J.P.; Findlay, J.; Barratt, S.; Hughes, Z.A.; Wilson, L.; Whiteley, E.; Popiolek, M.; Li, Y.; et al. FBXW7 regulates DISC1 stability via the ubiquitin-proteosome system. Mol. Psychiatry 2018, 23, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Ge, X.; Frank, C.L.; Madison, J.M.; Koehler, A.N.; Doud, M.K.; Tassa, C.; Berry, E.M.; Soda, T.; Singh, K.K.; et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell 2009, 136, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Khodosevich, K.; Monyer, H. Dendrite development regulated by the schizophrenia-associated gene FEZ1 involves the ubiquitin proteasome system. Cell Rep. 2014, 7, 552–564. [Google Scholar] [CrossRef]
- Srikanth, P.; Lagomarsino, V.N.; Pearse, R.V., 2nd; Liao, M.; Ghosh, S.; Nehme, R.; Seyfried, N.; Eggan, K.; Young-Pearse, T.L. Convergence of independent DISC1 mutations on impaired neurite growth via decreased UNC5D expression. Transl. Psychiatry 2018, 8, 245. [Google Scholar] [CrossRef] [PubMed]
- Salzberg, Y.; Pechuk, V.; Gat, A.; Setty, H.; Sela, S.; Oren-Suissa, M. Synaptic Protein Degradation Controls Sexually Dimorphic Circuits through Regulation of DCC/UNC-40. Curr. Biol. 2020, 30, 4128–4141.e5. [Google Scholar] [CrossRef] [PubMed]
- Kohlbrenner, E.A.; Shaskan, N.; Pietersen, C.Y.; Sonntag, K.C.; Woo, T.W. Gene expression profile associated with postnatal development of pyramidal neurons in the human prefrontal cortex implicates ubiquitin ligase E3 in the pathophysiology of schizophrenia onset. J. Psychiatr. Res. 2018, 102, 110–117. [Google Scholar] [CrossRef]
- Arion, D.; Corradi, J.P.; Tang, S.; Datta, D.; Boothe, F.; He, A.; Cacace, A.M.; Zaczek, R.; Albright, C.F.; Tseng, G.; et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol. Psychiatry 2015, 20, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.L.; Goodfellow, F.J.; Matosin, N.; Snelling, M.K.; Newell, K.A.; Huang, X.F.; Fernandez-Enright, F. Alterations of ubiquitin related proteins in the pathology and development of schizophrenia: Evidence from human and animal studies. J. Psychiatr. Res. 2017, 90, 31–39. [Google Scholar] [CrossRef]
- Bi, Y.; Ren, D.; Yuan, F.; Zhang, Z.; Zhou, D.; Yi, X.; Ji, L.; Li, K.; Yang, F.; Wu, X.; et al. TULP4, a novel E3 ligase gene, participates in neuronal migration as a candidate in schizophrenia. CNS Neurosci. Ther. 2023. [Google Scholar] [CrossRef] [PubMed]
- Vawter, M.P.; Barrett, T.; Cheadle, C.; Sokolov, B.P.; Wood, W.H., 3rd; Donovan, D.M.; Webster, M.; Freed, W.J.; Becker, K.G. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res. Bull. 2001, 55, 641–650. [Google Scholar] [CrossRef]
- Middleton, F.A.; Mirnics, K.; Pierri, J.N.; Lewis, D.A.; Levitt, P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J. Neurosci. 2002, 22, 2718–2729. [Google Scholar] [CrossRef] [PubMed]
- Novikova, S.I.; He, F.; Cutrufello, N.J.; Lidow, M.S. Identification of protein biomarkers for schizophrenia and bipolar disorder in the postmortem prefrontal cortex using SELDI-TOF-MS ProteinChip profiling combined with MALDI-TOF-PSD-MS analysis. Neurobiol. Dis. 2006, 23, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.R.; Rubio, M.D.; Haroutunian, V.; Meador-Woodruff, J.H. Protein Expression of Proteasome Subunits in Elderly Patients with Schizophrenia. Neuropsychopharmacology 2016, 41, 896–905. [Google Scholar] [CrossRef]
- Hertzberg, L.; Maggio, N.; Muler, I.; Yitzhaky, A.; Majer, M.; Haroutunian, V.; Zuk, O.; Katsel, P.; Domany, E.; Weiser, M. Comprehensive Gene Expression Analysis Detects Global Reduction of Proteasome Subunits in Schizophrenia. Schizophr. Bull. 2021, 47, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Bousman, C.A.; Luza, S.; Mancuso, S.G.; Kang, D.; Opazo, C.M.; Mostaid, M.S.; Cropley, V.; McGorry, P.; Shannon Weickert, C.; Pantelis, C.; et al. Elevated ubiquitinated proteins in brain and blood of individuals with schizophrenia. Sci. Rep. 2019, 9, 2307. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.D.; Wood, K.; Haroutunian, V.; Meador-Woodruff, J.H. Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacology 2013, 38, 1910–1920. [Google Scholar] [CrossRef]
- Scott, M.R.; Meador-Woodruff, J.H. Intracellular compartment-specific proteasome dysfunction in postmortem cortex in schizophrenia subjects. Mol. Psychiatry 2020, 25, 776–790. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Sun, C.; Chen, Q.; O’Neill, F.A.; Walsh, D.; Fanous, A.; Kendler, K.S. FBXL21 association with schizophrenia in Irish family and case-control samples. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008, 147, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Rouach, N.; Nicoll, R.A.; Bredt, D.S. Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc. Natl. Acad. Sci. USA 2005, 102, 5600–5605. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Cui, K.; Bi, X.; Wang, L.; Sun, M.; Yang, L.; Liu, L. Association between polymorphism of the NEDD4 gene and cognitive dysfunction of schizophrenia patients in Chinese Han population. BMC Psychiatry 2019, 19, 405. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Hou, Q.; Jarzylo, L.; Amato, S.; Gilbert, J.; Shang, F.; Man, H.Y. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J. Neurochem. 2011, 119, 27–39. [Google Scholar] [CrossRef]
- Cheng, Y.; Chen, X.; Zhang, X.Q.; Ju, P.J.; Wang, W.D.; Fang, Y.; Lin, G.N.; Cui, D.H. Interaction between RNF4 and SART3 is associated with the risk of schizophrenia. Heliyon 2024, 10, e32743. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.X.; et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Hasegawa, M.; Akiyama, H.; Ikeda, K.; Nonaka, T.; Mori, H.; Mann, D.; Tsuchiya, K.; Yoshida, M.; Hashizume, Y.; et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006, 351, 602–611. [Google Scholar] [CrossRef]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef]
- Kwiatkowski, T.J., Jr.; Bosco, D.A.; Leclerc, A.L.; Tamrazian, E.; Vanderburg, C.R.; Russ, C.; Davis, A.; Gilchrist, J.; Kasarskis, E.J.; Munsat, T.; et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009, 323, 1205–1208. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.; Rogelj, B.; Hortobágyi, T.; De Vos, K.J.; Nishimura, A.L.; Sreedharan, J.; Hu, X.; Smith, B.; Ruddy, D.; Wright, P.; et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009, 323, 1208–1211. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.K.; Wilcox, H.M.; Scott, R.W.; Siman, R. Proteasome inhibition enhances the stability of mouse Cu/Zn superoxide dismutase with mutations linked to familial amyotrophic lateral sclerosis. J. Neurol. Sci. 1996, 139, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.A.; Dalton, M.J.; Gurney, M.E.; Kopito, R.R. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2000, 97, 12571–12576. [Google Scholar] [CrossRef] [PubMed]
- Niwa, J.; Ishigaki, S.; Hishikawa, N.; Yamamoto, M.; Doyu, M.; Murata, S.; Tanaka, K.; Taniguchi, N.; Sobue, G. Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity. J. Biol. Chem. 2002, 277, 36793–36798. [Google Scholar] [CrossRef]
- Miyazaki, K.; Fujita, T.; Ozaki, T.; Kato, C.; Kurose, Y.; Sakamoto, M.; Kato, S.; Goto, T.; Itoyama, Y.; Aoki, M.; et al. NEDL1, a novel ubiquitin-protein isopeptide ligase for dishevelled-1, targets mutant superoxide dismutase-1. J. Biol. Chem. 2004, 279, 11327–11335. [Google Scholar] [CrossRef]
- Di Noto, L.; Whitson, L.J.; Cao, X.; Hart, P.J.; Levine, R.L. Proteasomal degradation of mutant superoxide dismutases linked to amyotrophic lateral sclerosis. J. Biol. Chem. 2005, 280, 39907–39913. [Google Scholar] [CrossRef]
- Kabuta, T.; Suzuki, Y.; Wada, K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J. Biol. Chem. 2006, 281, 30524–30533. [Google Scholar] [CrossRef] [PubMed]
- Ishigaki, S.; Niwa, J.; Ando, Y.; Yoshihara, T.; Sawada, K.; Doyu, M.; Yamamoto, M.; Kato, K.; Yotsumoto, Y.; Sobue, G. Differentially expressed genes in sporadic amyotrophic lateral sclerosis spinal cords-screening by molecular indexing and subsequent cDNA microarray analysis. FEBS Lett. 2002, 531, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Urushitani, M.; Kurisu, J.; Tsukita, K.; Takahashi, R. Proteasomal inhibition by misfolded mutant superoxide dismutase 1 induces selective motor neuron death in familial amyotrophic lateral sclerosis. J. Neurochem. 2002, 83, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.P.; Gerin, C.; Bindokas, V.P.; Miller, R.; Ghadge, G.; Roos, R.P. No correlation between aggregates of Cu/Zn superoxide dismutase and cell death in familial amyotrophic lateral sclerosis. J. Neurochem. 2002, 82, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Vlug, A.S.; Jaarsma, D. Long term proteasome inhibition does not preferentially afflict motor neurons in organotypical spinal cord cultures. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2004, 5, 16–21. [Google Scholar] [CrossRef]
- Aquilano, K.; Rotilio, G.; Ciriolo, M.R. Proteasome activation and nNOS down-regulation in neuroblastoma cells expressing a Cu,Zn superoxide dismutase mutant involved in familial ALS. J. Neurochem. 2003, 85, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Hyun, D.H.; Lee, M.; Halliwell, B.; Jenner, P. Proteasomal inhibition causes the formation of protein aggregates containing a wide range of proteins, including nitrated proteins. J. Neurochem. 2003, 86, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Cheroni, C.; Marino, M.; Tortarolo, M.; Veglianese, P.; De Biasi, S.; Fontana, E.; Zuccarello, L.V.; Maynard, C.J.; Dantuma, N.P.; Bendotti, C. Functional alterations of the ubiquitin-proteasome system in motor neurons of a mouse model of familial amyotrophic lateral sclerosis. Hum. Mol. Genet. 2009, 18, 82–96. [Google Scholar] [CrossRef]
- Ying, Z.; Wang, H.; Fan, H.; Zhu, X.; Zhou, J.; Fei, E.; Wang, G. Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Hum. Mol. Genet. 2009, 18, 4268–4281. [Google Scholar] [CrossRef]
- Dong, L.; Liu, L.; Li, Y.; Li, W.; Zhou, L.; Xia, Q. E3 ligase Smurf1 protects against misfolded SOD1 in neuronal cells by promoting its K63 ubiquitylation and aggresome formation. Hum. Mol. Genet. 2022, 31, 2035–2048. [Google Scholar] [CrossRef] [PubMed]
- Yonashiro, R.; Sugiura, A.; Miyachi, M.; Fukuda, T.; Matsushita, N.; Inatome, R.; Ogata, Y.; Suzuki, T.; Dohmae, N.; Yanagi, S. Mitochondrial ubiquitin ligase MITOL ubiquitinates mutant SOD1 and attenuates mutant SOD1-induced reactive oxygen species generation. Mol. Biol. Cell 2009, 20, 4524–4530. [Google Scholar] [CrossRef] [PubMed]
- Hebron, M.L.; Lonskaya, I.; Sharpe, K.; Weerasinghe, P.P.; Algarzae, N.K.; Shekoyan, A.R.; Moussa, C.E. Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6). J. Biol. Chem. 2013, 288, 4103–4115. [Google Scholar] [CrossRef] [PubMed]
- Rafiee, M.R.; Rohban, S.; Davey, K.; Ule, J.; Luscombe, N.M. RNA polymerase II-associated proteins reveal pathways affected in VCP-related amyotrophic lateral sclerosis. Brain 2023, 146, 2547–2556. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.; Huang, W.C.; Lin, J.H.; Kao, T.J.; Lin, H.C.; Lee, K.H.; Lin, H.C.; Shen, C.J.; Chang, W.C.; Huang, C.C. Znf179 E3 ligase-mediated TDP-43 polyubiquitination is involved in TDP-43- ubiquitinated inclusions (UBI) (+)-related neurodegenerative pathology. J. Biomed. Sci. 2018, 25, 76. [Google Scholar] [CrossRef] [PubMed]
- Watabe, K.; Kato, Y.; Sakuma, M.; Murata, M.; Niida-Kawaguchi, M.; Takemura, T.; Hanagata, N.; Tada, M.; Kakita, A.; Shibata, N. Praja1 RING-finger E3 ubiquitin ligase suppresses neuronal cytoplasmic TDP-43 aggregate formation. Neuropathology 2020, 40, 570–586. [Google Scholar] [CrossRef]
- Hans, F.; Fiesel, F.C.; Strong, J.C.; Jäckel, S.; Rasse, T.M.; Geisler, S.; Springer, W.; Schulz, J.B.; Voigt, A.; Kahle, P.J. UBE2E ubiquitin-conjugating enzymes and ubiquitin isopeptidase Y regulate TDP-43 protein ubiquitination. J. Biol. Chem. 2014, 289, 19164–19179. [Google Scholar] [CrossRef]
- Ma, P.; Li, Y.; Wang, H.; Mao, B. Haploinsufficiency of the TDP43 ubiquitin E3 ligase RNF220 leads to ALS-like motor neuron defects in the mouse. J. Mol. Cell Biol. 2021, 13, 374–382. [Google Scholar] [CrossRef]
- Wang, I.F.; Wu, L.S.; Shen, C.K. TDP-43: An emerging new player in neurodegenerative diseases. Trends Mol. Med. 2008, 14, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Buratti, E.; Baralle, F.E. The molecular links between TDP-43 dysfunction and neurodegeneration. Adv. Genet. 2009, 66, 1–34. [Google Scholar] [CrossRef]
- Farrawell, N.E.; McAlary, L.; Lum, J.S.; Chisholm, C.G.; Warraich, S.T.; Blair, I.P.; Vine, K.L.; Saunders, D.N.; Yerbury, J.J. Ubiquitin Homeostasis Is Disrupted in TDP-43 and FUS Cell Models of ALS. iScience 2020, 23, 101700. [Google Scholar] [CrossRef] [PubMed]
- The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Bence, N.F.; Sampat, R.M.; Kopito, R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2001, 292, 1552–1555. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E.J.; Shaler, T.A.; Woodman, B.; Ryu, K.Y.; Zaitseva, T.S.; Becker, C.H.; Bates, G.P.; Schulman, H.; Kopito, R.R. Global changes to the ubiquitin system in Huntington’s disease. Nature 2007, 448, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.; Yan, S.; Wang, C.E.; Li, S.; Li, X.J. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. USA 2014, 111, 5706–5711. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, M.; Shekhar, S.; Singh, B.K.; Jamal, I.; Vatsa, N.; Kumar, V.; Sharma, A.; Jana, N.R. Deficiency of Ube3a in Huntington’s disease mice brain increases aggregate load and accelerates disease pathology. Hum. Mol. Genet. 2014, 23, 6235–6245. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Dikshit, P.; Purkayastha, S.; Sharma, J.; Nukina, N.; Jana, N.R. E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J. Biol. Chem. 2008, 283, 7648–7656. [Google Scholar] [CrossRef]
- Lee, J.M.; Wheeler, V.C.; Chao, M.J.; Vonsattel, J.P.G.; Pinto, R.M.; Lucente, D.; Abu-Elneel, K.; Ramos, E.M.; Mysore, J.S.; Gillis, T.; et al. Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell 2015, 162, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Koyuncu, S.; Saez, I.; Lee, H.J.; Gutierrez-Garcia, R.; Pokrzywa, W.; Fatima, A.; Hoppe, T.; Vilchez, D. The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington’s disease patients. Nat. Commun. 2018, 9, 2886. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhong, X.; Ballar, P.; Luo, S.; Shen, Y.; Rubinsztein, D.C.; Monteiro, M.J.; Fang, S. Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp. Cell. Res. 2007, 313, 538–550. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef]
- Bonifati, V.; Rizzu, P.; van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 2003, 299, 256–259. [Google Scholar] [CrossRef]
- Paisán-Ruíz, C.; Jain, S.; Evans, E.W.; Gilks, W.P.; Simón, J.; van der Brug, M.; López de Munain, A.; Aparicio, S.; Gil, A.M.; Khan, N.; et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 2004, 44, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.; Heimbach, A.; Gründemann, J.; Stiller, B.; Hampshire, D.; Cid, L.P.; Goebel, I.; Mubaidin, A.F.; Wriekat, A.L.; Roeper, J.; et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006, 38, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Fujiwara, H.; Nonaka, T.; Wakabayashi, K.; Takahashi, H.; Lee, V.M.; Trojanowski, J.Q.; Mann, D.; Iwatsubo, T. Phosphorylated alpha-synuclein is ubiquitinated in alpha-synucleinopathy lesions. J. Biol. Chem. 2002, 277, 49071–49076. [Google Scholar] [CrossRef] [PubMed]
- Upadhya, S.C.; Hegde, A.N. A potential proteasome-interacting motif within the ubiquitin-like domain of parkin and other proteins. Trends Biochem. Sci. 2003, 28, 280–283. [Google Scholar] [CrossRef]
- Shinbo, Y.; Niki, T.; Taira, T.; Ooe, H.; Takahashi-Niki, K.; Maita, C.; Seino, C.; Iguchi-Ariga, S.M.; Ariga, H. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006, 13, 96–108. [Google Scholar] [CrossRef]
- Ding, X.; Goldberg, M.S. Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS ONE 2009, 4, e5949. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, I.N.; Kaganovich, A.; Langston, R.G.; Beilina, A.; Ndukwe, K.; Kumaran, R.; Dillman, A.A.; Chia, R.; Cookson, M.R. The G2385R risk factor for Parkinson’s disease enhances CHIP-dependent intracellular degradation of LRRK2. Biochem. J. 2017, 474, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Stevens, D.A.; Kang, S.U.; Jiang, H.; Lee, Y.I.; Ko, H.S.; Scarffe, L.A.; Umanah, G.E.; Kang, H.; Ham, S.; et al. PINK1 Primes Parkin-Mediated Ubiquitination of PARIS in Dopaminergic Neuronal Survival. Cell Rep. 2017, 18, 918–932. [Google Scholar] [CrossRef]
- Si, J.; Van den Haute, C.; Lobbestael, E.; Martin, S.; van Veen, S.; Vangheluwe, P.; Baekelandt, V. ATP13A2 Regulates Cellular α-Synuclein Multimerization, Membrane Association, and Externalization. Int. J. Mol. Sci. 2021, 22, 2689. [Google Scholar] [CrossRef] [PubMed]
- Leroy, E.; Boyer, R.; Auburger, G.; Leube, B.; Ulm, G.; Mezey, E.; Harta, G.; Brownstein, M.J.; Jonnalagada, S.; Chernova, T.; et al. The ubiquitin pathway in Parkinson’s disease. Nature 1998, 395, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Jangir, D.K.; Verma, G.; Shekhar, S.; Hanpude, P.; Kumar, S.; Kumari, R.; Singh, N.; Sarovar Bhavesh, N.; Ranjan Jana, N.; et al. S-nitrosylation of UCHL1 induces its structural instability and promotes α-synuclein aggregation. Sci. Rep. 2017, 7, 44558. [Google Scholar] [CrossRef]
- Andersson, F.I.; Werrell, E.F.; McMorran, L.; Crone, W.J.; Das, C.; Hsu, S.T.; Jackson, S.E. The effect of Parkinson’s-disease-associated mutations on the deubiquitinating enzyme UCH-L1. J. Mol. Biol. 2011, 407, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.L.; Dawson, V.L.; Dawson, T.M. Parkin-mediated lysine 63-linked polyubiquitination: A link to protein inclusions formation in Parkinson’s and other conformational diseases? Neurobiol. Aging 2006, 27, 524–529. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Chin, L.S. Parkin-mediated K63-linked polyubiquitination: A signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy 2008, 4, 85–87. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.S.; Olzmann, J.A.; Li, L. Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem. Soc. Trans. 2010, 38, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Ordureau, A.; Sarraf, S.A.; Duda, D.M.; Heo, J.M.; Jedrychowski, M.P.; Sviderskiy, V.O.; Olszewski, J.L.; Koerber, J.T.; Xie, T.; Beausoleil, S.A.; et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 2014, 56, 360–375. [Google Scholar] [CrossRef]
- Ordureau, A.; Heo, J.M.; Duda, D.M.; Paulo, J.A.; Olszewski, J.L.; Yanishevski, D.; Rinehart, J.; Schulman, B.A.; Harper, J.W. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl. Acad. Sci. USA 2015, 112, 6637–6642. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.C.; Fishman, P.S.; Thakor, N.V.; Oyler, G.A. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 2003, 278, 22044–22055. [Google Scholar] [CrossRef] [PubMed]
- Morishima, Y.; Wang, A.M.; Yu, Z.; Pratt, W.B.; Osawa, Y.; Lieberman, A.P. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum. Mol. Genet. 2008, 17, 3942–3952. [Google Scholar] [CrossRef] [PubMed]
- Winborn, B.J.; Travis, S.M.; Todi, S.V.; Scaglione, K.M.; Xu, P.; Williams, A.J.; Cohen, R.E.; Peng, J.; Paulson, H.L. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 2008, 283, 26436–26443. [Google Scholar] [CrossRef] [PubMed]
- Todi, S.V.; Winborn, B.J.; Scaglione, K.M.; Blount, J.R.; Travis, S.M.; Paulson, H.L. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009, 28, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A.; Li, L.; Chin, L.S. Aggresome formation and neurodegenerative diseases: Therapeutic implications. Curr. Med. Chem. 2008, 15, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Buhmann, C.; Bussopulos, A.; Oechsner, M. Dopaminergic response in Parkinsonian phenotype of Machado-Joseph disease. Mov. Disord. 2003, 18, 219–221. [Google Scholar] [CrossRef] [PubMed]
- Bedford, L.; Hay, D.; Devoy, A.; Paine, S.; Powe, D.G.; Seth, R.; Gray, T.; Topham, I.; Fone, K.; Rezvani, N.; et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J. Neurosci. 2008, 28, 8189–8198. [Google Scholar] [CrossRef] [PubMed]
- Wahl, C.; Kautzmann, S.; Krebiehl, G.; Strauss, K.; Woitalla, D.; Müller, T.; Bauer, P.; Riess, O.; Krüger, R. A comprehensive genetic study of the proteasomal subunit S6 ATPase in German Parkinson’s disease patients. J. Neural. Transm. 2008, 115, 1141–1148. [Google Scholar] [CrossRef]
- Fernández-Cruz, I.; Sánchez-Díaz, I.; Narváez-Padilla, V.; Reynaud, E. Rpt2 proteasome subunit reduction causes Parkinson’s disease like symptoms in Drosophila. IBRO Rep. 2020, 9, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, Y.; Goto, J.; Hattori, M.; Toyoda, A.; Ishii, K.; Jeong, S.Y.; Hashida, H.; Masuda, N.; Ogata, K.; Kasai, F.; et al. The genomic structure and expression of MJD, the Machado-Joseph disease gene. J. Hum. Genet. 2001, 46, 413–422. [Google Scholar] [CrossRef]
- Schmitt, I.; Linden, M.; Khazneh, H.; Evert, B.O.; Breuer, P.; Klockgether, T.; Wuellner, U. Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochem. Biophys. Res. Commun. 2007, 362, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Pittman, R.N. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 2006, 15, 2409–2420. [Google Scholar] [CrossRef] [PubMed]
- Ristic, G.; Sutton, J.R.; Libohova, K.; Todi, S.V. Toxicity and aggregation of the polyglutamine disease protein, ataxin-3 is regulated by its binding to VCP/p97 in Drosophila melanogaster. Neurobiol. Dis. 2018, 116, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.M.; Bhattacharyya, B.; Rachel, R.A.; Coppola, V.; Tessarollo, L.; Householder, D.B.; Fletcher, C.F.; Miller, R.J.; Copeland, N.G.; Jenkins, N.A. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat. Genet. 2002, 32, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.; Crimmins, S.; Wilson, J.A.; Korbel, G.A.; Ploegh, H.L.; Wilson, S.M. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J. Neurochem. 2005, 95, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Crimmins, S.; Jin, Y.; Wheeler, C.; Huffman, A.K.; Chapman, C.; Dobrunz, L.E.; Levey, A.; Roth, K.A.; Wilson, J.A.; Wilson, S.M. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J. Neurosci. 2006, 26, 11423–11431. [Google Scholar] [CrossRef]
- Chen, P.C.; Qin, L.N.; Li, X.M.; Walters, B.J.; Wilson, J.A.; Mei, L.; Wilson, S.M. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J. Neurosci. 2009, 29, 10909–10919. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hegde, A.N.; Timm, L.E.; Sivley, C.J.; Ramiyaramcharankarthic, S.; Lowrimore, O.J.; Hendrix, B.J.; Grozdanov, T.G.; Anderson, W.J. Ubiquitin-Proteasome-Mediated Protein Degradation and Disorders of the Central Nervous System. Int. J. Mol. Sci. 2025, 26, 966. https://doi.org/10.3390/ijms26030966
Hegde AN, Timm LE, Sivley CJ, Ramiyaramcharankarthic S, Lowrimore OJ, Hendrix BJ, Grozdanov TG, Anderson WJ. Ubiquitin-Proteasome-Mediated Protein Degradation and Disorders of the Central Nervous System. International Journal of Molecular Sciences. 2025; 26(3):966. https://doi.org/10.3390/ijms26030966
Chicago/Turabian StyleHegde, Ashok N., Logan E. Timm, Connor J. Sivley, Shrenik Ramiyaramcharankarthic, Olivia J. Lowrimore, Brenna J. Hendrix, Teodora G. Grozdanov, and William J. Anderson. 2025. "Ubiquitin-Proteasome-Mediated Protein Degradation and Disorders of the Central Nervous System" International Journal of Molecular Sciences 26, no. 3: 966. https://doi.org/10.3390/ijms26030966
APA StyleHegde, A. N., Timm, L. E., Sivley, C. J., Ramiyaramcharankarthic, S., Lowrimore, O. J., Hendrix, B. J., Grozdanov, T. G., & Anderson, W. J. (2025). Ubiquitin-Proteasome-Mediated Protein Degradation and Disorders of the Central Nervous System. International Journal of Molecular Sciences, 26(3), 966. https://doi.org/10.3390/ijms26030966



