Machine Learning-Aided Optimization of In Vitro Tetraploid Induction in Cannabis
Abstract
:1. Introduction
2. Results
2.1. Effects of Different Concentrations of Oryzalin and Different Exposure Times on Tetraploid Induction
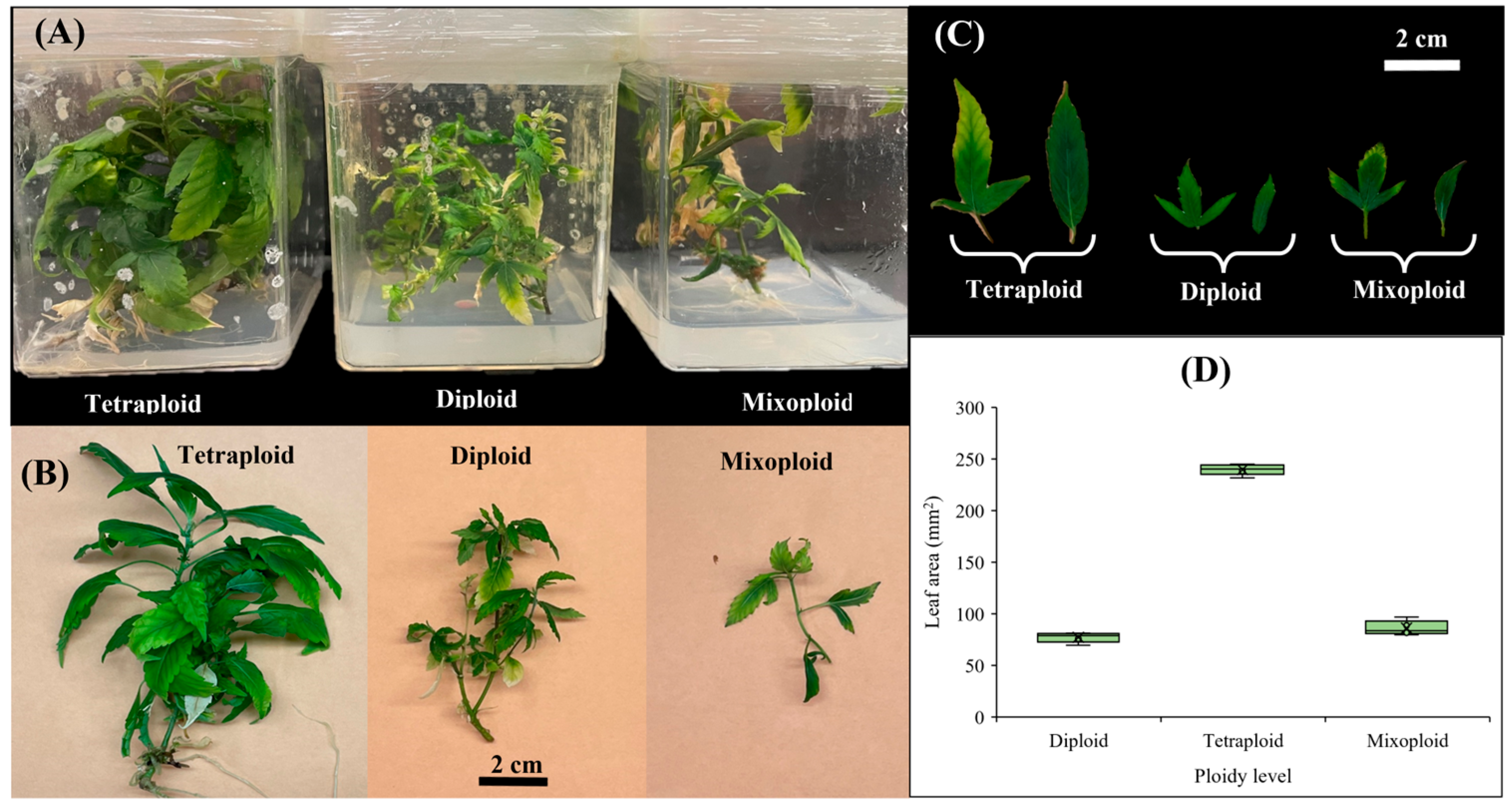
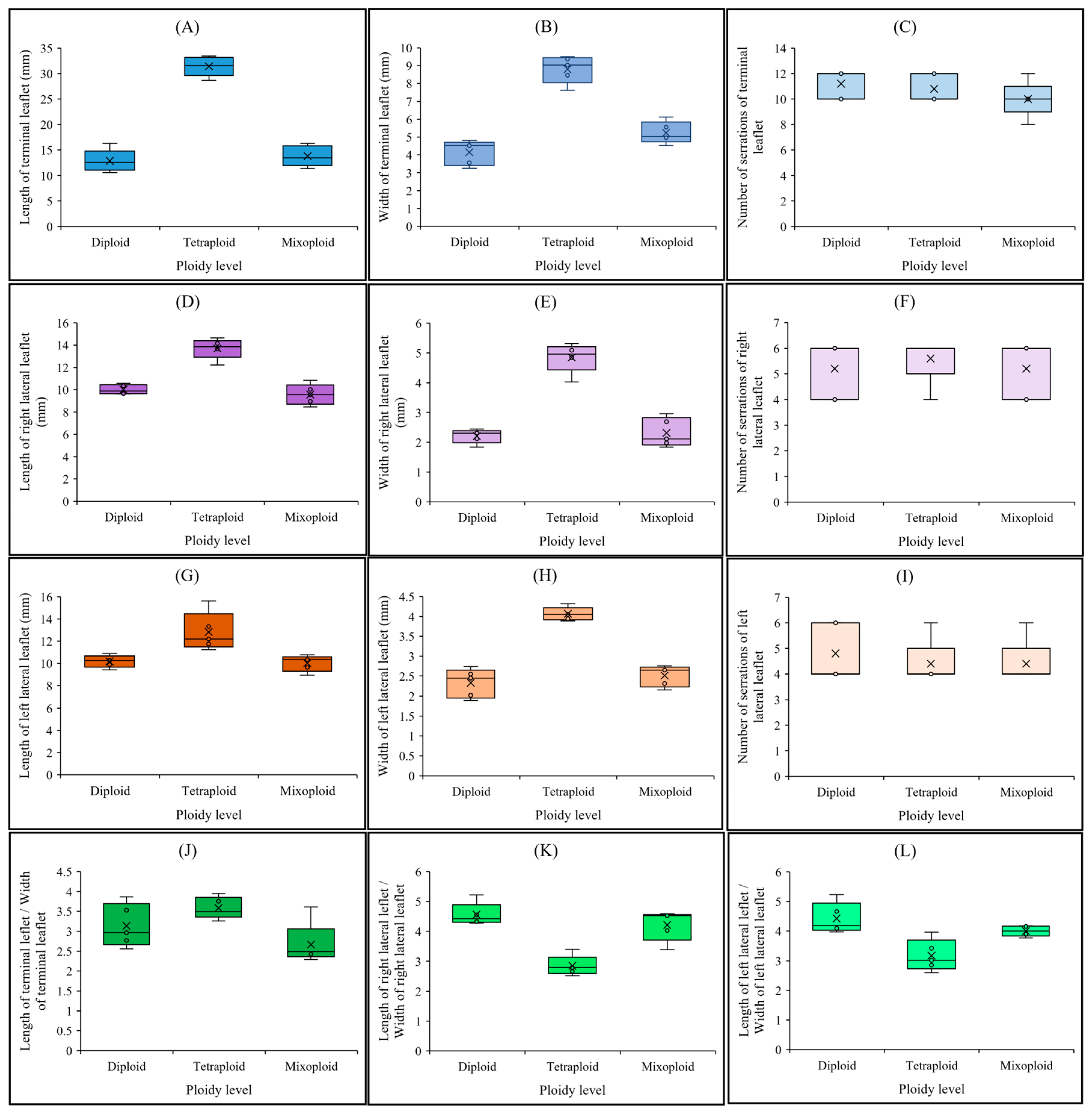
2.2. Leaf-Related Morphological Traits in Diploid, Mixoploid, and Tetraploid Plants
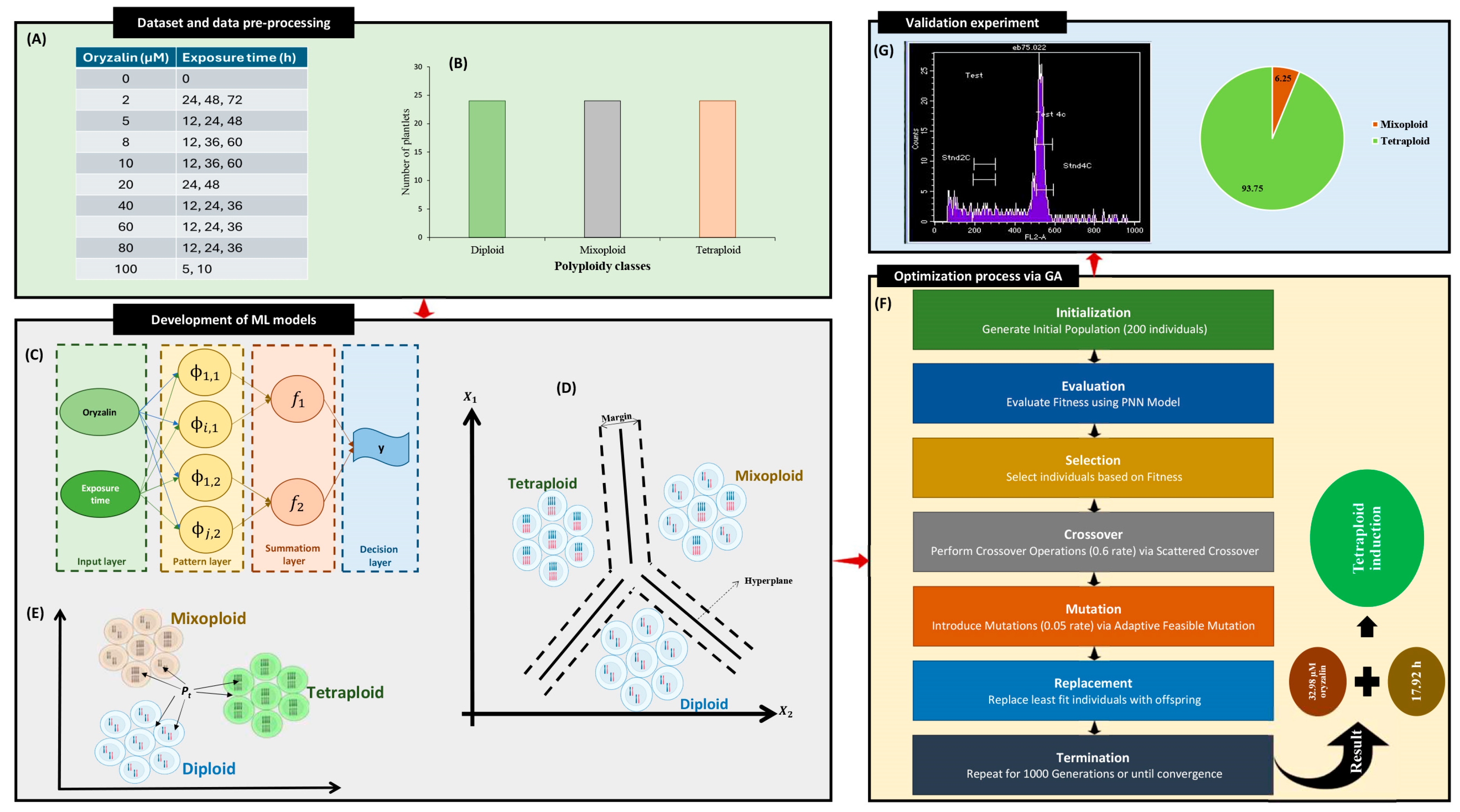
2.3. Evaluation and Comparison of the Developed Machine Learning Models
2.4. Optimization Process and Experimental Confirmation of Predicted-Optimized Conditions
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Leaf-Related Morphological Traits in Diploid, Mixoploid, and Tetraploid Plants
4.3. Dataset Description
4.4. Machine Learning Algorithms
4.4.1. Probabilistic Neural Network (PNN)
4.4.2. Support Vector Classification (SVC)
4.4.3. K-Nearest Neighbors (KNNs)
4.5. Model Performance
4.6. Genetic Optimization Algorithm
4.7. Validation Experiment
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heslop-Harrison, J.S.; Schwarzacher, T.; Liu, Q. Polyploidy: Its consequences and enabling role in plant diversification and evolution. Ann. Bot. 2023, 131, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, B.; Yang, M.; Hu, F.; Qi, H.; Zhang, H.; Jia, Y.; Li, Y.; Wang, Z.; Wang, X. Deciphering recursive polyploidization in Lamiales and reconstructing their chromosome evolutionary trajectories. Plant Physiol. 2024, 195, 2143–2157. [Google Scholar] [CrossRef]
- Morris, J.P.; Baslan, T.; Soltis, D.E.; Soltis, P.S.; Fox, D.T. Integrating the Study of Polyploidy Across Organisms, Tissues, and Disease. Annu. Rev. Genet. 2024, 58, 297–318. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Jung, K.; Masuda, K.; Shimizu, K.K. Polyploidy before and after domestication of crop species. Curr. Opin. Plant Biol. 2022, 69, 102255. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, S.; Cheng, Z.; Chang, X.; Yun, Y.; Jiang, M.; Chen, X.; Wen, X.; Li, H.; Zhu, W.; et al. Origin and evolution of the triploid cultivated banana genome. Nat. Genet. 2024, 56, 136–142. [Google Scholar] [CrossRef]
- Bradshaw, J.E. A brief history of the impact of potato genetics on the breeding of tetraploid potato cultivars for tuber propagation. Potato Res. 2022, 65, 461–501. [Google Scholar] [CrossRef]
- Juery, C.; Concia, L.; De Oliveira, R.; Papon, N.; Ramírez-González, R.; Benhamed, M.; Uauy, C.; Choulet, F.; Paux, E. New insights into homoeologous copy number variations in the hexaploid wheat genome. Plant Genome 2021, 14, e20069. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, V.M.; Knapp, S.J.; Hardigan, M.A.; Edger, P.P.; Slovin, J.P.; V Bassil, N.; Hytönen, T.; Mackenzie, K.K.; Lee, S.; Jung, S.; et al. A roadmap for research in octoploid strawberry. Hortic. Res. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Baiton, A.; Alizadeh, M.; Pepe, M.; Torkamaneh, D.; Jones, A.M. Advances and perspectives in tissue culture and genetic engineering of cannabis. Int. J. Mol. Sci. 2021, 22, 5671. [Google Scholar] [CrossRef]
- Suchoff, D.H.; Inoa, S.H.; Stack, G.M.; Wares, A.J.; Snyder, S.I.; Murdock, M.J.; Rose, J.K.C.; Smart, L.B.; Caton, T.A.; Pearce, R.C. Characterization of agronomic performance and sterility in triploid and diploid cannabinoid hemp. Agron. J. 2024, 116, 2470–2482. [Google Scholar] [CrossRef]
- Crawford, S.; Rojas, B.M.; Crawford, E.; Otten, M.; Schoenenberger, T.A.; Garfinkel, A.R.; Chen, H. Characteristics of the diploid, triploid, and tetraploid versions of a cannabigerol-dominant F1 hybrid industrial hemp cultivar, Cannabis sativa ‘Stem Cell CBG’. Genes 2021, 12, 923. [Google Scholar] [CrossRef] [PubMed]
- Philbrook, R.; Jafari, M.; Gerstenberg, S.; Say, K.L.; Warren, J.; Jones, A.M. Naturally occurring triploidy in cannabis. Plants 2023, 12, 3927. [Google Scholar] [CrossRef]
- Balant, M.; Rodríguez González, R.; Garcia, S.; Garnatje, T.; Pellicer, J.; Vallès, J.; Vitales, D.; Hidalgo, O. Novel insights into the nature of intraspecific genome size diversity in Cannabis sativa L. Plants 2022, 11, 2736. [Google Scholar] [CrossRef]
- Sharma, V.; Srivastava, D.K.; Gupta, R.C.; Singh, B. Abnormal meiosis in tetraploid (4x) Cannabis sativa (L.) from Lahaul-Spiti (cold desert higher altitude Himalayas): A neglected but important herb. J. Biol. Chem. Chron 2015, 2, 38–42. [Google Scholar]
- Bagheri, M.; Mansouri, H. Effect of induced polyploidy on some biochemical parameters in Cannabis sativa L. Appl. Biochem. Biotechnol. 2015, 175, 2366–2375. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, L.E.; Brand, M.H.; Lubell-Brand, J.D. Production of tetraploid and triploid hemp. HortScience 2020, 55, 1703–1707. [Google Scholar] [CrossRef]
- McLeod, A.; Contreras, R.; Halstead, M.; Vining, K. In vivo and in vitro chromosome doubling of ‘I3’ hemp. HortScience 2023, 58, 1018–1022. [Google Scholar] [CrossRef]
- Parsons, J.L.; Martin, S.L.; James, T.; Golenia, G.; Boudko, E.A.; Hepworth, S.R. Polyploidization for the genetic improvement of Cannabis sativa. Front. Plant Sci. 2019, 10, 476. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, M.A.; Gennadievna, N.E.; Mikhailovna, S.O.; Mikhailovna, K.E. Effect of Induced Polyploidy on Morphology, Antioxidant Activity, and Dissolved Sugars in Allium cepa L. Horticulturae 2025, 11, 154. [Google Scholar] [CrossRef]
- Madani, H.; Escrich, A.; Hosseini, B.; Sanchez-Muñoz, R.; Khojasteh, A.; Palazon, J. Effect of Polyploidy Induction on Natural Metabolite Production in Medicinal Plants. Biomolecules 2021, 11, 899. [Google Scholar] [CrossRef]
- Gantait, S.; Mukherjee, E. Induced autopolyploidy—A promising approach for enhanced biosynthesis of plant secondary metabolites: An insight. J. Genet. Eng. Biotechnol. 2021, 19, 4. [Google Scholar] [CrossRef]
- Niazian, M.; Nalousi, A.M. Artificial polyploidy induction for improvement of ornamental and medicinal plants. Plant Cell Tissue Organ Cult. 2020, 142, 447–469. [Google Scholar] [CrossRef]
- Del Pozo, J.C.; Ramirez-Parra, E. Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant Cell Environ. 2014, 37, 2722–2737. [Google Scholar] [CrossRef] [PubMed]
- Harun, A.; Fang, Z.; Chen, C. The contributions of cytogenetics, genetics, and epigenetics to the stability of plants polyploidy. Discov. Plants 2024, 1, 11. [Google Scholar] [CrossRef]
- Tossi, V.E.; Martínez Tosar, L.J.; Laino, L.E.; Iannicelli, J.; Regalado, J.J.; Escandón, A.S.; Baroli, I.; Causin, H.F.; Pitta-Álvarez, S.I. Impact of polyploidy on plant tolerance to abiotic and biotic stresses. Front. Plant Sci. 2022, 13, 869423. [Google Scholar] [CrossRef]
- Yoosefzadeh Najafabadi, M.; Hesami, M.; Rajcan, I. Unveiling the mysteries of non-mendelian heredity in plant breeding. Plants 2023, 12, 1956. [Google Scholar] [CrossRef] [PubMed]
- Basit, A.; Lim, K.-B. Systematic approach of polyploidy as an evolutionary genetic and genomic phenomenon in horticultural crops. Plant Sci. 2024, 348, 112236. [Google Scholar] [CrossRef] [PubMed]
- Trojak-Goluch, A.; Kawka-Lipińska, M.; Wielgusz, K.; Praczyk, M. Polyploidy in industrial crops: Applications and perspectives in plant breeding. Agronomy 2021, 11, 2574. [Google Scholar] [CrossRef]
- Chu, Y.; Lyrene, P.M. Artificial Induction of Polyploidy in Blueberry Breeding: A Review. HortScience 2025, 60, 100–110. [Google Scholar] [CrossRef]
- Obando-González, R.I.; Martínez-Hernández, L.E.; Núñez-Muñoz, L.A.; Calderón-Pérez, B.; Ruiz-Medrano, R.; Ramírez-Pool, J.A.; Xoconostle-Cázares, B. Plant growth Enhancement in Colchicine-Treated Tomato Seeds without Polyploidy Induction. Plant Mol. Biol. 2024, 115, 3. [Google Scholar] [CrossRef] [PubMed]
- Fomicheva, M.; Kulakov, Y.; Alyokhina, K.; Domblides, E. Spontaneous and Chemically Induced Genome Doubling and Polyploidization in Vegetable Crops. Horticulturae 2024, 10, 551. [Google Scholar] [CrossRef]
- Liu, B.; Lee, Y.-R.J. Spindle Assembly and Mitosis in Plants. Annu. Rev. Plant Biol. 2022, 73, 227–254. [Google Scholar] [CrossRef] [PubMed]
- Neenu, M.G.; Aswathi, A.; Prasath, D. Synthetic polyploidy in spice crops: A review. Crop Sci. 2024, 64, 2–23. [Google Scholar] [CrossRef]
- Kunakh, V.A.; Adonin, V.I.; Ozheredov, S.P.; Blyum, Y.B. Mixoploidy in wild and cultivated species of Cruciferae capable of hybridizing with rapeseed Brassica napus. Cytol. Genet. 2008, 42, 204–209. [Google Scholar] [CrossRef]
- Eng, W.-H.; Ho, W.-S. Polyploidization using colchicine in horticultural plants: A review. Sci. Hortic. 2019, 246, 604–617. [Google Scholar] [CrossRef]
- Vilcherrez-Atoche, J.A.; Iiyama, C.M.; Cardoso, J.C. Polyploidization in Orchids: From Cellular Changes to Breeding Applications. Plants 2022, 11, 469. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Jones, A.M.P. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl. Microbiol. Biotechnol. 2020, 104, 9449–9485. [Google Scholar] [CrossRef]
- Jafari, M.; Shahsavar, A. The application of artificial neural networks in modeling and predicting the effects of melatonin on morphological responses of citrus to drought stress. PLoS ONE 2020, 15, e0240427. [Google Scholar] [CrossRef]
- Ramezanpour, M.R.; Farajpour, M. Application of artificial neural networks and genetic algorithm to predict and optimize greenhouse banana fruit yield through nitrogen, potassium and magnesium. PLoS ONE 2022, 17, e0264040. [Google Scholar] [CrossRef]
- Hesami, M.; Alizadeh, M.; Jones, A.M.P.; Torkamaneh, D. Machine learning: Its challenges and opportunities in plant system biology. Appl. Microbiol. Biotechnol. 2022, 106, 3507–3530. [Google Scholar] [CrossRef]
- Niazian, M.; Niedbała, G. Machine learning for plant breeding and biotechnology. Agriculture 2020, 10, 436. [Google Scholar] [CrossRef]
- Rezaei, H.; Mirzaie-asl, A.; Abdollahi, M.R.; Tohidfar, M. Comparative analysis of different artificial neural networks for predicting and optimizing in vitro seed germination and sterilization of petunia. PLoS ONE 2023, 18, e0285657. [Google Scholar] [CrossRef]
- Özcan, E.; Atar, H.H.; Ali, S.A.; Aasim, M. Artificial neural network and decision tree–based models for prediction and validation of in vitro organogenesis of two hydrophytes—Hemianthus callitrichoides and Riccia fluitans. Vitr. Cell. Dev. Biol. Plant 2023, 59, 547–562. [Google Scholar] [CrossRef]
- Aasim, M.; Katırcı, R.; Acar, A.Ş.; Ali, S.A. A comparative and practical approach using quantum machine learning (QML) and support vector classifier (SVC) for Light emitting diodes mediated in vitro micropropagation of black mulberry (Morus nigra L.). Ind. Crops Prod. 2024, 213, 118397. [Google Scholar] [CrossRef]
- Rezaei, H.; Mirzaie-asl, A.; Abdollahi, M.R.; Tohidfar, M. Enhancing petunia tissue culture efficiency with machine learning: A pathway to improved callogenesis. PLoS ONE 2023, 18, e0293754. [Google Scholar] [CrossRef]
- Niazian, M.; Shariatpanahi, M.E.; Abdipour, M.; Oroojloo, M. Modeling callus induction and regeneration in an anther culture of tomato (Lycopersicon esculentum L.) using image processing and artificial neural network method. Protoplasma 2019, 256, 1317–1332. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Naderi, R.; Tohidfar, M. Introducing a hybrid artificial intelligence method for high-throughput modeling and optimizing plant tissue culture processes: The establishment of a new embryogenesis medium for chrysanthemum, as a case study. Appl. Microbiol. Biotechnol. 2020, 104, 10249–10263. [Google Scholar] [CrossRef] [PubMed]
- Jafari, M.; Daneshvar, M.H.; Jafari, S.; Hesami, M. Machine learning-assisted in vitro rooting optimization in Passiflora caerulea. Forests 2022, 13, 2020. [Google Scholar] [CrossRef]
- Hesami, M.; Alizadeh, M.; Naderi, R.; Tohidfar, M. Forecasting and optimizing Agrobacterium-mediated genetic transformation via ensemble model- fruit fly optimization algorithm: A data mining approach using chrysanthemum databases. PLoS ONE 2020, 15, e0239901. [Google Scholar] [CrossRef]
- Becker, F.W.; Oberlander, K.C.; Trávníček, P.; Dreyer, L.L. Inconsistent expression of the gigas effect in polyploid Oxalis. Am. J. Bot. 2022, 109, 1607–1621. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Jones, A.M.P. Modeling and optimizing callus growth and development in Cannabis sativa using random forest and support vector machine in combination with a genetic algorithm. Appl. Microbiol. Biotechnol. 2021, 105, 5201–5212. [Google Scholar] [CrossRef] [PubMed]
- Pepe, M.; Hesami, M.; Jones, A.M. Machine learning-mediated development and optimization of disinfection protocol and scarification method for improved in vitro germination of cannabis seeds. Plants 2021, 10, 2397. [Google Scholar] [CrossRef]
- Pepe, M.; Hesami, M.; Small, F.; Jones, A.M.P. Comparative analysis of machine learning and evolutionary optimization algorithms for precision micropropagation of Cannabis sativa: Prediction and validation of in vitro shoot growth and development based on the optimization of light and carbohydrate sources. Front. Plant Sci. 2021, 12, 757869. [Google Scholar] [CrossRef]
- Hesami, M.; Pepe, M.; Monthony, A.S.; Baiton, A.; Phineas Jones, A.M. Modeling and optimizing in vitro seed germination of industrial hemp (Cannabis sativa L.). Ind. Crops Prod. 2021, 170, 113753. [Google Scholar] [CrossRef]
- Aasim, M.; Katırcı, R.; Akgur, O.; Yildirim, B.; Mustafa, Z.; Nadeem, M.A.; Baloch, F.S.; Karakoy, T.; Yılmaz, G. Machine learning (ML) algorithms and artificial neural network for optimizing in vitro germination and growth indices of industrial hemp (Cannabis sativa L.). Ind. Crops Prod. 2022, 181, 114801. [Google Scholar] [CrossRef]
- Aasim, M.; Yıldırım, B.; Say, A.; Ali, S.A.; Aytaç, S.; Nadeem, M.A. Artificial intelligence models for validating and predicting the impact of chemical priming of hydrogen peroxide (H2O2) and light emitting diodes on in vitro grown industrial hemp (Cannabis sativa L.). Plant Mol. Biol. 2024, 114, 33. [Google Scholar] [CrossRef] [PubMed]
- Vieira, R.G.; Dhimish, M.; de Araújo, F.M.U.; da Silva Guerra, M.I. Comparing multilayer perceptron and probabilistic neural network for PV systems fault detection. Expert Syst. Appl. 2022, 201, 117248. [Google Scholar] [CrossRef]
- Uddin, S.; Haque, I.; Lu, H.; Moni, M.A.; Gide, E. Comparative performance analysis of K-nearest neighbour (KNN) algorithm and its different variants for disease prediction. Sci. Rep. 2022, 12, 6256. [Google Scholar] [CrossRef] [PubMed]
- Sadat-Hosseini, M.; Arab, M.M.; Soltani, M.; Eftekhari, M.; Soleimani, A.; Vahdati, K. Predictive modeling of Persian walnut (Juglans regia L.) in vitro proliferation media using machine learning approaches: A comparative study of ANN, KNN and GEP models. Plant Methods 2022, 18, 48. [Google Scholar] [CrossRef] [PubMed]
- Aasim, M.; Ali, S.A.; Bekiş, P.; Nadeem, M.A. Light-emitting diodes induced in vitro regeneration of Alternanthera reineckii mini and validation via machine learning algorithms. In Vitro Cell. Dev. Biol. Plant 2022, 58, 816–825. [Google Scholar] [CrossRef]
- Vinothkumar, T.; Deepa, S.N.; Raj, F.V.A. Adaptive probabilistic neural network based on hybrid PSO–ALO for predicting wind speed in different regions. Neural Comput. Appl. 2023, 35, 19997–20011. [Google Scholar] [CrossRef]
- Omer, N.; Samak, A.H.; Taloba, A.I.; Abd El-Aziz, R.M. A novel optimized probabilistic neural network approach for intrusion detection and categorization. Alex. Eng. J. 2023, 72, 351–361. [Google Scholar] [CrossRef]
- Sadat-Hosseini, M.; Arab, M.M.; Soltani, M.; Eftekhari, M.; Soleimani, A. Applicability of soft computing techniques for in vitro micropropagation media simulation and optimization: A comparative study on Salvia macrosiphon Boiss. Ind. Crops Prod. 2023, 199, 116750. [Google Scholar] [CrossRef]
- Arab, M.M.; Yadollahi, A.; Ahmadi, H.; Eftekhari, M.; Maleki, M. Mathematical modeling and optimizing of in vitro hormonal combination for G × N15 vegetative rootstock proliferation using artificial neural network-genetic algorithm (ANN-GA). Front. Plant Sci. 2017, 8, 1853. [Google Scholar] [CrossRef]
- Jafari, M.; Daneshvar, M.H. Machine learning-mediated Passiflora caerulea callogenesis optimization. PLoS ONE 2024, 19, e0292359. [Google Scholar] [CrossRef]
- Jafari, M.; Daneshvar, M.H. Prediction and optimization of indirect shoot regeneration of Passiflora caerulea using machine learning and optimization algorithms. BMC Biotechnol. 2023, 23, 27. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Zhang, B.; Tian, J.-R.; Chen, M.-M.; Zhang, Y.-Y.; Zhang, Z.-H.; Ma, Y. Comparison of the morphology, growth and development of diploid and autotetraploid ‘Hanfu’ apple trees. Sci. Hortic. 2017, 225, 277–285. [Google Scholar] [CrossRef]
- Fernandes, H.P.; Choi, Y.H.; Vrieling, K.; de Bresser, M.; Sewalt, B.; Tonolo, F. Cultivar-dependent phenotypic and chemotypic responses of drug-type Cannabis sativa L. to polyploidization. Front. Plant Sci. 2023, 14, 1233191. [Google Scholar] [CrossRef]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Tadeo, F.; Froelicher, Y.; Talon, M.; Navarro, L.; Ollitrault, P.; Morillon, R. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J. Exp. Bot. 2011, 62, 2507–2519. [Google Scholar] [CrossRef]
- Zahumenická, P.; Fernández, E.; Šedivá, J.; Žiarovská, J.; Ros-Santaella, J.L.; Martínez-Fernández, D.; Russo, D.; Milella, L. Morphological, physiological and genomic comparisons between diploids and induced tetraploids in Anemone sylvestris L. Plant Cell Tissue Organ Cult. 2018, 132, 317–327. [Google Scholar] [CrossRef]
- Baker, R.L.; Yarkhunova, Y.; Vidal, K.; Ewers, B.E.; Weinig, C. Polyploidy and the relationship between leaf structure and function: Implications for correlated evolution of anatomy, morphology, and physiology in Brassica. BMC Plant Biol. 2017, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Di, Z.; Zhang, R.; Mu, Y.; Sun, T.; Tian, Z.; Lu, Y.; Zheng, J. Tetraploid induction with leaf morphology and sunburn variation in Sorbus pohuashanensis (Hance) Hedl. Forests 2023, 14, 1589. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, K.; Wang, W.; Liu, G.; Yang, C.; Jiang, J. Differences in leaf morphology and related gene expression between diploid and tetraploid birch (Betula pendula). Int. J. Mol. Sci. 2022, 23, 12966. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, H.; He, T.; Gao, R.; Guo, G.; Lu, R.; Chen, Z.; Liu, C. Comparative analysis of morphology, photosynthetic physiology, and transcriptome between diploid and tetraploid barley derived from microspore culture. Front. Plant Sci. 2021, 12, 626916. [Google Scholar] [CrossRef]
- Šímová, I.; Herben, T. Geometrical constraints in the scaling relationships between genome size, cell size and cell cycle length in herbaceous plants. Proc. R. Soc. B Biol. Sci. 2011, 279, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.; Ito, M.; Matsunaga, S.; Fukui, K. An upper limit of the ratio of DNA volume to nuclear volume exists in plants. Genes Genet. Syst. 2005, 80, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Driver, J.A.; Kuniyuki, A.H. In vitro propagation of paradox walnut rootstock. HortScience 1984, 19, 507–509. [Google Scholar] [CrossRef]
- Dpooležel, J.; Binarová, P.; Lcretti, S. Analysis of Nuclear DNA content in plant cells by Flow cytometry. Biol. Plant. 1989, 31, 113–120. [Google Scholar] [CrossRef]
- Hesami, M.; Pepe, M.; Jones, A.M. Morphological characterization of Cannabis sativa L. Throughout its complete life cycle. Plants 2023, 12, 3646. [Google Scholar] [CrossRef]



| Performance Criteria | Training Set | Testing Set | ||||
|---|---|---|---|---|---|---|
| KNN | PNN | SVC | KNN | PNN | SVC | |
| Accuracy | 92.9825% | 96.4912% | 80.7018% | 80% | 86.6667% | 80% |
| Error rate | 7.0175% | 3.5088% | 19.2982% | 20% | 13.3333% | 20% |
| Precision | 0.91238 | 0.95238 | 0.91273 | 0.8 | 0.93254 | 0.89599 |
| Recall | 0.87238 | 0.95238 | 0.74074 | 0.8 | 0.89725 | 0.57143 |
| F1 Score | 0.89193 | 0.95238 | 0.81779 | 0.8 | 0.91514 | 0.69782 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jafari, M.; Paul, N.; Hesami, M.; Jones, A.M.P. Machine Learning-Aided Optimization of In Vitro Tetraploid Induction in Cannabis. Int. J. Mol. Sci. 2025, 26, 1746. https://doi.org/10.3390/ijms26041746
Jafari M, Paul N, Hesami M, Jones AMP. Machine Learning-Aided Optimization of In Vitro Tetraploid Induction in Cannabis. International Journal of Molecular Sciences. 2025; 26(4):1746. https://doi.org/10.3390/ijms26041746
Chicago/Turabian StyleJafari, Marzieh, Nathan Paul, Mohsen Hesami, and Andrew Maxwell Phineas Jones. 2025. "Machine Learning-Aided Optimization of In Vitro Tetraploid Induction in Cannabis" International Journal of Molecular Sciences 26, no. 4: 1746. https://doi.org/10.3390/ijms26041746
APA StyleJafari, M., Paul, N., Hesami, M., & Jones, A. M. P. (2025). Machine Learning-Aided Optimization of In Vitro Tetraploid Induction in Cannabis. International Journal of Molecular Sciences, 26(4), 1746. https://doi.org/10.3390/ijms26041746







