The Potential Impact of Edible Fruit Extracts on Bacterial Nucleases in Preliminary Research—In Silico and In Vitro Insight
Abstract
:1. Introduction
2. Results and Discussion
2.1. Molecular Docking to Colicin E9
| H-Bond | Cation-π | Salt Bridge | Figure | |
|---|---|---|---|---|
| Procyanidin B2 1 | ASP D:504, ASN E:670, PHE E:686, LYS E:689, TYR E;699 | LYS D:416 | Figure S6 | |
| Cornuside | ASP B:104, GLU B:112, ASP B:117, LYS C:297, ASN C:272, SER C:274 | LYS C:289 | LYS B:16, LYS C:289 | Figure S8 |
| Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside | ASP B:104, HIE B: 102, ILE B:130, ARG B:132, ASP C:224 | Figure S3 | ||
| Isorhamnetin-3-O-rutinoside | SER B:30, ARG B: 132, LYS C:221, ASP C:224, GLY E:627, GLN E:691, LYS E:725, ASP E:729 | ARG B:126 | Figure S4 | |
| Loganic acid | ASP B:104, ASN C:272, THR C:287, GLN C:292, | LYS C:289 | Figure S7 | |
| Isorhamnetin 3-O-β-D-glucoside | ASP C:220, LYS C:221, LEU C:223, ASP C:224, PHE C:286 | Figure S5 | ||
| Procyanidin C1 | LYS D:404, SER D:508, ASP E:729, GLY E:733 | Figure S2 | ||
| Epicatechin | MET B:1, SER B:3, SER B:108, GLY D:495, | ARG D:496 | Figure S9 |
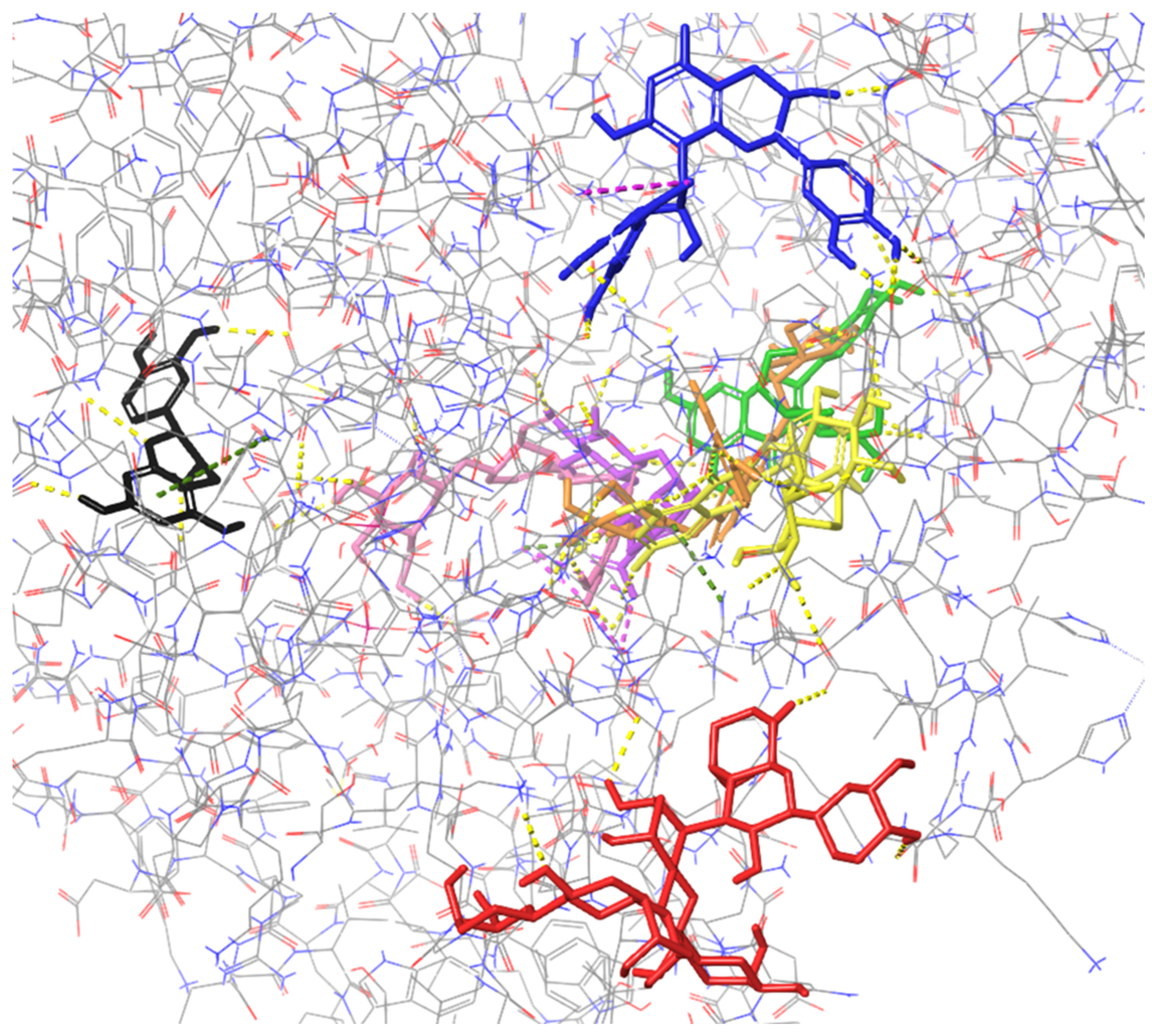
2.2. Molecular Docking to Endonuclease 1
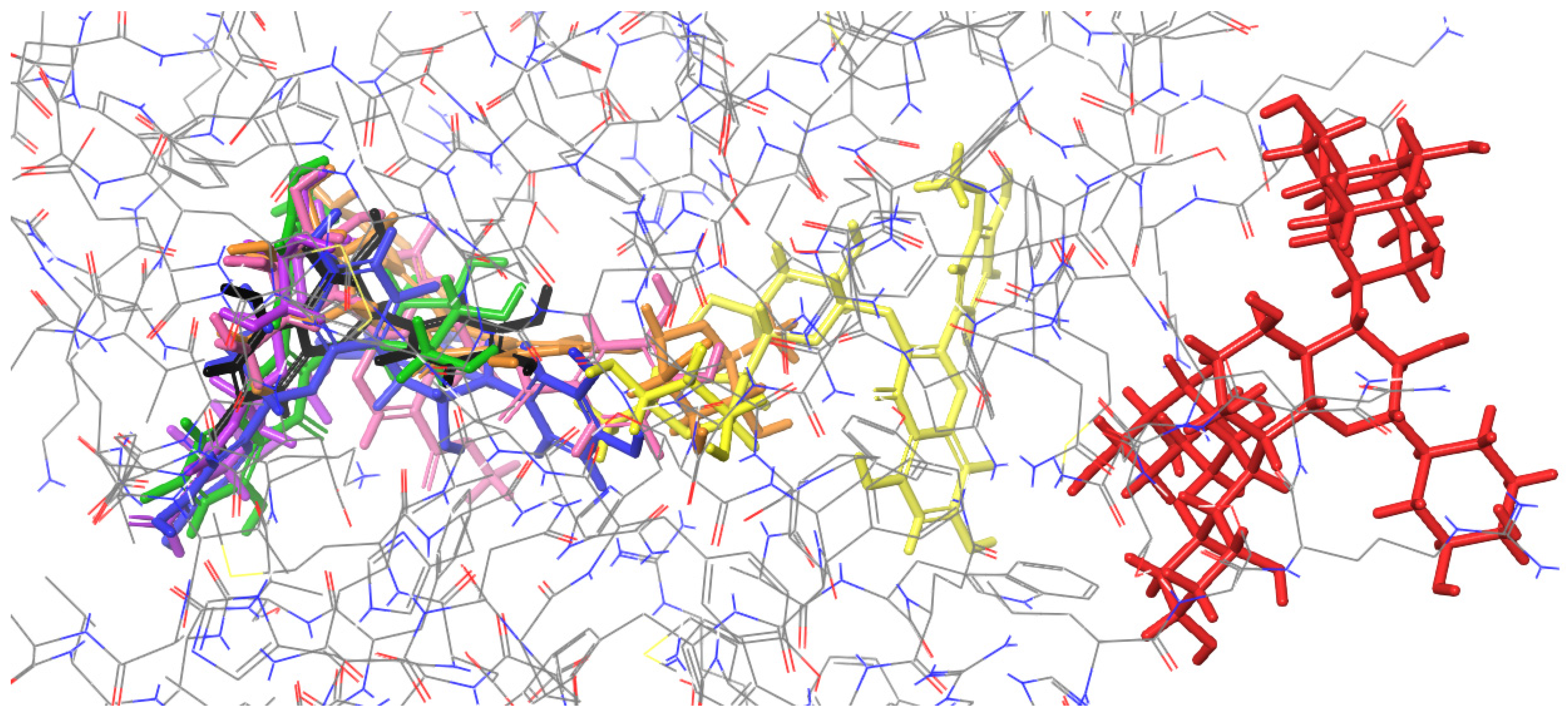
| H-Bond | Cation-π | Salt Bridge | π–π Stacking | Figure | |
|---|---|---|---|---|---|
| Isorhamnetin-3-O-rutinoside 1 | ARG 76, TRP 82, GLU 83, HIE 84 ARG 103, ASN 130, GLY 131 | Figure S10 | |||
| Cornuside | LYS 57, GLY 59, VAL 80, TRP 82, ASN 130 | ARG 103 | Figure S16 | ||
| Loganic acid | LYS 57, VAL 80, TRP 82 | LYS 57 | Figure S17 | ||
| Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside | LYS 57, GLY 59 VAL 80, TRP 82, ASN 130 | ARG 79 | Figure S11 | ||
| Procyanidin C1 | SER 139, GLN 190 | Figure S14 | |||
| Isorhamnetin 3-O-β-D-glucoside | GLY 59, VAL 80, GLU 81 | LYS 32 | LYS 57 | Figure S12 | |
| Procyanidin B2 | SER 29, LYS 32, LYS 57, VAL 80, TRP 82, ARG 103 | ARG 79 | TRP 82 | Figure S13 | |
| Epicatechin | LYS 57, VAL 80, GLU 81, TRP 82 | Figure S15 |
2.3. Molecular Docking to Ribonuclease H
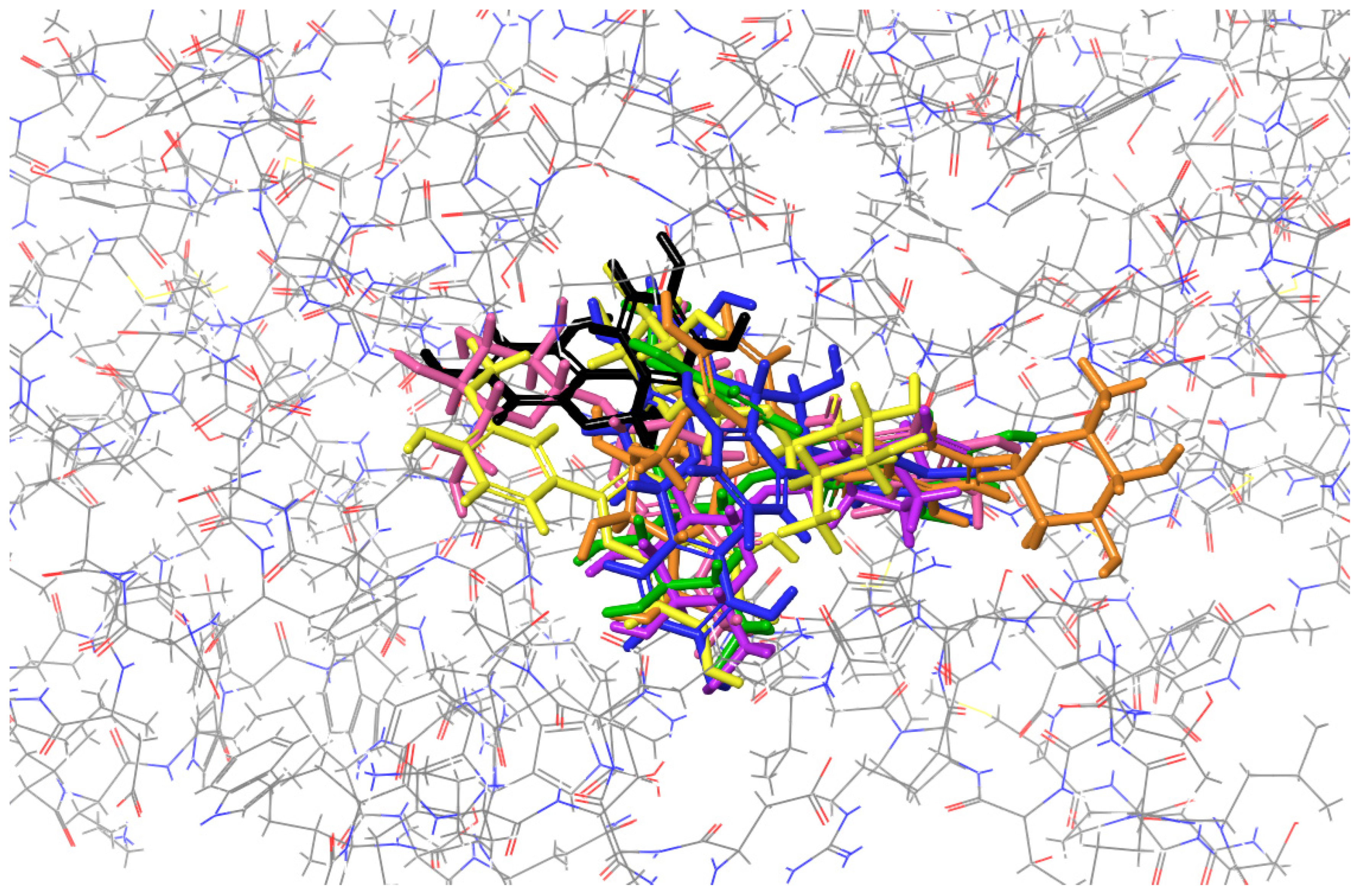
| H-Bond | Cation-π | Salt Bridge | Figure | |
|---|---|---|---|---|
| Procyanidin B2 1 | GLN A: 152, PRO B: 97, ASP B: 102, GLY B: 150, GLN B: 152, VAL B:153 | LYS A:96 | Figure S21 | |
| Cornuside | VAL A: 96, ASN A: 100, GLN A: 152, ASP B: 102, GLY B: 150, GLN B: 152 | Figure S24 | ||
| Isorhamnetin-3-O-rutinoside | THR A: 92, GLN A: 152, VAL B: 98, GLN B: 152 | Figure S19 | ||
| Isorhamnetin 3-O-β-D-glucoside | PRO A: 97, ASP B: 102, VAL B: 153 | Figure S18 | ||
| Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside | PRO A: 97, GLY B: 150 | Figure S20 | ||
| Loganic acid | PRO A: 97, VAL B: 153 | Figure S22 | ||
| Epicatechin | VAL A: 153, GLU A: 154, PRO B: 97 | LYS B: 96 | Figure S23 | |
| Procyanidin C1 | The interaction has not been established. | |||
2.4. Molecular Docking to Thermonuclease
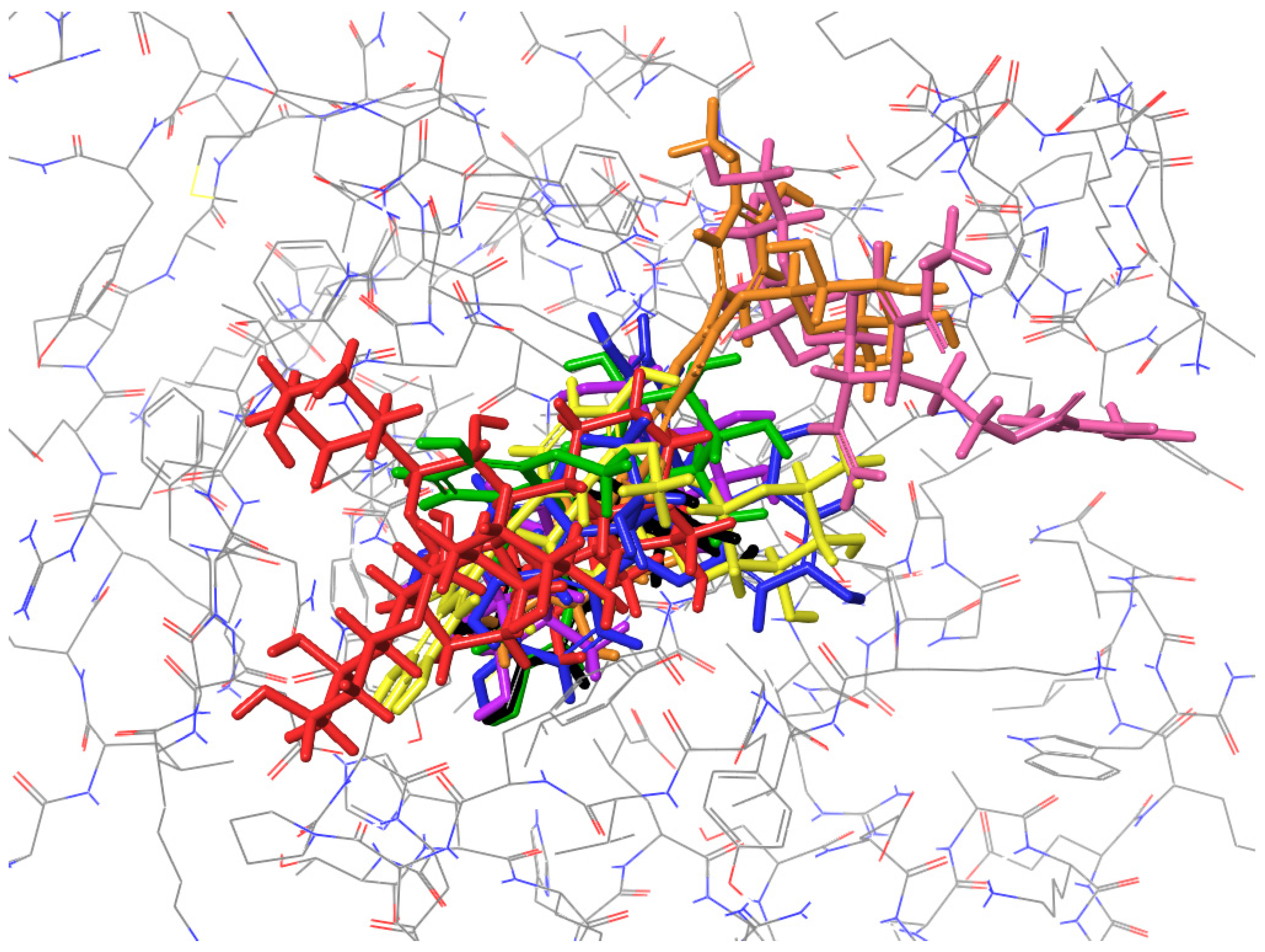
| H-Bond | Salt Bridge | π–π Stacking | Figure | |
|---|---|---|---|---|
| Isorhamnetin-3-O-rutinoside 1 | ASP 40, ASN 118 | Figure S32 | ||
| Epicatechin | LEU 36, ASP 40, TYR 115, ASN 118 | Figure S37 | ||
| Isorhamnetin 3-O-β-D-glucoside | LEU 38, ASP 40, ASP 83, TYR 115, ASN 118 | TYR 115 | Figure S33 | |
| Loganic acid | ASP 40, ASP 83, TYR 115, ASN 118 | Figure S38 | ||
| Procyanidin B2 | ASP 40, LYS 84, ASN 118 | ARG 35, ARG 87 | TYR 115 | Figure S35 |
| Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside | LEU 36, GLU 43, LYS 84, TYR 85, TYR 113 | Figure S34 | ||
| Procyanidin C1 | GLN 80, LYS 84 | Figure S36 | ||
| Cornuside | ASP 21, ARG 35, GLU 43, GLU 52, ARG 87 | LYS 49 | HIE 46 | Figure S39 |
2.5. Molecular Docking to Nuclease SbcCD Subunit C
| H-Bond | Cation-π | Figure | |
|---|---|---|---|
| Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside 1 | PRO 152, PHE 156, ASP 948, GLY 915 | Figure S25 | |
| Isorhamnetin 3-O-β-D-glucoside | PHE 175, LYS 179, TYR 860, GLN 936, SER 937 | Figure S27 | |
| Epicatechin | PRO 152, GLY 915 | Figure S29 | |
| Isorhamnetin-3-O-rutinoside | PRO 152, GLN 153, PHE 156, LYS 157, GLY 915, ASP 948, GLU 949, GLY 950, GLY 952 | LYS 157 | Figure S26 |
| Procyanidin B2 | LEU 174, ASP 176, LYS 179, TYR 860, GLN 936 | LYS 179 | Figure S28 |
| Cornuside | GLU 58, GLU 155, LYS 157, GLY 915 | Figure S30 | |
| Loganic acid | GLY 139, GLN 144, LEU 174, PHE 175, GLN 936 | Figure S31 | |
| Procyanidin C1 | The interaction has not been established. | ||
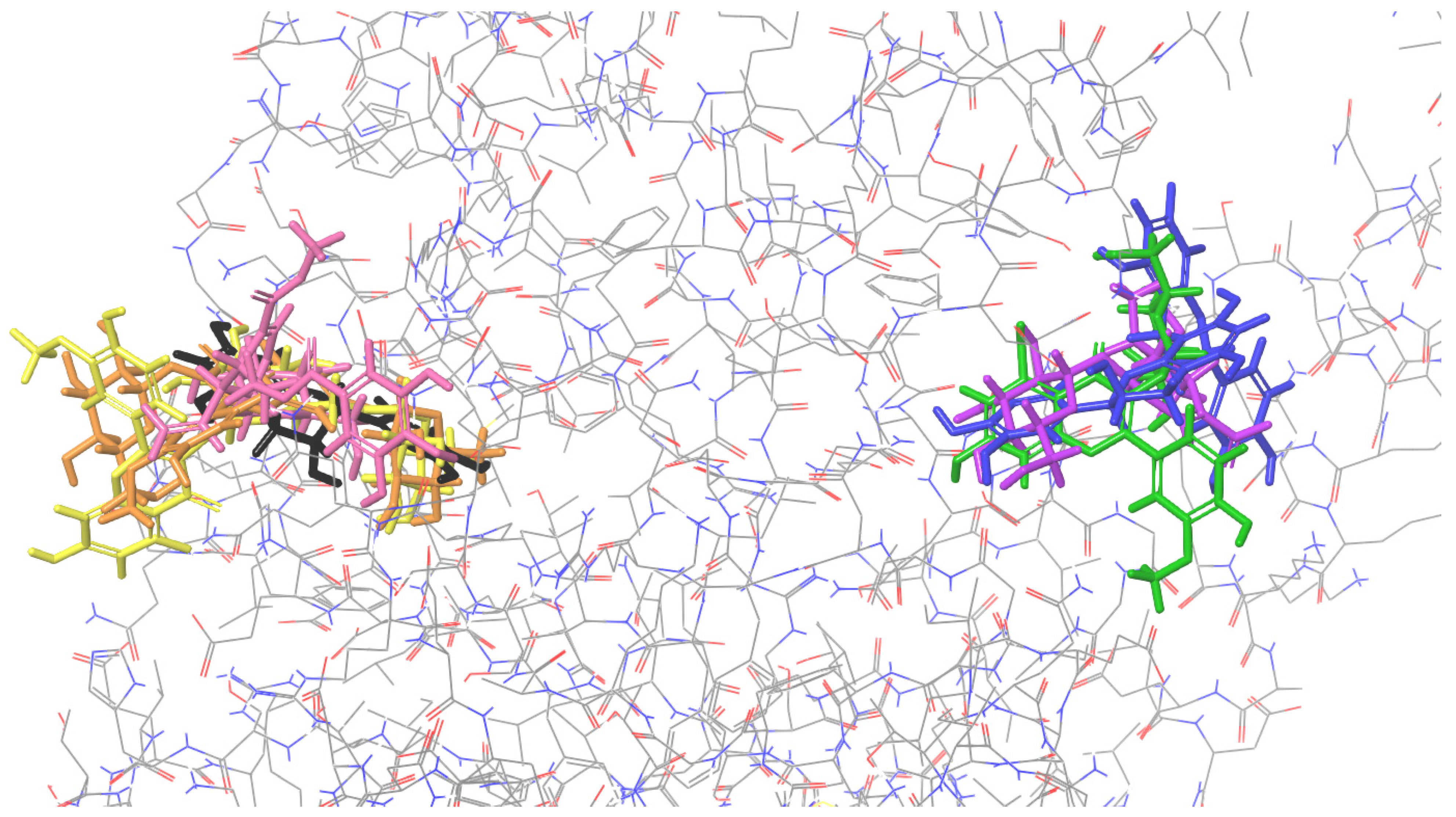
2.6. Summary of the Docking Results
| Colicin E9 | Endonuclease 1 | Ribonuclease H | Thermonuclease | Nuclease SbcCD Subunit C | |
|---|---|---|---|---|---|
| 1 | Procyanidin B2 1 | Isorhamnetin-3-O-rutinoside | Procyanidin B2 | Isorhamnetin-3-O-rutinoside | Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside |
| 2 | Cornuside | Cornuside | Cornuside | Epicatechin | Isorhamnetin 3-O-β-D-glucoside |
| 3 | Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside | Loganic acid | Isorhamnetin-3-O-rutinoside | Isorhamnetin 3-O-β-D-glucoside | Epicatechin |
| 4 | Isorhamnetin-3-O-rutinoside | Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside | Isorhamnetin 3-O-β-D-glucoside | Loganic acid | Isorhamnetin-3-O-rutinoside |
| 5 | Loganic acid | Procyanidin C1 | Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside | Procyanidin B2 | Procyanidin B2 |
| 6 | Isorhamnetin 3-O-β-D-glucoside | Isorhamnetin 3-O-β-D-glucoside | Loganic acid | Isorhamnetin-3-O-β-D-glucosyl-7-O-α-L-rhamnoside | Cornuside |
| 7 | Procyanidin C1 | Procyanidin B2 | Epicatechin | Procyanidin C1 | Loganic acid |
| 8 | Epicatechin | Epicatechin | Procyanidin C1 | Cornuside | Procyanidin C1 |
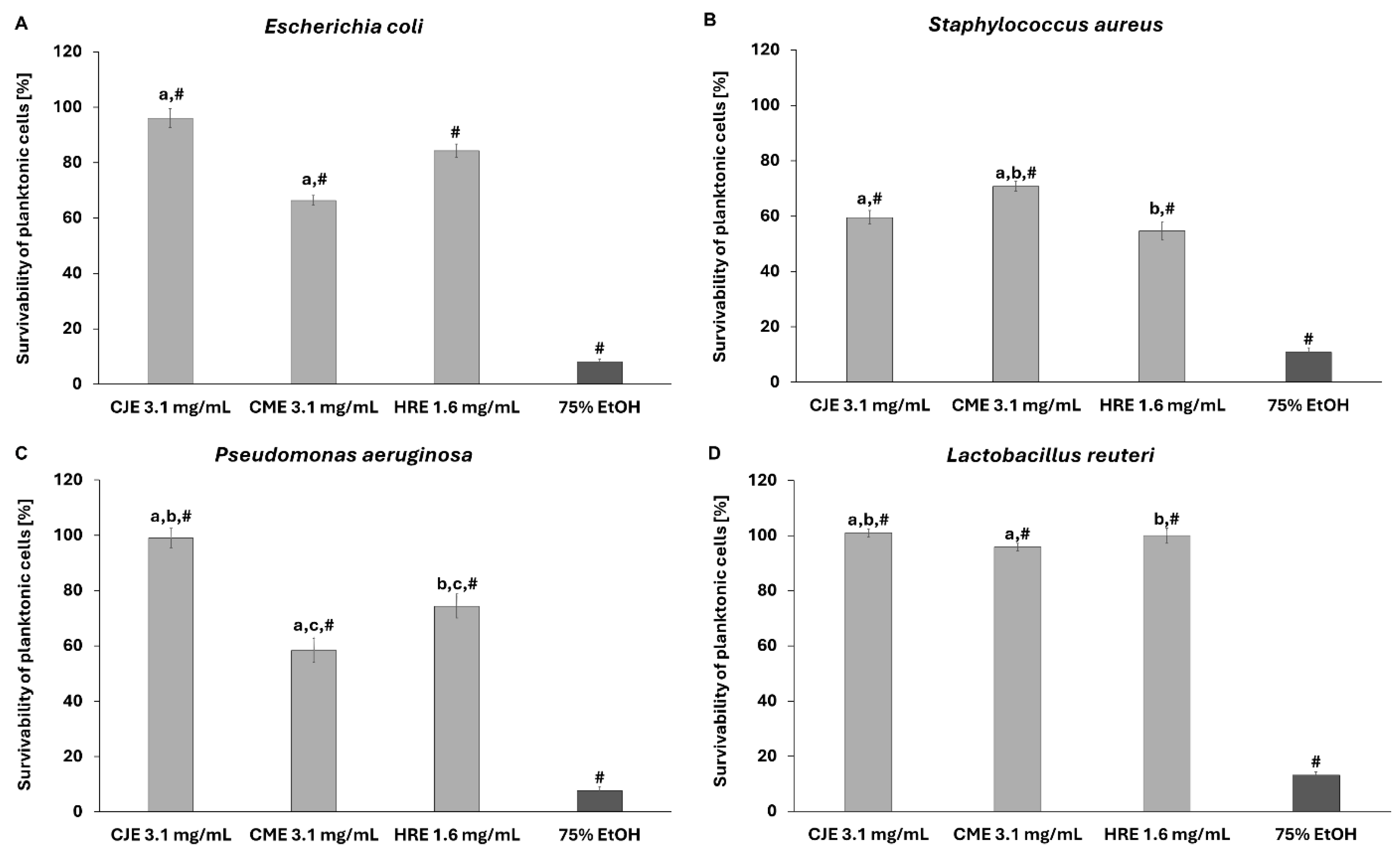
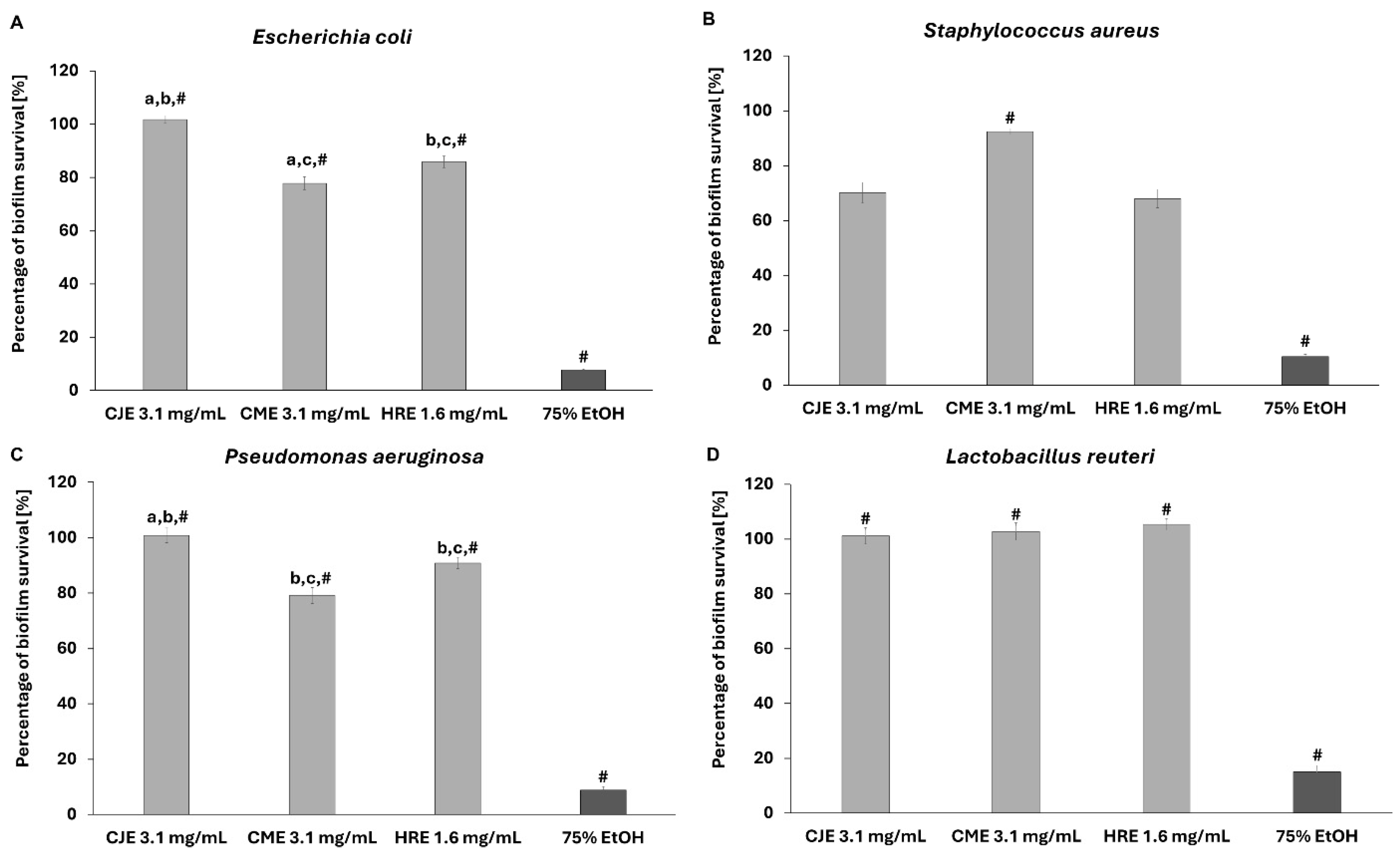
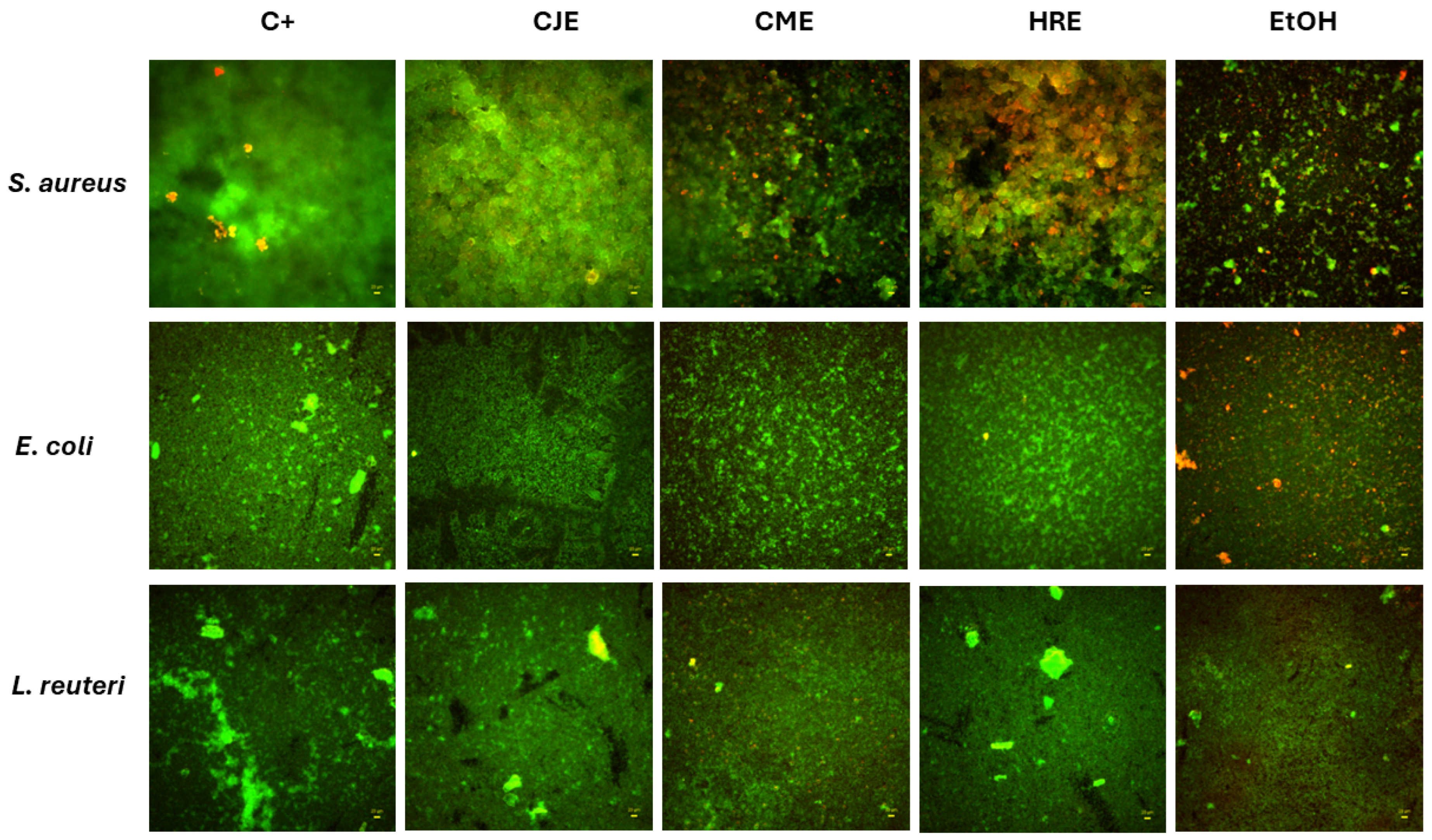
2.7. Effect of Extracts on Fibroblast and Intestinal Cell Lines’ Viability and Toward Pathogenic and Probiotic Microbial Strain
3. Materials and Methods
3.1. Structures Preparation
3.2. Molecular Docking
3.2.1. Active Site Identification and Grid Generation
3.2.2. Compounds Preparation
3.2.3. Glide XP–Compound Docking
3.2.4. MM-GBSA Calculations
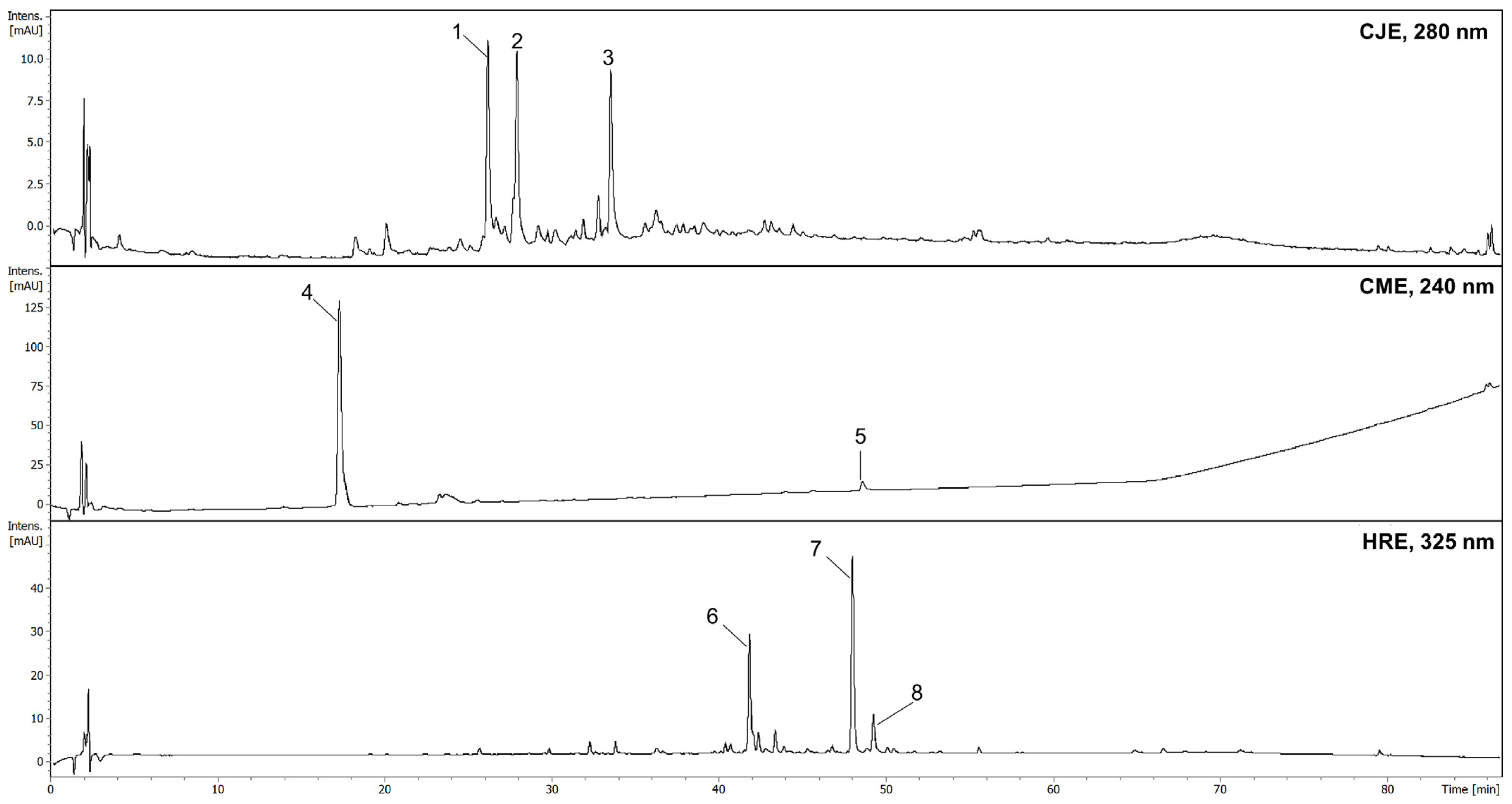
3.3. In Vitro Studies of Extracts Rich in Tested Compounds
3.3.1. Plant Materials and Extract Preparation
3.3.2. Chromatographic Conditions
3.3.3. Evaluation of Extracts’ Cytotoxicity Towards Fibroblast and Caco-2 Cell Line
3.3.4. Evaluation of Extracts’ Activity Towards Pathogens and Probiotic Strain
3.3.5. Spectrophotometric Evaluation of Extracts Activity Against Biofilm Formed by Pathogens and Probiotic Strain
3.3.6. Microscopic Evaluation of Extracts Activity Against Biofilm Formed by Pathogens and Probiotic Strain
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dudek-Wicher, R.K.; Junka, A.; Bartoszewicz, M. The influence of antibiotics and dietary components on gut microbiota. Gastroenterol. Rev. 2018, 13, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Aboobacker, P.A.; Ragunathan, L.; Sanjeevi, T.; Manoharan, A.; Sasi, A.C.; Chandran, V.; Kannaiyan, K.; Samuel, M.S. Synthetic biology’s latest trends in antimicrobial resistance and biofilm. J. Pure Appl. Microbiol. 2023, 17, 23–34. [Google Scholar] [CrossRef]
- Pereira, F.C.; Berry, D. Microbial nutrient niches in the gut. Environ. Microbiol. 2017, 19, 1366–1378. [Google Scholar] [CrossRef] [PubMed]
- Siegień, J.; Buchholz, T.; Popowski, D.; Granica, S.; Osińska, E.; Melzig, M.F.; Czerwińska, M.E. Pancreatic lipase and α-amylase inhibitory activity of extracts from selected plant materials after gastrointestinal digestion in vitro. Food Chem. 2021, 355, 129414. [Google Scholar] [CrossRef] [PubMed]
- Świerczewska, A.; Buchholz, T.; Melzig, M.F.; Czerwińska, M.E. In vitro α-amylase and pancreatic lipase inhibitory activity of Cornus mas L. and Cornus alba L. fruit extracts. J. Food Drug Anal. 2019, 27, 249–258. [Google Scholar] [CrossRef]
- Zhang, L.; Reha-Krantz, L.J. Nuclease. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 118–123. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Tan, M.H.; Koh, E.; Ho, C.L.; Poh, C.L.; Chang, M.W. Reprogramming Microbes to Be Pathogen-Seeking Killers. ACS Synth. Biol. 2014, 3, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Kavutcu, M.; Melzig, M.F. In vitro effects of selected flavonoids on the 5′-nucleotidase activity. Pharmazie 1999, 54, 457–459. [Google Scholar]
- Ma, L.; Cao, X.; Wang, H.; Lu, K.; Wang, Y.; Tu, C.; Dai, Y.; Meng, Y.; Li, Y.; Yu, P.; et al. Discovery of myricetin as a potent inhibitor of human flap endonuclease 1, which potentially can be used as sensitizing agent against HT-29 human colon cancer cells. J. Agric. Food Chem. 2019, 67, 1656–1665. [Google Scholar] [CrossRef]
- Kanakis, C.D.; Nafisi, S.; Rajabi, M.; Shadaloi, A.; Tarantilis, P.A.; Polissiou, M.G.; Bariyanga, J.; Tajmir-Riahi, H.A. Structural analysis of DNA and RNA interactions with antioxidant flavonoids. Spectroscopy 2009, 23, 154321. [Google Scholar] [CrossRef]
- Srivastava, S.; Somasagara, R.R.; Hegde, M.; Nishana, M.; Tadi, S.K.; Srivastava, M.; Choudhary, B.; Raghavan, S.C. Quercetin, a natural flavonoid interacts with DNA, arrests cell cycle and causes tumor regression by activating mitochondrial pathway of apoptosis. Sci. Rep. 2016, 6, 24049. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.; Rocca, R.; Moraca, F.; Talarico, C.; Romeo, I.; Ortuso, F.; Alcaro, S.; Artese, A. A comparative docking strategy to identify polyphenolic derivatives as promising antineoplastic binders of G-quadruplex DNA c-myc and bcl-2 sequences. Mol. Inf. 2016, 35, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.; Karhadkar, S.; Acharya, B.; Banerjee, A.; De, S.; Dasgupta, S. Enhancement of angiogenin inhibition by polyphenol-capped gold nanoparticles. Biopolymers 2021, 112, e23429. [Google Scholar] [CrossRef]
- Ghosh, K.S.; Debnath, J.; Pathak, T.; Dasgupta, S. Using proton nuclear magnetic resonance to study the mode of ribonuclease A inhibition by competitive and noncompetitive inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 5503–5506. [Google Scholar] [CrossRef]
- Ghosh, K.S.; Maiti, T.K.; Debnath, J.; Dasgupta, S. Inhibition of Ribonuclease A by polyphenols present in green tea. Proteins 2007, 69, 566–580. [Google Scholar] [CrossRef]
- Dutta, S.; Basak, A.; Dasgupta, S. Synthesis and ribonuclease A inhibition activity of resorcinol and phloroglucinol derivatives of catechin and epicatechin: Importance of hydroxyl groups. Bioorg. Med. Chem. 2010, 18, 6538–6546. [Google Scholar] [CrossRef]
- Shinozuka, K.; Kikuchi, Y.; Nishino, C.; Mori, A.; Tawata, S. Inhibitory effect of flavonoids on DNA-dependent DNA and RNA polymerases. Experientia 1988, 44, 882–885. [Google Scholar] [CrossRef] [PubMed]
- Turkiewicz, I.P.; Wojdyło, A.; Tkacz, K.; Nowicka, P. UPLC/ESI-Q-TOF-MS analysis of (poly)phenols, tocols and amino acids in Chaenomeles leaves versus in vitro anti-enzyme activities. Ind. Crops Prod. 2022, 181, 114829. [Google Scholar] [CrossRef]
- Olędzka, A.; Cichocka, K.; Woliński, K.; Melzig, M.F.; Czerwińska, M.E. Potentially bio-accessible metabolites from an extract of Cornus mas fruit after gastrointestinal digestion in vitro and gut microbiota ex vivo treatment. Nutrients 2022, 14, 2287. [Google Scholar] [CrossRef]
- Laskowska, A.K.; Wilczak, A.; Skowrońska, W.; Michel, P.; Melzig, M.F.; Czerwińska, M.E. Fruits of Hippophaë rhamnoides in human leukocytes and Caco-2 cell monolayer models—A question about their preventive role in lipopolysaccharide leakage and cytokine secretion in endotoxemia. Front. Pharmacol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.-P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334. [Google Scholar] [CrossRef]
- Nohynek, L.J.; Alakomi, H.L.; Kähkönen, M.P.; Heinonen, M.; Helander, I.M.; Oksman-Caldentey, K.M.; Puupponen-Pimiä, R.H. Berry phenolics: Antimicrobial properties and mechanisms of action against severe human pathogens. Nutr. Cancer 2006, 54, 18–32. [Google Scholar] [CrossRef]
- Szandruk-Bender, M.; Rutkowska, M.; Merwid-Ląd, A.; Wiatrak, B.; Szeląg, A.; Dzimira, S.; Sobieszczańska, B.; Krzystek-Korpacka, M.; Kucharska, A.Z.; Matuszewska, A.; et al. Cornelian cherry iridoid-polyphenolic extract improves mucosal epithelial barrier integrity in rat experimental colitis and exerts antimicrobial and antiadhesive activities in vitro. Oxid. Med. Cell Longev. 2020, 2020, 7697851. [Google Scholar] [CrossRef]
- Hanhineva, K.; Törrönen, R.; Bondia-Pons, I.; Pekkinen, J.; Kolehmainen, M.; Mykkänen, H.; Poutanen, K. Impact of dietary polyphenols on carbohydrate metabolism. Int. J. Mol. Sci. 2010, 11, 1365–1402. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Chen, J.; Ye, X.; Pan, H.; Chen, S. Improving regulation of polymeric proanthocyanidins and tea polyphenols against postprandial hyperglycemia via acid-catalyzed transformation. J. Agric. Food Chem. 2022, 70, 5218–5227. [Google Scholar] [CrossRef]
- Wei, M.; Chai, W.M.; Yang, Q.; Wang, R.; Peng, Y. Novel insights into the inhibitory effect and mechanism of proanthocyanidins from Pyracantha fortuneana fruit on α-glucosidase. J. Food Sci. 2017, 82, 2260–2268. [Google Scholar] [CrossRef]
- Han, L.; Zhang, L.; Ma, W.; Li, D.; Shi, R.; Wang, M. Proanthocyanidin B(2) attenuates postprandial blood glucose and its inhibitory effect on alpha-glucosidase: Analysis by kinetics, fluorescence spectroscopy, atomic force microscopy and molecular docking. Food Funct. 2018, 9, 4673–4682. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Chen, Y.; Ye, X.; Wang, L.; Shao, J.; Jing, H.; Jiang, C.; Wang, H.; Ma, C. Three flavanols delay starch digestion by inhibiting α-amylase and binding with starch. Int. J. Biol. Macromol. 2021, 172, 503–514. [Google Scholar] [CrossRef]
- Tian, J.-L.; Si, X.; Wang, Y.-H.; Gong, E.-S.; Xie, X.; Zhang, Y.; Li, B.; Shu, C. Bioactive flavonoids from Rubus corchorifolius inhibit α-glucosidase and α-amylase to improve postprandial hyperglycemia. Food Chem. 2021, 341, 128149. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ma, Y.; Cheng, G.; Zhang, Y.; Cai, S. Comparative study of dietary flavonoids with different structures as α-glucosidase inhibitors and insulin sensitizers. J. Agric. Food Chem. 2019, 67, 10521–10533. [Google Scholar] [CrossRef]
- Mironova, N.L.; Zenkova, M.A. Vertebrate RNase 1 homologs: Potential regulators of extracellular RNAs. Biotarget 2018, 2, 3. [Google Scholar] [CrossRef]
- Mileo, A.M.; Di Venere, D.; Mardente, S.; Miccadei, S. Artichoke polyphenols sensitize human breast cancer cells to chemotherapeutic drugs via a ROS-mediated downregulation of flap endonuclease 1. Oxid. Med. Cell Longev. 2020, 2020, 7965435. [Google Scholar] [CrossRef]
- Tripathy, D.R.; Panda, A.; Dinda, A.K.; Dasgupta, S. Positional preferences in flavonoids for inhibition of ribonuclease A: Where “—OH” where? Proteins 2021, 89, 577–587. [Google Scholar] [CrossRef]
- Sultana, M.F.; Suzuki, M.; Yamasaki, F.; Kubota, W.; Takahashi, K.; Abo, H.; Kawashima, H. Identification of crucial amino acid eesidues for antimicrobial activity of angiogenin 4 and its modulation of gut microbiota in mice. Front. Microbiol. 2022, 13, 900948. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2015, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Kryszak, B.; Biernat, M.; Tymowicz-Grzyb, P.; Junka, A.; Brożyna, M.; Worek, M.; Dzienny, P.; Antończak, A.; Szustakiewicz, K. The effect of extrusion and injection molding on physical, chemical, and biological properties of PLLA/HAp whiskers composites. Polymer 2023, 287, 126428. [Google Scholar] [CrossRef]
- ISO 10993-5:2009; Biological Evaluation of medical Devices. Part 5: Tests for In Vitro Cytotoxicity. International Organization for Standardization: Geneva, Switzerland, 2009.
- Brożyna, M.; Paleczny, J.; Kozłowska, W.; Ciecholewska-Juśko, D.; Parfieńczyk, A.; Chodaczek, G.; Junka, A. Chemical composition and antibacterial activity of liquid and volatile phase of essential oils against planktonic and biofilm-forming cells of Pseudomonas aeruginosa. Molecules 2022, 27, 4096. [Google Scholar] [CrossRef]
- Paleczny, J.; Junka, A.F.; Krzyżek, P.; Czajkowska, J.; Kramer, A.; Benkhai, H.; Żyfka-Zagrodzińska, E.; Bartoszewicz, M. Comparison of antibiofilm activity of low-concentrated hypochlorites vs polyhexanide-containing antiseptic. Front. Cell Infect. Microbiol. 2023, 13, 1119188. [Google Scholar] [CrossRef]
- Paleczny, J.; Brożyna, M.; Dudek-Wicher, R.; Dydak, K.; Oleksy-Wawrzyniak, M.; Madziała, M.; Bartoszewicz, M.; Junka, A. The medium composition impacts Staphylococcus aureus biofilm formation and susceptibility to antibiotics applied in the treatment of bone infections. Int. J. Mol. Sci. 2022, 23, 11564. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szeleszczuk, Ł.; Brożyna, M.; Dudek, B.; Czarnecki, M.; Junka, A.; Czerwińska, M.E. The Potential Impact of Edible Fruit Extracts on Bacterial Nucleases in Preliminary Research—In Silico and In Vitro Insight. Int. J. Mol. Sci. 2025, 26, 1757. https://doi.org/10.3390/ijms26041757
Szeleszczuk Ł, Brożyna M, Dudek B, Czarnecki M, Junka A, Czerwińska ME. The Potential Impact of Edible Fruit Extracts on Bacterial Nucleases in Preliminary Research—In Silico and In Vitro Insight. International Journal of Molecular Sciences. 2025; 26(4):1757. https://doi.org/10.3390/ijms26041757
Chicago/Turabian StyleSzeleszczuk, Łukasz, Malwina Brożyna, Bartłomiej Dudek, Marcin Czarnecki, Adam Junka, and Monika E. Czerwińska. 2025. "The Potential Impact of Edible Fruit Extracts on Bacterial Nucleases in Preliminary Research—In Silico and In Vitro Insight" International Journal of Molecular Sciences 26, no. 4: 1757. https://doi.org/10.3390/ijms26041757
APA StyleSzeleszczuk, Ł., Brożyna, M., Dudek, B., Czarnecki, M., Junka, A., & Czerwińska, M. E. (2025). The Potential Impact of Edible Fruit Extracts on Bacterial Nucleases in Preliminary Research—In Silico and In Vitro Insight. International Journal of Molecular Sciences, 26(4), 1757. https://doi.org/10.3390/ijms26041757








