Comprehensive Analysis and Comparison of Amino Acid Levels in Cerebrospinal Fluid and Plasma of Children with Leukemia by the LC-MS Technique
Abstract
:1. Introduction
2. Results and Discussion
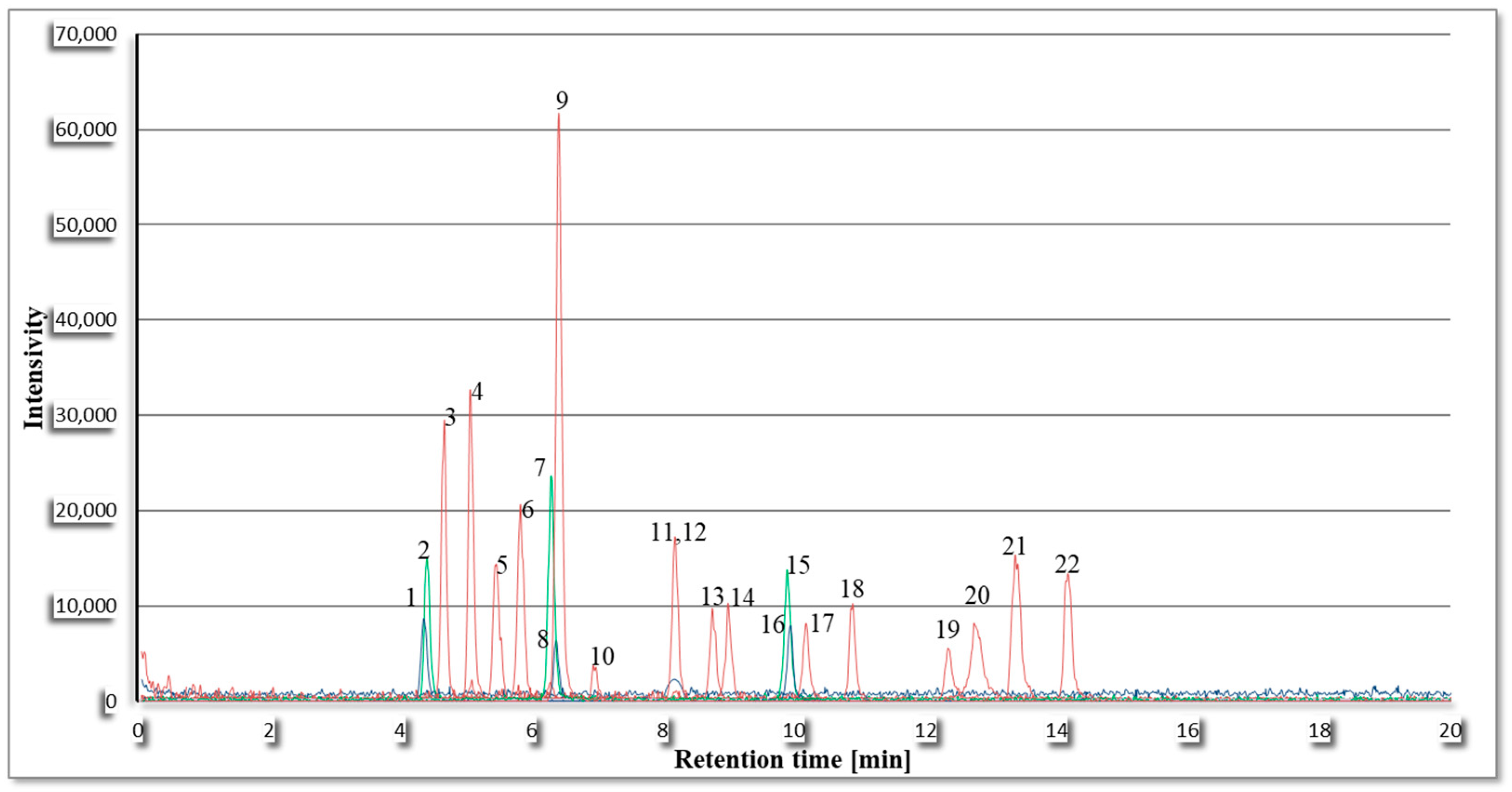
2.1. Optimization of Chromatographic and Mass Spectrometer Analysis Parameters
2.2. Sample Preparation Procedure
2.2.1. Solid-Phase Extraction
2.2.2. Liquid–Liquid Extraction
2.3. Method Validation
2.3.1. Calibration Curve
2.3.2. Specificity
2.3.3. Linearity
2.3.4. Limit of Detection and Limit of Quantification
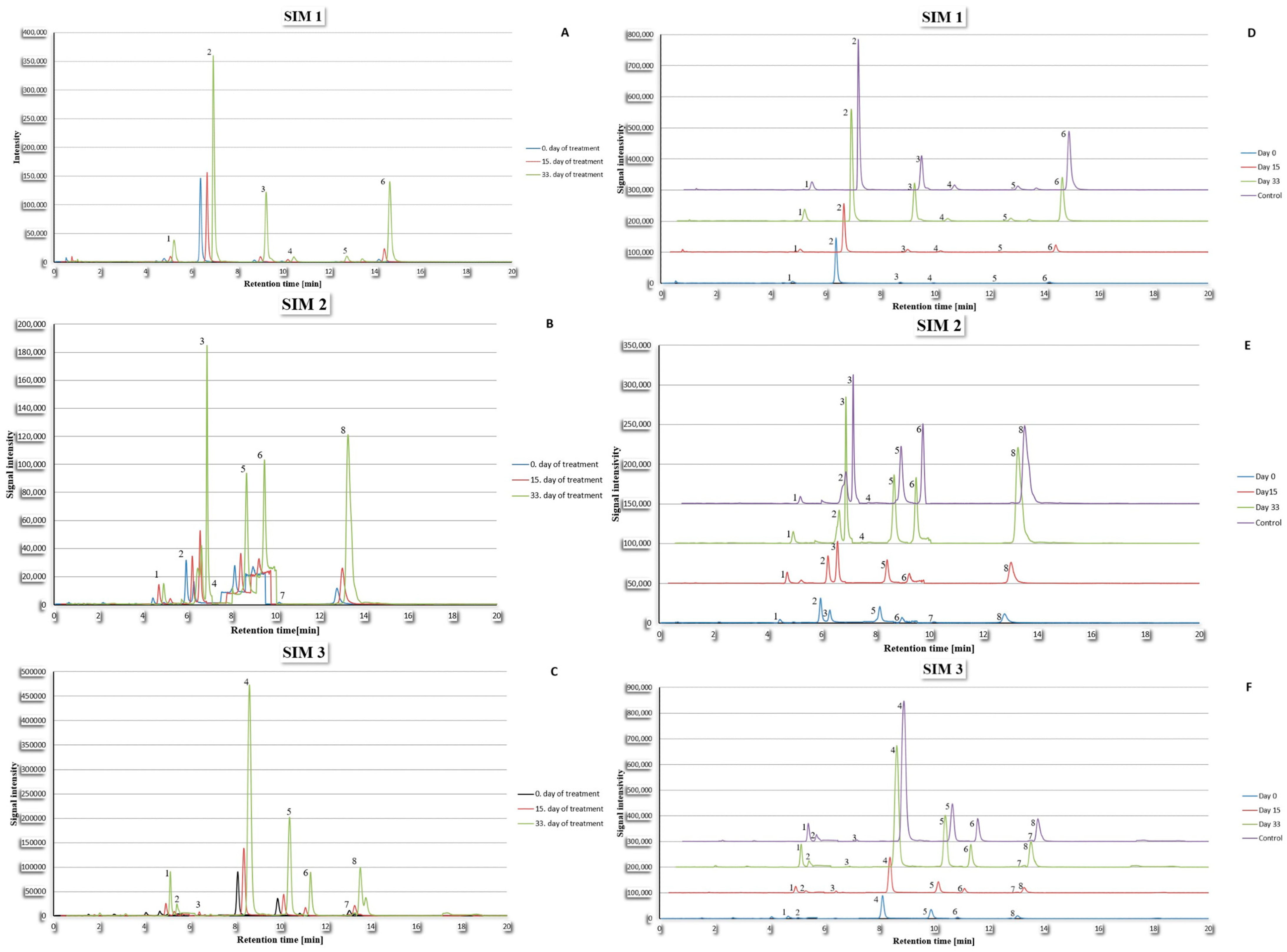
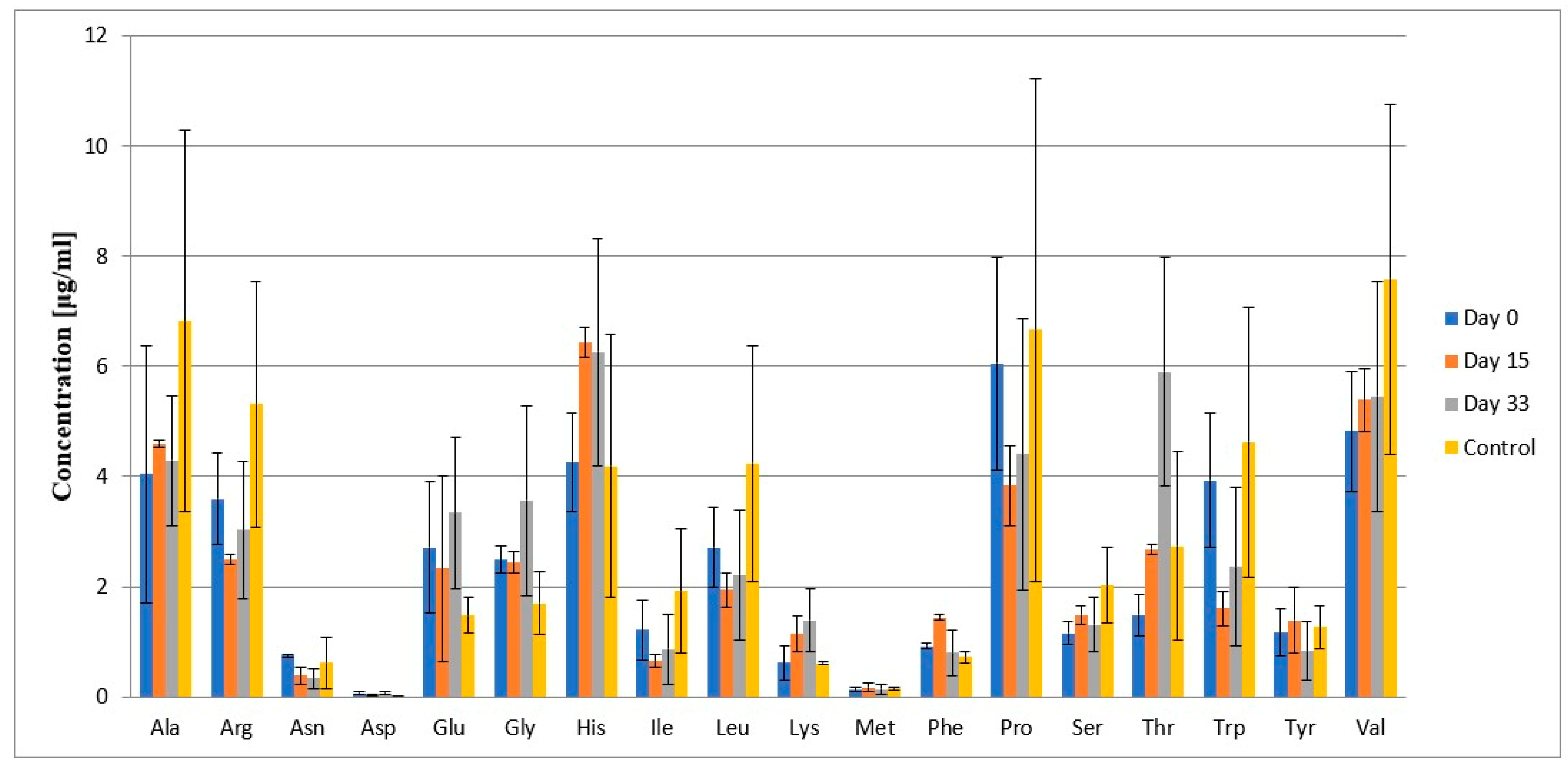
2.4. Results
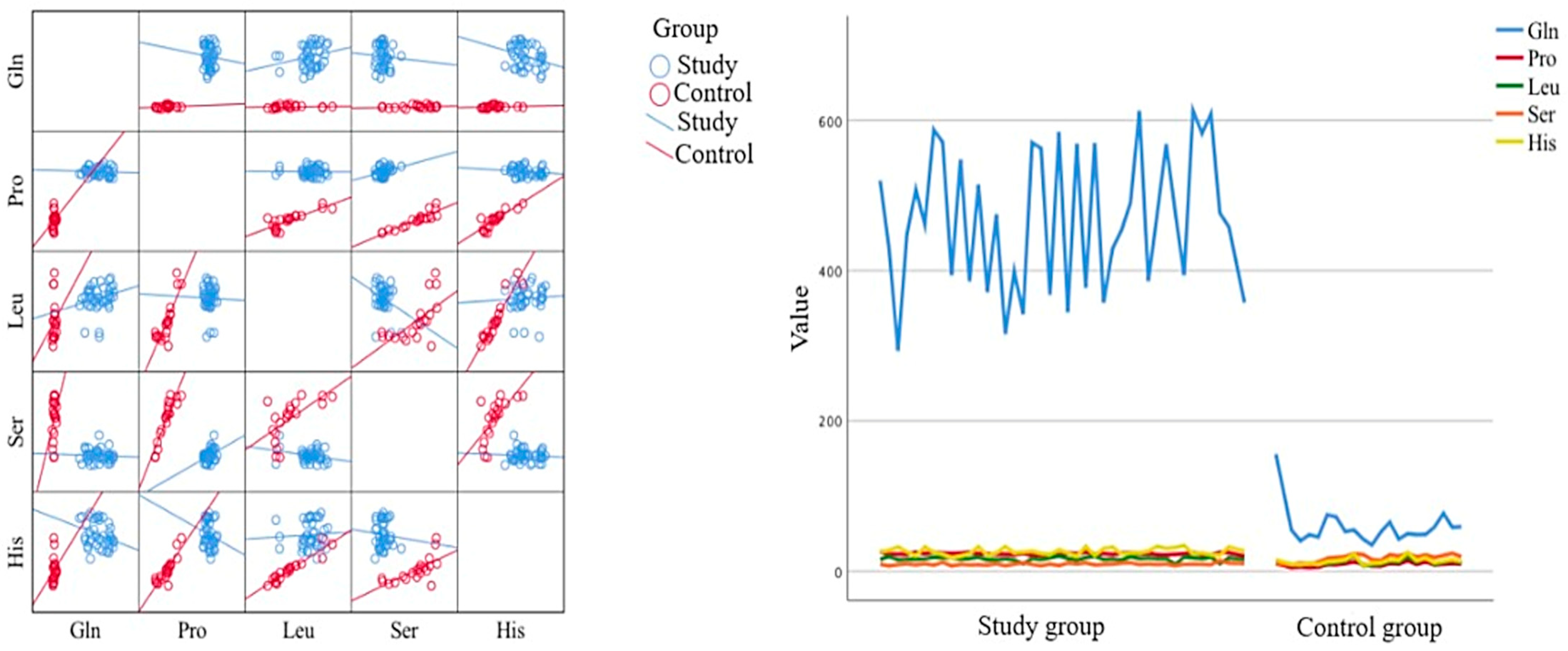
2.5. Statistical Analysis
2.6. Discussion
3. Materials and Methods
3.1. Chemicals
3.2. Instrumentation
3.3. Preparation of Standard Solutions
3.4. Sample Collection
3.5. Sample Preparation
3.6. Final Chromatographic Analysis Parameters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CSF | Cerebrospinal fluid |
| ESI-MS | Electrospray ionization mass spectrometry |
| SIM | Selected Ion Monitoring |
| CE | Collision energy |
| SPE | Solid-phase extraction |
| LOD | Limit of detection |
| LOQ | Limit of quantification |
References
- Wishart, D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef] [PubMed]
- Schraw, J.W.; Woodhouse, J.P.; Bernhardt, M.B.; Taylor, O.A.; Horton, T.M.; Scheurer, M.E.; Okcu, M.F.; Rabin, K.R.; Lupo, P.J.; Brown, A.L. Comparison of the blood, bone marrow, and cerebrospinal fluid metabolomes in children with b-cell acute lymphoblastic leukemia. Sci. Rep. 2021, 11, 19613. [Google Scholar] [CrossRef]
- Brown, A.L.; Sok, P.; Taylor, O.; Woodhouse, P.; Bernhardt, M.B.; Raghubar, K.P.; Kahalley, L.S.; Lupo, P.J.; Hockenberry, M.J.; Scheurer, M.E. Cerebrospinal fluid metabolomic profiles associated with fatigue during treatment for pediatric acute lymphoblastic leukemia. J. Pain Symptom Manag. 2020, 63, 464–473. [Google Scholar] [CrossRef]
- Wu, J.L.; Yu, S.Y.; Wu, S.H.; Bao, A.M. A sensitive and practical RP-HPLC-FLD for determination of the low neuroactive amino acid levels in body fluids and its application in depression. Neurosci. Lett. 2016, 616, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Mingot, M.; Alcatraz, L.A.; MacIntyre, D.A.; Jimenez, B.; Pineda-Lucena, A.; Montiel, V.; Banks, C.E.; Iniesta, J. Development of a novel analytical approach combining the quantification of amino acids, organic acids and glucose using HPLC-UV-Vis and LC-MS/MS with screening via NMR. Anal. Methods 2012, 4, 284. [Google Scholar] [CrossRef]
- Waterval, W.A.; Scheijen, J.L.J.M.; Ortmans-Ploemen, M.J.C.; Habets-van der Poel, C.D.; Bierau, J. Quantitative UHLC-MS/MS analysis of underivatised amino acids in body fluids is a reliable tool for the diagnosis and follow-up of patients with inborn error of metabolism. Clin. Chim. Acta 2009, 407, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Virág, D.; Kiralym, M.; Drahos, L.; Edes, A.E.; Gecse, K.; Bagdy, G.; Juhasz, G.; Antal, I.; Klebovich, I.; Kiss, B.D.; et al. Development, validation and application of LC-MS/MS method for quantification of amino acids, kynurenine and serotonin in human plasma. J. Pharm. Biomed. Anal. 2020, 180, 113018. [Google Scholar] [CrossRef] [PubMed]
- Ohman, A.; Forsgern, L. NMR metabolomics of cerebrospinal fluid distinguishes between Parkinson’s disease and controls. Neurosci. Lett. 2015, 594, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Menkov, T.; D’Alessandro, A.; Reisz, J.A. Metabolic underpinnings of leukemia pathology and treatment. Cancer Rep. 2019, 2, e1139. [Google Scholar] [CrossRef]
- Vettore, L.; Westvrook, R.L.; Tennant, D.A. New aspects of amino acid metabolism in cancer. Br. J. Cancer 2020, 122, 150–156. [Google Scholar] [CrossRef]
- Song, Y.; Xu, C.; Kuroki, H.; Liao, Y.; Tsunoda, M. Recent trends in analytical methods for the determination of amino acids in biological samples. J. Pharm. Biomed. Anal. 2018, 147, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.; Clarke, R.T. Acute leukemia in children: Diagnosis and management. Br. Med. J. 2008, 338, b2285. [Google Scholar] [CrossRef]
- Blau, O.; Hofmann, W.K.; Baldus, C.D.; Thiel, G.; Nolte, F.; Schuemann, E.; Benlasfer, O.; Sindram, A.; Thiel, E.; Blau, I.W. Mesenchimal stroma cells from patients with myelodysplastic syndrome and acute myeloid leukemia show distinct cytogenic and DNA-mutation data as compared with bone marrow leukemic blasts. Blood 2009, 114, 4697. [Google Scholar] [CrossRef]
- Akiyama, T.; Tada, H.; Shiokwa, T.; Kobayashi, K.; Yoshinaga, H. Total folate and 5-methyltetrahydrofolate in cerebrospinal fluid of children: Correlation and reference values. Clin. Chem. Lab. Med. 2015, 53, 2009–2014. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.T.; Wu, K.H.; Lan, S.J.; Tsai, J.J.; Tsai, F.J.; Tsai, C.H. Amino acid concentrations in cerebrospinal fluid in children with acute lymphoblastic leukemia undergoing chemotherapy. Eur. J. Cancer 2005, 41, 1158–1163. [Google Scholar] [CrossRef]
- Protas, P.T.; Muszynska-Roslan, K.; Holownia, A.; Grabowska, A.; Krawczuk-Rybak, M.; Braszko, J.J. Cerebrospinal Fluid Changes in the Excitatory Amino Acids Concentration Caused by the Standard Treatment of Acute Lymphoblastic Leukaemia in Children do not Correlate with their Later Cognitive Functioning. Neuropediatrics 2009, 40, 295–297. [Google Scholar] [CrossRef]
- Nuzzo, T.; Mancini, A.; Miroballo, M.; Casamass, A.; Di Maio, A.; Donati, G.; Snsone, G.; Gaetani, L.; Paoletti, F.P.; Isidori, A.; et al. High performance liquid chromatography determination of L-glutamate, L-glutamine and glycine content in brain, cerebrospinal fluid and blood serum of patients affected by Alzheimer’s disease. Amino Acids 2021, 53, 435–449. [Google Scholar] [CrossRef] [PubMed]


| Amino Acid | Recovery [%] | |
|---|---|---|
| Cerebrospinal Fluid | Plasma | |
| Alanine | 96.65 (94.7–98.5) | 95.36 (93.2–98.1) |
| Arginine | 95.22 (93.26–97.2) | 94.43 (92.3–96.5) |
| Asparagine | 93.78 (91.7–95.66) | 94.22 (92.5–96.1) |
| Cysteine | 95.47 (93.5–97.74) | 93.29 (91.43–95.3) |
| Phenylalanine | 98.62 (96.91–100.28) | 99.36 (97.47–101.25) |
| Glycine | 97.55 (95.83–99.34) | 95.24 (93.19–96.98) |
| Glutamine | 99.96 (98.12–101.7) | 100.25 (98.32–102.18) |
| Histidine | 98.88 (96.97–100.45) | 96.33 (94.6–98.33) |
| Homoarginine | 97.66 (95.36–99.56) | 97.25 (95.76–98.97) |
| Hydroxyproline | 100.23 (98.56–101.88) | 99.99 (98.13–102.23) |
| Isoleucine | 101.58 (99.14–103.78) | 100.56 (98.57–102.31) |
| Aspartic acid | 96.33 (95.02–98.21) | 95.37 (93.71–97.13) |
| Glutamic acid | 97.26 (95.2–99.11) | 98.21 (96.5–99.91) |
| Leucine | 96.28 (94.67–98.15) | 95.14 (93.59–96.83) |
| Lysine | 94.55 (92.66–96.32) | 96.32 (94.21–98.38) |
| Methionine | 93.12 (92.01–95.23) | 95.25 (93.49–97.2) |
| Norvaline | 99.52 (97.62–101.6) | 100.01 (98.39–101.75) |
| Proline | 100.02 (98.63–101.81) | 99.83 (96.93–101.2) |
| Serine | 93.55 (92.13–95.17) | 95.18 (93.48–97.08) |
| Threonine | 96.64 (94.9–98.53) | 98.21 (96.33–100.05) |
| Tryptophan | 95.19 (93.17–97.14) | 95.14 (94.03–96.15) |
| Tyrosine | 94.49 (93.17–95.88) | 95.98 (94.12–97.98) |
| Valine | 98.36 (96.8–99.92) | 98.74 (96.64–100.34) |
| Amino Acid | Calibration Curve y = ax + b | R2 * | s ** | |||
|---|---|---|---|---|---|---|
| a | (±SD) 1 | b | (±SD) 2 | |||
| Alanine | 0.23768 | 0.000221 | 0.00432 | 0.017908 | 0.999483 | 0.041639 |
| Arginine | 0.033134 | 0.016347 | 0.010492 | 0.132593 | 0.998403 | 0.308293 |
| Asparagine | 0.130382 | 0.000459 | −0.00026 | 0.003327 | 0.999901 | 0.009052 |
| Phenylalanine | 0.294056 | 0.002133 | 0.017366 | 0.016311 | 0.999632 | 0.041272 |
| Glycine | 0.148241 | 0.001787 | −0.00204 | 0.016742 | 0.999419 | 0.030993 |
| Glutamine | 0.020109 | 0.000104 | −0.00114 | 0.003187 | 0.999814 | 0.007881 |
| Histidine | 0.26369 | 0.000219 | 0.001032 | 0.020489 | 0.999725 | 0.037931 |
| Hydroxyproline | 0.037831 | 0.000274 | 0.001557 | 0.00799 | 0.999582 | 0.021309 |
| Isoleucine | 0.494622 | 0.001565 | −0.00228 | 0.011351 | 0.99992 | 0.030882 |
| Aspartic acid | 0.106095 | 0.000313 | 0.029904 | 0.00239 | 0.999939 | 0.006048 |
| Glutamic acid | 0.173118 | 0.000746 | −0.00321 | 0.005701 | 0.99987 | 0.014426 |
| Leucine | 0.37182 | 0.000214 | 0.021159 | 0.017393 | 0.9998 | 0.040442 |
| Lysine | 0.236996 | 0.001738 | −0.008 | 0.014094 | 0.999678 | 0.032769 |
| Methionine | 0.277166 | 0.000898 | −0.00291 | 0.00687 | 0.999926 | 0.017384 |
| Proline | 1,120,977 | 0.004009 | −0.00327 | 0.029084 | 0.999898 | 0.079128 |
| Serine | 0.087556 | 0.001281 | 0.048316 | 0.009297 | 0.998289 | 0.025293 |
| Threonine | 0.113684 | 0.000758 | 0.044726 | 0.0058 | 0.999689 | 0.014677 |
| Tryptophan | 0.211539 | 0.001083 | −0.00296 | 0.008282 | 0.9998 | 0.020957 |
| Tyrosine | 0.066164 | 0.000435 | 0.003175 | 0.003326 | 0.999698 | 0.008415 |
| Valine | 0.297504 | 0.002691 | −0.00091 | 0.020578 | 0.999428 | 0.052068 |
| Amino Acid | Patients During Treatment | Control Group | ||||||
|---|---|---|---|---|---|---|---|---|
| Day 0 | Day 15 | Day 33 | ||||||
| Average Concentration [μg/mL] | ±SD ** | Average Concentration [μg/mL] | ±SD ** | Average Concentration [μg/mL] | ±SD ** | Average Concentration [μg/mL] | ±SD ** | |
| Ala | 0.62 | ±0.15 | 0.3 | ±0.10 | 0.53 | ±0.28 | 0.49 | ±0.20 |
| Arg | 0.14 | ±0.11 | 0.11 | ±0.07 | 0.19 | ±0.11 | 0.36 | ±0.17 |
| Asn | 0.07 | ±0.03 | 0.12 | ±0.07 | 0.12 | ±0.07 | 0.13 | ±0.07 |
| Asp | 0.17 | ±0.06 | 0.11 | ±0.06 | 0.2 | ±0.04 | 0.22 | ±0.02 |
| Gln | 456.93 | ±97.11 | 256.29 | ±17.15 | 139.01 | ±11.78 | 80.26 | ±12.8 |
| Glu | 0.28 | ±0.11 | 0.5 | ±0.32 | 0.61 | ±0.35 | 0.18 | ±0.03 |
| Gly | 0.18 | ±0.07 | 0.1 | ±0.03 | 0.08 | ±0.03 | 0.11 | ±0.04 |
| His | 1.61 | ±0.30 | 1.3 | ±0.33 | 1.09 | ±0.34 | 1.65 | ±0.40 |
| Hpro | 391.5 | ±75.67 | 103.71 | ±25.22 | 64.11 | ±11.81 | 50.19 | ±11.70 |
| Ile | 0.09 | ±0.01 | 0.05 | ±0.01 | 0.1 | ±0.04 | 0.07 | ±0.01 |
| Leu | 0.15 | ±0.06 | 0.06 | ±0.02 | 0.18 | ±0.09 | 0.19 | ±0.08 |
| Lys | 1.31 | ±0.37 | 1.49 | ±0.48 | 1.1 | ±0.40 | 1.45 | ±0.79 |
| Phe | 0.06 | ±0.04 | 0.06 | ±0.04 | 0.07 | ±0.05 | 0.05 | ±0.03 |
| Pro | 0.39 | ±0.18 | 0.06 | ±0.04 | 0.15 | ±0.09 | 0.11 | ±0.08 |
| Ser | 0.35 | ±0.04 | 0.34 | ±0.05 | 0.25 | ±0.06 | 0.21 | ±0.07 |
| Thr | 0.67 | ±0.32 | 0.28 | ±0.07 | 0.47 | ±0.14 | 0.26 | ±0.10 |
| Trp | 0.11 | ±0.06 | 0.05 | ±0.03 | 0.09 | ±0.05 | 0.07 | ±0.04 |
| Tyr | 0.1 | ±0.07 | 0.06 | ±0.03 | 0.11 | ±0.03 | 0.13 | ±0.04 |
| Val | 1.71 | ±0.43 | 2.79 | ±1.09 | 0.46 | ±0.15 | 0.53 | ±0.10 |
| Amino Acid | Patients During Treatment | Control Group | ||||||
|---|---|---|---|---|---|---|---|---|
| Day 0 | Day 15 | Day 33 | ||||||
| Average Concentration [μg/mL] | ±SD ** | Average Concentration [μg/mL] | ±SD ** | Average Concentration [μg/mL] | ±SD ** | Average Concentration [μg/mL] | ±SD ** | |
| Ala | 4.05 | ±2.34 | 4.6 | ±0.07 | 4.29 | ±1.17 | 6.83 | ±3.47 |
| Arg | 3.59 | ±0.83 | 2.49 | ±0.09 | 3.03 | ±1.25 | 5.31 | ±2.23 |
| Asn | 0.75 | ±0.04 | 0.39 | ±0.15 | 0.33 | ±0.19 | 0.62 | ±0.46 |
| Asp | 0.06 | ±0.03 | 0.03 | ±0.01 | 0.07 | ±0.02 | 0.01 | ±0.008 |
| Gln | 47.22 | ±23.68 | 95.87 | ±28.83 | 129.03 | ±34.84 | 40.03 | ±15.52 |
| Glu | 2.71 | ±1.19 | 2.33 | ±1.69 | 3.34 | ±1.37 | 1.48 | ±0.33 |
| Gly | 2.49 | ±0.24 | 2.44 | ±0.19 | 3.56 | ±1.73 | 1.7 | ±0.57 |
| His | 4.26 | ±0.90 | 6.43 | ±0.27 | 6.26 | ±2.05 | 4.19 | ±2.38 |
| Hpro | 76.18 | ±10.68 | 142.79 | ±21.06 | 248.24 | ±70.74 | 33.64 | ±15.29 |
| Ile | 1.22 | ±0.54 | 0.65 | ±0.13 | 0.86 | ±0.63 | 1.92 | ±1.13 |
| Leu | 2.71 | ±0.73 | 1.95 | ±0.31 | 2.21 | ±1.18 | 4.24 | ±2.13 |
| Lys | 0.612 | ±0.31 | 1.15 | ±0.32 | 1.38 | ±0.57 | 0.6 | ±0.03 |
| Met | 0.13 | ±0.03 | 0.17 | ±0.08 | 0.14 | ±0.08 | 0.16 | ±0.03 |
| Phe | 0.92 | ±0.05 | 1.44 | ±0.05 | 0.8 | ±0.42 | 0.72 | ±0.11 |
| Pro | 6.05 | ±1.93 | 3.84 | ±0.72 | 4.4 | ±2.46 | 6.66 | ±4.56 |
| Ser | 1.15 | ±0.21 | 1.49 | ±0.18 | 1.31 | ±0.50 | 2.03 | ±0.69 |
| Thr | 1.49 | ±0.37 | 2.69 | ±0.09 | 5.9 | ±2.07 | 2.74 | ±1.71 |
| Trp | 3.93 | ±1.22 | 1.61 | ±0.32 | 2.36 | ±1.43 | 4.63 | ±2.45 |
| Tyr | 1.17 | ±0.43 | 1.39 | ±0.59 | 0.83 | ±0.53 | 1.27 | ±0.39 |
| Val | 4.82 | ±1.09 | 5.39 | ±0.57 | 5.46 | ±2.08 | 7.58 | ±3.182 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaliszewska, A.; Struczyński, P.; Bączek, T.; Niedźwiecki, M.; Konieczna, L. Comprehensive Analysis and Comparison of Amino Acid Levels in Cerebrospinal Fluid and Plasma of Children with Leukemia by the LC-MS Technique. Int. J. Mol. Sci. 2025, 26, 1888. https://doi.org/10.3390/ijms26051888
Kaliszewska A, Struczyński P, Bączek T, Niedźwiecki M, Konieczna L. Comprehensive Analysis and Comparison of Amino Acid Levels in Cerebrospinal Fluid and Plasma of Children with Leukemia by the LC-MS Technique. International Journal of Molecular Sciences. 2025; 26(5):1888. https://doi.org/10.3390/ijms26051888
Chicago/Turabian StyleKaliszewska, Anna, Piotr Struczyński, Tomasz Bączek, Maciej Niedźwiecki, and Lucyna Konieczna. 2025. "Comprehensive Analysis and Comparison of Amino Acid Levels in Cerebrospinal Fluid and Plasma of Children with Leukemia by the LC-MS Technique" International Journal of Molecular Sciences 26, no. 5: 1888. https://doi.org/10.3390/ijms26051888
APA StyleKaliszewska, A., Struczyński, P., Bączek, T., Niedźwiecki, M., & Konieczna, L. (2025). Comprehensive Analysis and Comparison of Amino Acid Levels in Cerebrospinal Fluid and Plasma of Children with Leukemia by the LC-MS Technique. International Journal of Molecular Sciences, 26(5), 1888. https://doi.org/10.3390/ijms26051888






