Genome-Wide Analysis of the Aspartate Aminotransferase Family in Brassica rapa and the Role of BraASP1 in Response to Nitrogen Starvation
Abstract
:1. Introduction
2. Results
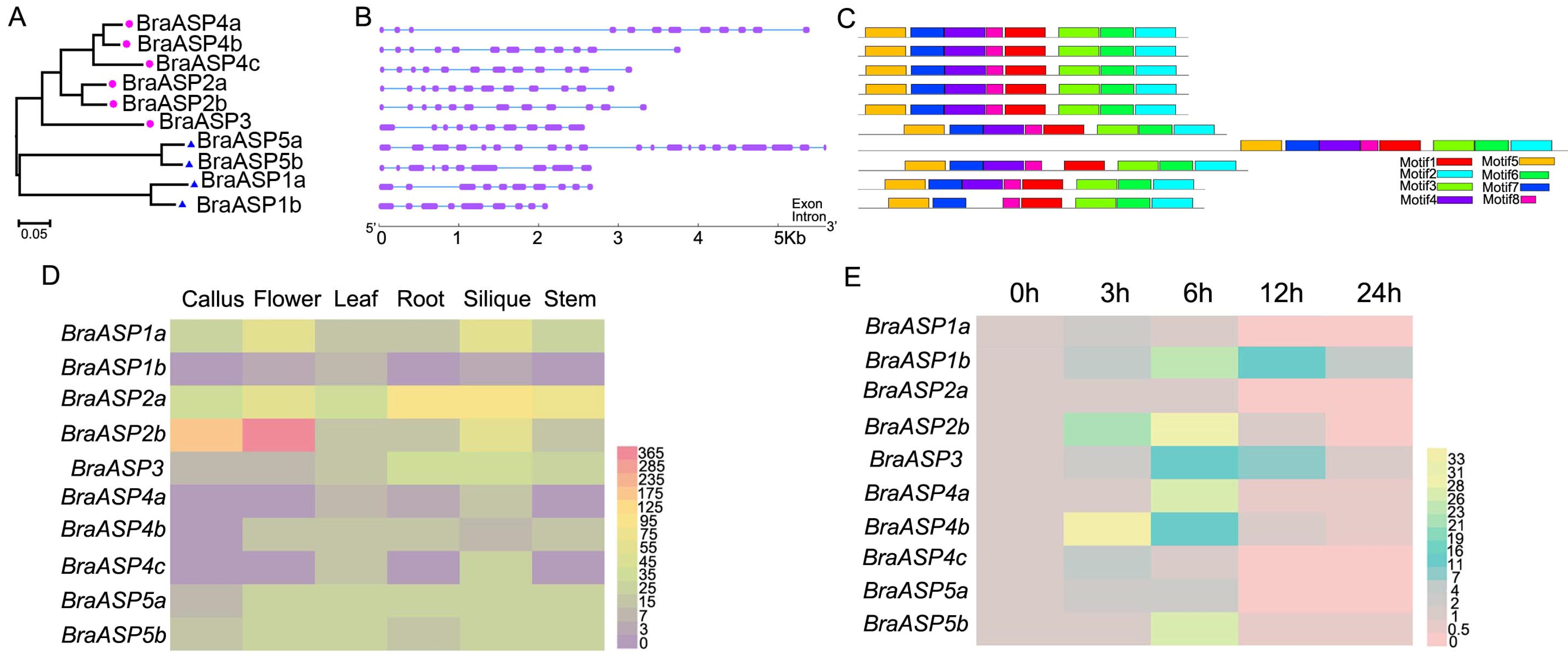
2.1. The Phylogenetic Associations and Collinearity Analysis of ASP Members Among B. rapa, A. thaliana, O. sativa, B. napus, and B. oleracea
2.2. The Cis-Acting Regulatory Elements of Promoters in BraASPs
2.3. The Structures and Expression Levels of BraASP Members
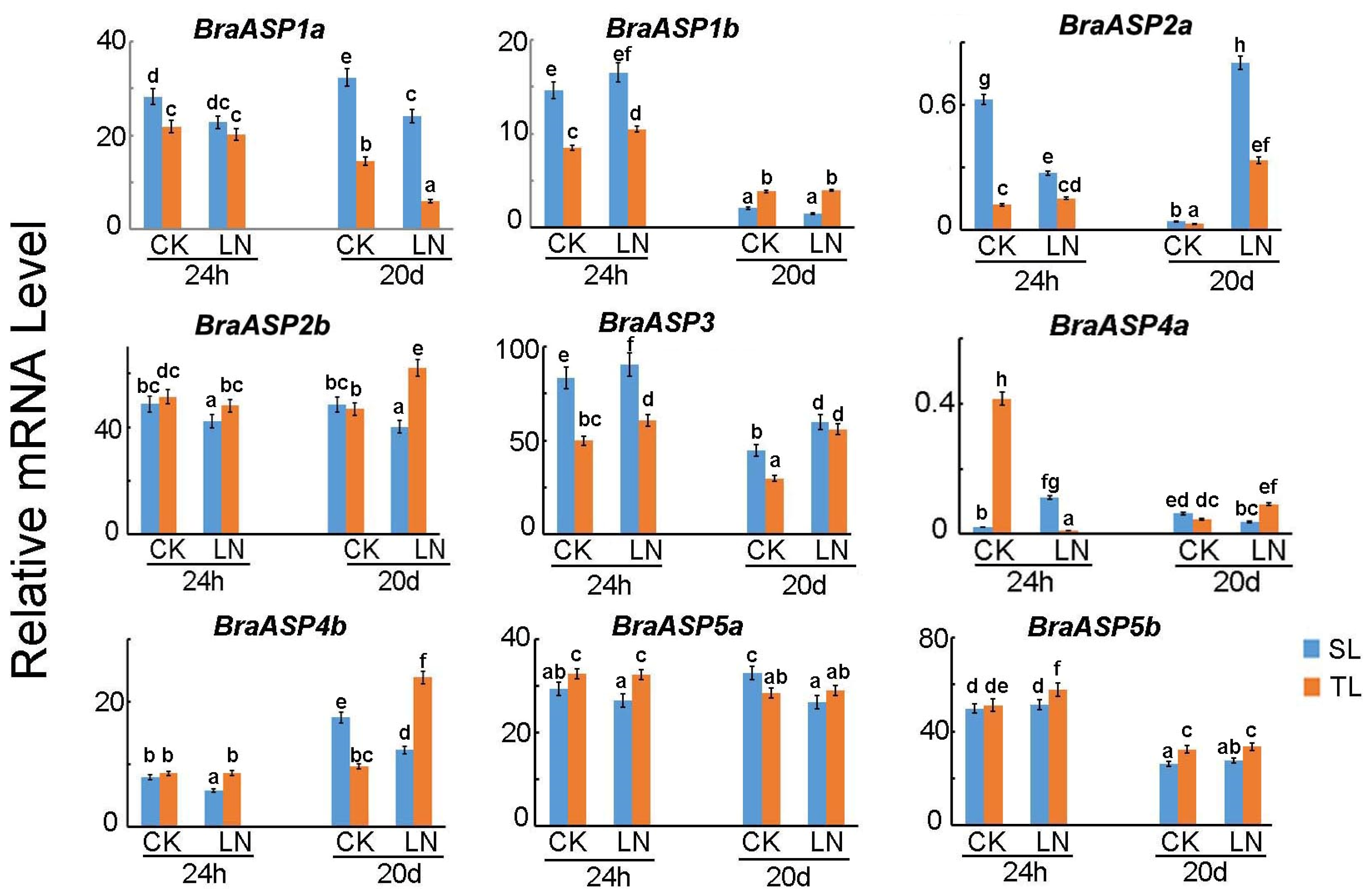
2.4. The Expressions of BraASPs in Sensitive and Insensitive Chinese Cabbage Inbred Lines Under Low N Stresses
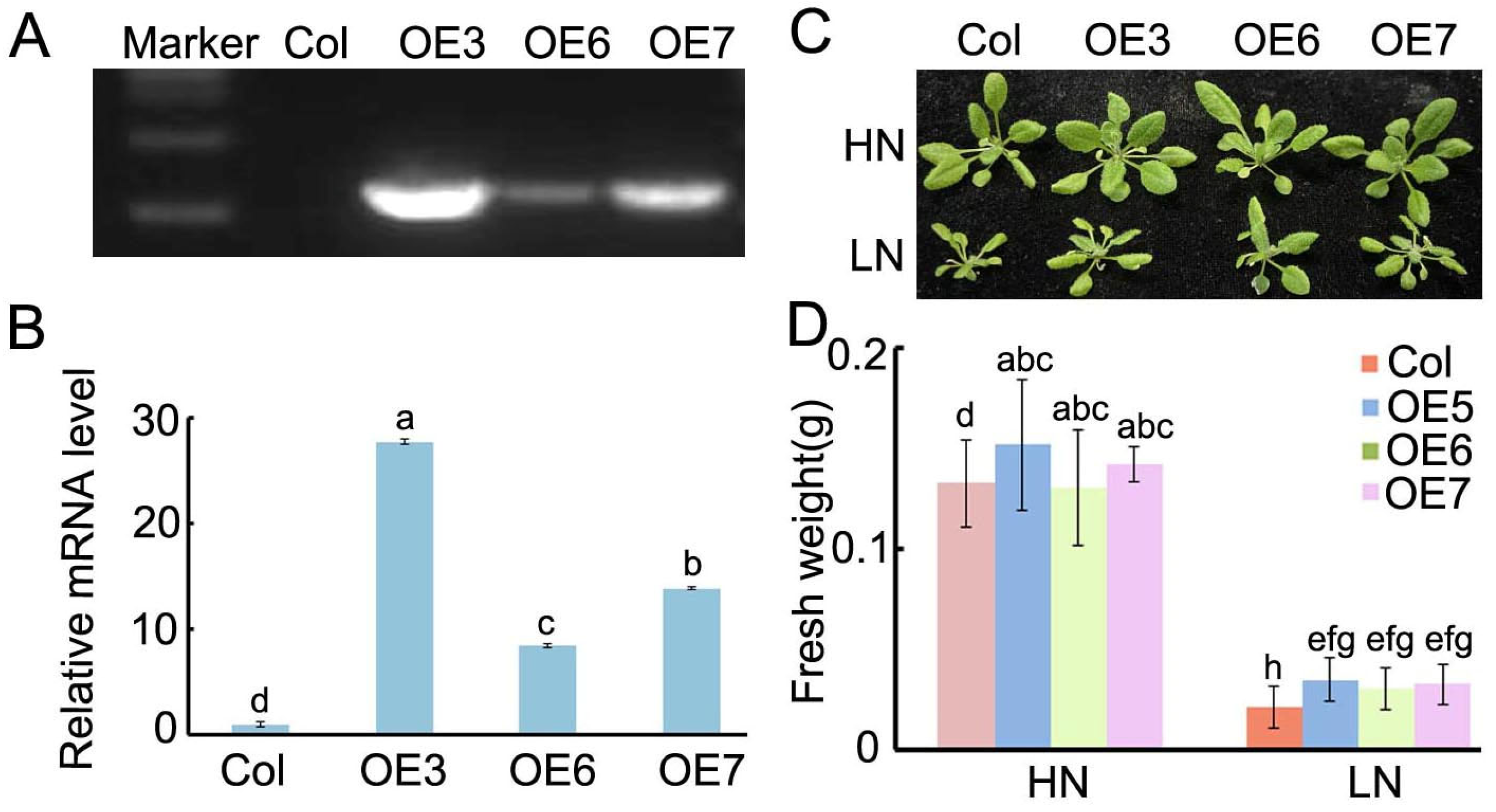
2.5. The Overexpression of BraASP1b in A. thaliana Under Low N Treatments
2.6. The Subcellular Localization of BraASP1b-GFP in Nicotiana benthamiana (N. benthamiana)
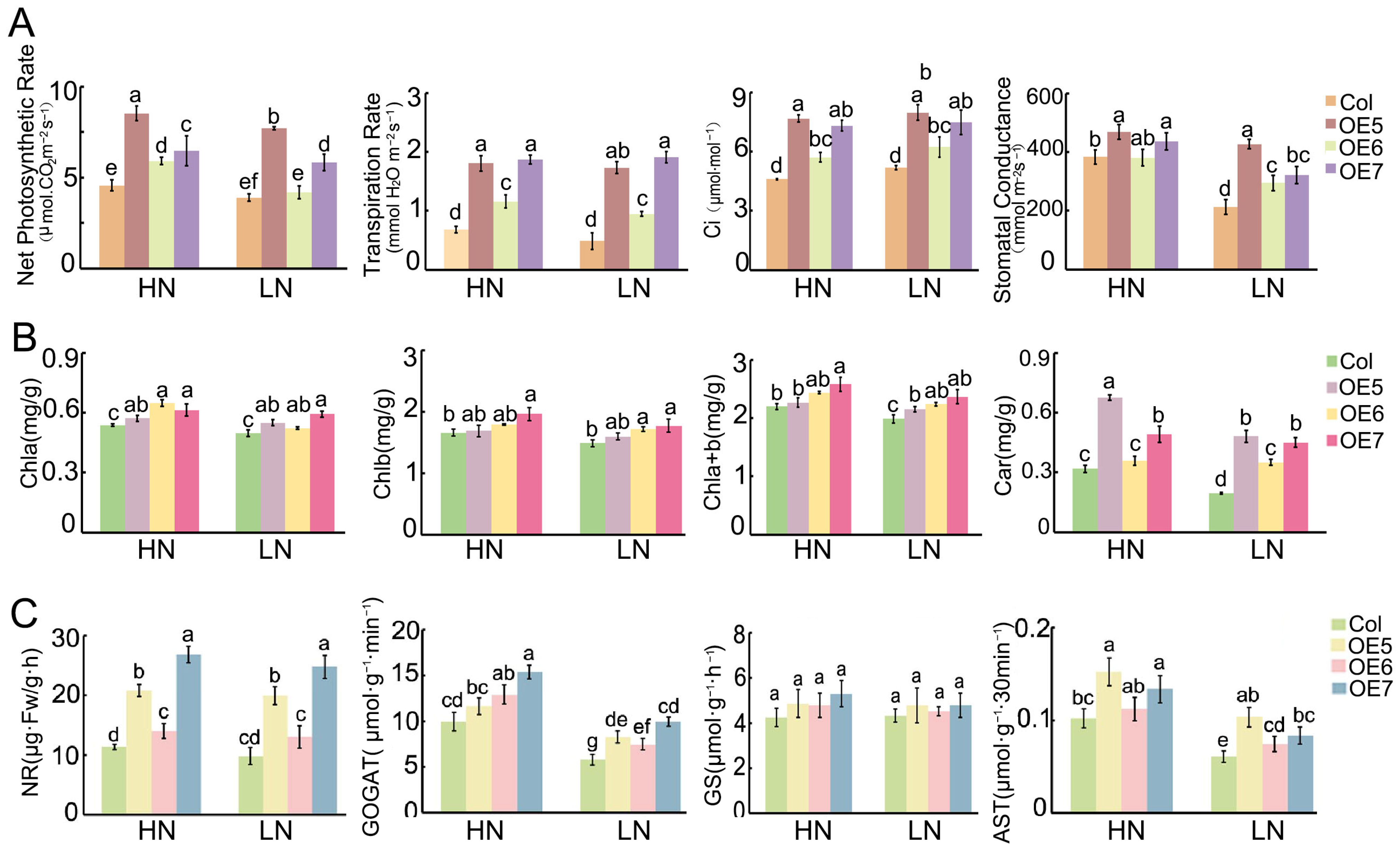
2.7. The Physiological Characteristics of BraASP1b Overexpressed Transgenic A. thaliana Lines Under Low N Stresses
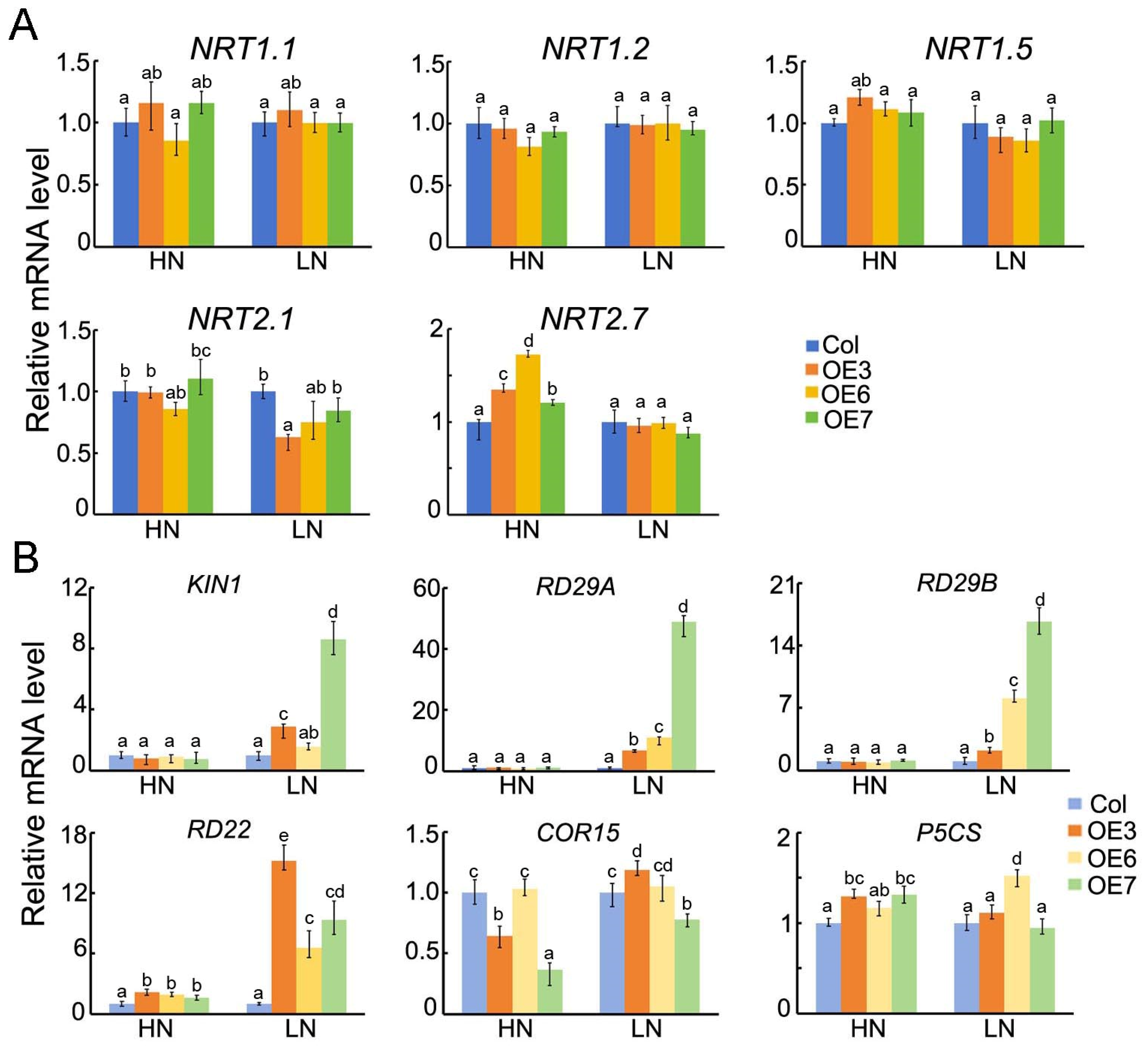
2.8. The Expression Levels of Abiotic Stress and N-Metabolism-Related Genes in Transgenic A. thaliana Lines of Overexpressed BraASP1b Under Low N Stresses
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of ASP in Chinese Cabbage
4.3. The Cis-Acting Elements in Promoters, Gene Structures, and Collinearity Analysis
4.4. Phylogenetic Tree and Motifs Analysis of ASP Protein
4.5. Total RNA Extraction and Real-Time PCR Analysis
4.6. Plasmid Construction and Plant Transformation
4.7. Physiological Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ren, T.; Christie, P.; Wang, J.H.; Chen, Q.; Zhang, F.S. Root zone soil nitrogen management to maintain high tomato yields and minimum nitrogen losses to the environment. Sci. Hortic. 2010, 125, 25–33. [Google Scholar] [CrossRef]
- Lu, C.Q.; Tian, H.Q. Global nitrogen and phosphorus fertilizer use for agriculture production in the past half century: Shifted hot spots and nutrient imbalance. Earth Syst. Sci. Data 2017, 9, 181–192. [Google Scholar] [CrossRef]
- Xuan, W.; Beeckman, T.; Xu, G.H. Plant nitrogen nutrition: Sensing and signaling. Curr. Opin. Plant Biol. 2017, 39, 57–65. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate Transport, Signaling, and Use Efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef]
- Gojon, A.; Krouk, G.; Perrine-Walker, F.; Laugier, E. Nitrate transceptor(s) in plants. J. Exp. Bot. 2011, 62, 2299–2308. [Google Scholar] [CrossRef]
- Kant, S. Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Semin. Cell Dev. Biol. 2018, 74, 89–96. [Google Scholar] [CrossRef]
- The, S.V.; Snyder, R.; Tegeder, M. Targeting Nitrogen Metabolism and Transport Processes to Improve Plant Nitrogen Use Efficiency. Front. Plant Sci. 2020, 11, 628366. [Google Scholar] [CrossRef]
- Kishorekumar, R.; Bulle, M.; Wany, A.; Gupta, K.J. An Overview of Important Enzymes Involved in Nitrogen Assimilation of Plants. Methods Mol. Biol. 2020, 2057, 1–13. [Google Scholar]
- Lebedev, V.G.; Popova, A.A.; Shestibratov, K.A. Genetic Engineering and Genome Editing for Improving Nitrogen Use Efficiency in Plants. Cells 2021, 10, 3303. [Google Scholar] [CrossRef]
- de la Torre, F.; El-Azaz, J.; Avila, C.; Cánovas, F.M. Deciphering the Role of Aspartate and Prephenate Aminotransferase Activities in Plastid Nitrogen Metabolism. Plant Physiol. 2014, 164, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Brauc, S.; De Vooght, E.; Claeys, M.; Höfte, M.; Angenon, G. Influence of over-expression of cytosolic aspartate aminotransferase on amino acid metabolism and defence responses against infection in. J. Plant Physiol. 2011, 168, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cai, H.M.; Xiao, J.H.; Li, X.H.; Zhang, Q.F.; Lian, X.M. Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 2009, 118, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Chandra, H.M.; Mark, W.; Allen, G.G. The impact on nitrogen-efficient phenotypes when aspartate aminotransferase is expressed tissue-specifically in Brassica napus. New Negat. Plant Sci. 2016, 3, 1–9. [Google Scholar]
- Liu, Y.; Guan, X.Y.; Liu, S.N.; Yang, M.; Ren, J.H.; Guo, M.; Huang, Z.H.; Zhang, Y.W. Genome-Wide Identification and Analysis of TCP Transcription Factors Involved in the Formation of Leafy Head in Chinese Cabbage. Int. J. Mol. Sci. 2018, 19, 847. [Google Scholar] [CrossRef]
- Padilla, F.M.; Gallardo, M.; Peña-Fleitas, M.T.; de Souza, R.; Thompson, R.B. Proximal Optical Sensors for Nitrogen Management of Vegetable Crops: A Review. Sensors 2018, 18, 2083. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Zhang, X.F.; Zhang, H.C.; Ren, Y.N.; Gu, R.; Zhang, Y.W. Dynamic transcriptome and coexpression network analysis of Chinese cabbage seedling responses to low-nitrogen stress. Sci. Hortic. 2024, 328, 112866. [Google Scholar] [CrossRef]
- Miesak, B.H.; Coruzzi, G.M. Molecular and physiological analysis of Arabidopsis mutants defective in cytosolic or chloroplastic aspartate aminotransferase. Plant Physiol. 2002, 129, 650–660. [Google Scholar] [CrossRef]
- Schultz, C.J.; Coruzzi, G.M. The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments. Plant J. Cell Mol. Biol. 1995, 7, 61–75. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef]
- Hwang, I.S.; An, S.H.; Hwang, B.K. Pepper is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 2011, 67, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Jing, W.; Cao, C.J.; Sun, M.F.; Chi, W.C.; Zhao, S.L.; Dai, J.Y.; Shi, X.Y.; Wu, Q.; Zhang, B.L.; et al. Transcriptional repressor RST1 controls salt tolerance and grain yield in rice by regulating gene expression of asparagine synthetase. Proc. Natl. Acad. Sci. USA 2022, 119, e2210338119. [Google Scholar] [CrossRef] [PubMed]
- Maaroufi-Dguimi, H.; Debouba, M.; Gaufichon, L.; Clement, G.; Gouia, H.; Hajjaji, A.; Suzuki, A. An Arabidopsis mutant disrupted in ASN2 encoding asparagine synthetase 2 exhibits low salt stress tolerance. Plant Physiol. Biochem. PPB 2011, 49, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Wang, M.L.; Zhang, F.X.; Xu, Y.D.; Chen, X.H.; Qin, X.L.; Wen, X.X. Effect of post-silking drought on nitrogen partitioning and gene expression patterns of glutamine synthetase and asparagine synthetase in two maize varieties. Plant Physiol. Biochem. 2016, 102, 62–69. [Google Scholar] [CrossRef]
- Yang, M.; Geng, M.Y.; Shen, P.F.; Chen, X.H.; Li, Y.J.; Wen, X.X. Effect of post-silking drought stress on the expression profiles of genes involved in carbon and nitrogen metabolism during leaf senescence in maize. Plant Physiol. Biochem. 2019, 135, 304–309. [Google Scholar] [CrossRef]
- Msanne, J.; Lin, J.S.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef]
- Neves-Borges, A.C.; Guimaraes-Dias, F.; Cruz, F.; Mesquita, R.O.; Nepomuceno, A.L.; Romano, E.; Loureiro, M.E.; Grossi-de-Sá, M.D.; Alves-Ferreira, M. Expression pattern of drought stress marker genes in soybean roots under two water deficit systems. Genet. Mol. Biol. 2012, 35, 212–221. [Google Scholar] [CrossRef]
- Zhu, D.; Bai, X.; Zhu, Y.M.; Cai, H.; Li, Y.; Ji, W.; Chen, C.; An, L.; Zhu, Y. Isolation and functional analysis of GsTIFY11b relevant to salt and alkaline stress from Glycine soja. Yi Chuan = Hereditas 2012, 34, 230–239. [Google Scholar] [CrossRef]
- Artus, N.N.; Uemura, M.; Steponkus, P.L.; Gilmour, S.J.; Lin, C.; Thomashow, M.F. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. USA 1996, 93, 13404–13409. [Google Scholar] [CrossRef]
- Strizhov, N.; Abraham, E.; Okresz, L.; Blickling, S.; Zilberstein, A.; Schell, J.; Koncz, C.; Szabados, L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. Cell Mol. Biol. 1997, 12, 557–569. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.L.; Ziegler, J.; Guevara, D.; Abel, S.; Klösgen, R.B.; Mathur, J.; Rothstein, S.J.; Schattat, M.H. Agrobacterium-derived cytokinin influences plastid morphology and starch accumulation in Nicotiana benthamiana during transient assays. BMC Plant Biol. 2014, 14, 127. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.D.; Qin, R.; Li, M.; Alamin, M.; Jin, X.L.; Liu, Y.; Shi, C.H. The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Mol. Genet. Genom. MGG 2017, 292, 385–395. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Gao, Z.; Liang, C.; Wei, Y.; Li, Y.; Zhang, Y.; Zhang, Y. Genome-Wide Analysis of the Aspartate Aminotransferase Family in Brassica rapa and the Role of BraASP1 in Response to Nitrogen Starvation. Int. J. Mol. Sci. 2025, 26, 1586. https://doi.org/10.3390/ijms26041586
Liu Y, Gao Z, Liang C, Wei Y, Li Y, Zhang Y, Zhang Y. Genome-Wide Analysis of the Aspartate Aminotransferase Family in Brassica rapa and the Role of BraASP1 in Response to Nitrogen Starvation. International Journal of Molecular Sciences. 2025; 26(4):1586. https://doi.org/10.3390/ijms26041586
Chicago/Turabian StyleLiu, Yan, Zihan Gao, Chuang Liang, Yuting Wei, Yuge Li, Yan Zhang, and Yaowei Zhang. 2025. "Genome-Wide Analysis of the Aspartate Aminotransferase Family in Brassica rapa and the Role of BraASP1 in Response to Nitrogen Starvation" International Journal of Molecular Sciences 26, no. 4: 1586. https://doi.org/10.3390/ijms26041586
APA StyleLiu, Y., Gao, Z., Liang, C., Wei, Y., Li, Y., Zhang, Y., & Zhang, Y. (2025). Genome-Wide Analysis of the Aspartate Aminotransferase Family in Brassica rapa and the Role of BraASP1 in Response to Nitrogen Starvation. International Journal of Molecular Sciences, 26(4), 1586. https://doi.org/10.3390/ijms26041586






