Calcium Signaling Is a Universal Carbon Source Signal Transducer and Effects an Ionic Memory of Past Carbon Sources
Abstract
:1. Introduction
2. Results
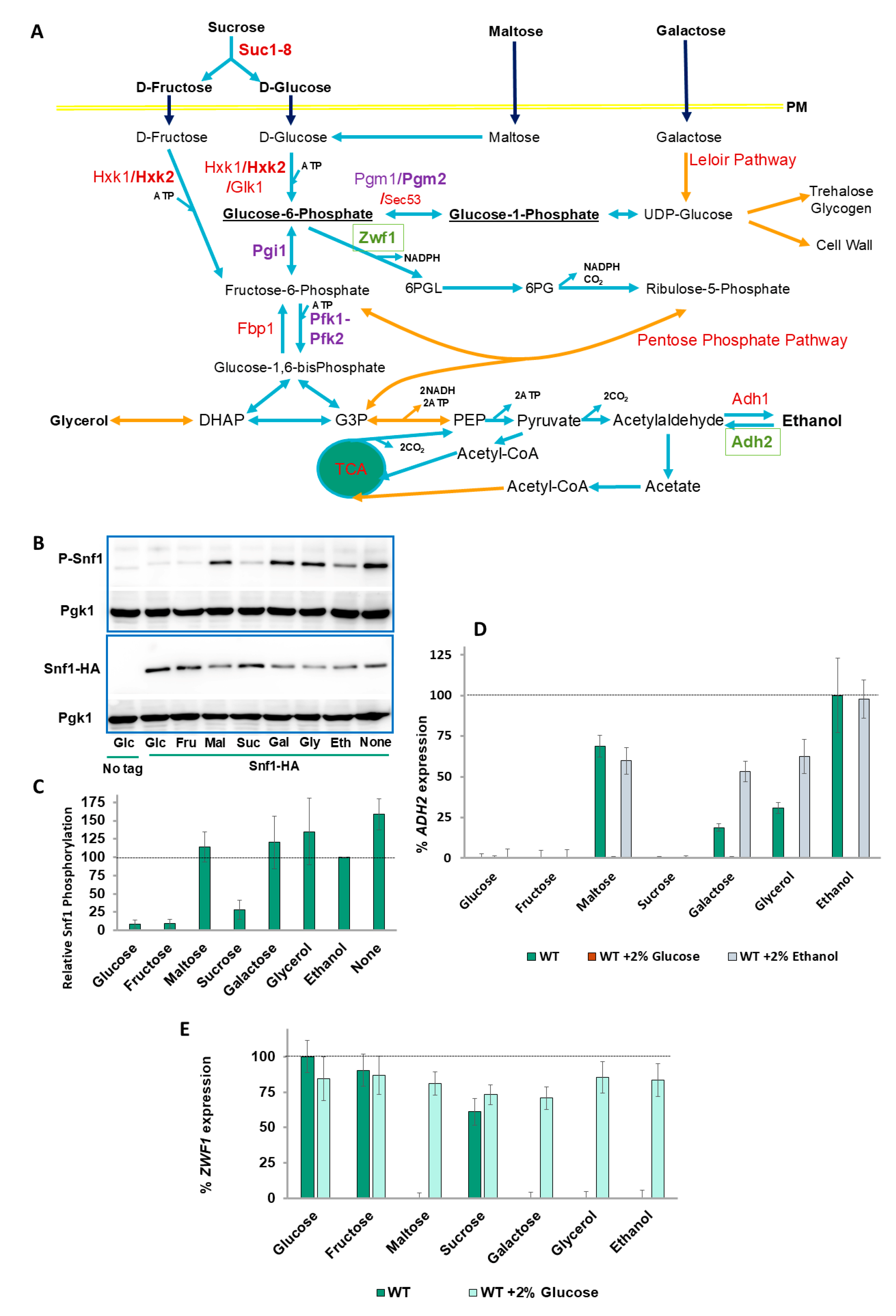
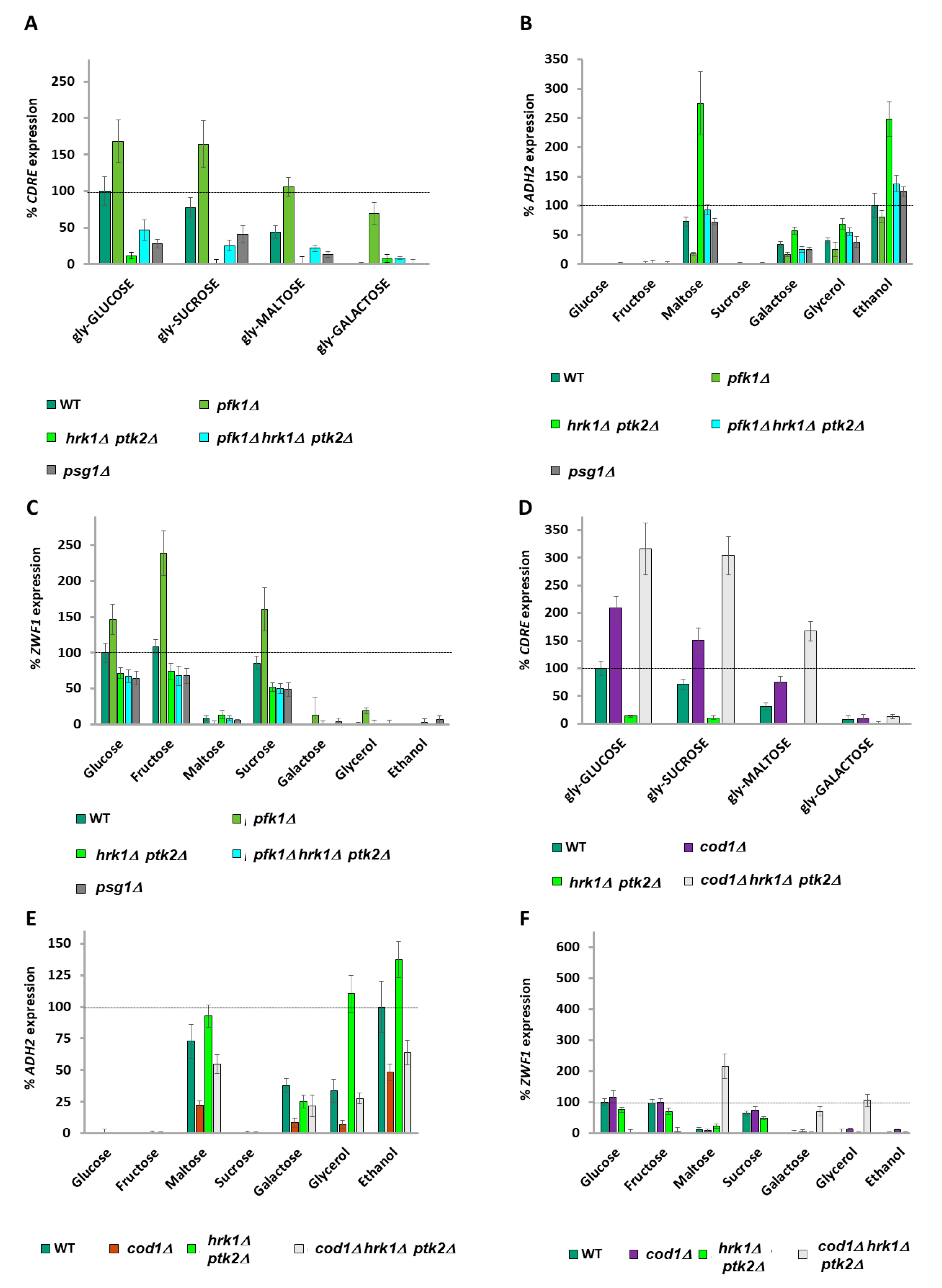
2.1. Snf1 Phosphorylation Levels Do Not Correspond with ADH2 Expression
2.2. ZWF1 Also Shows Differential Regulation by Carbon Source
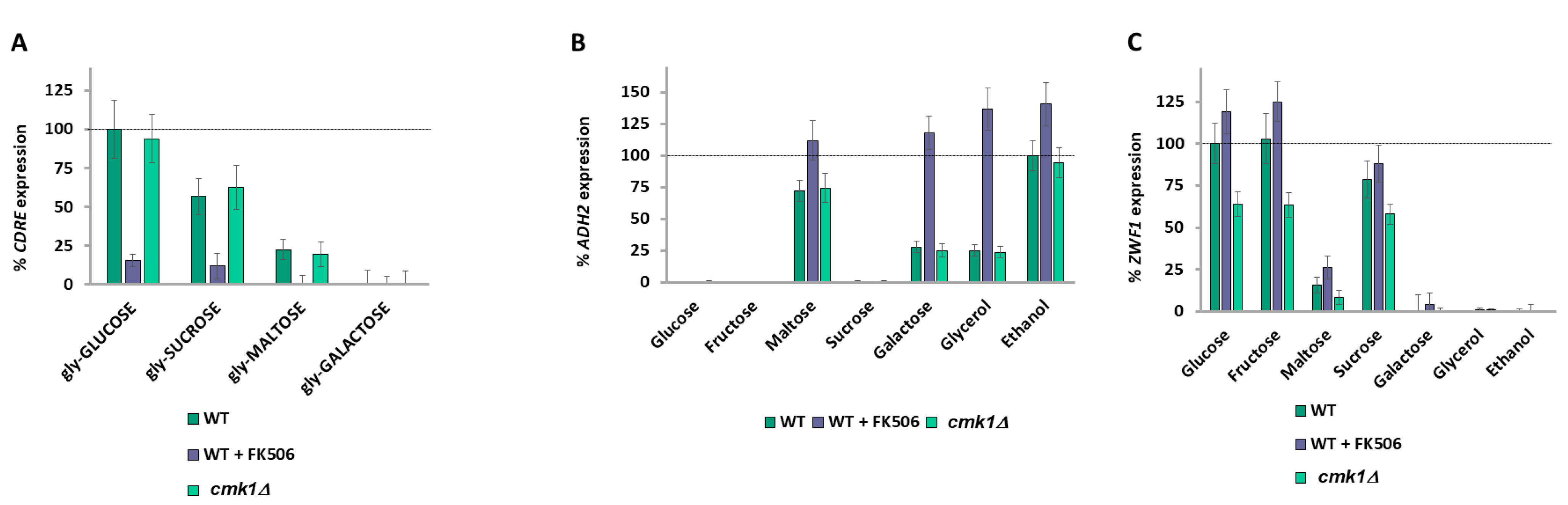
2.3. Snf1, PKA, and the SRR Pathways Do Not Regulate the Carbon Source Differential ADH2 and ZWF1 Expression Levels
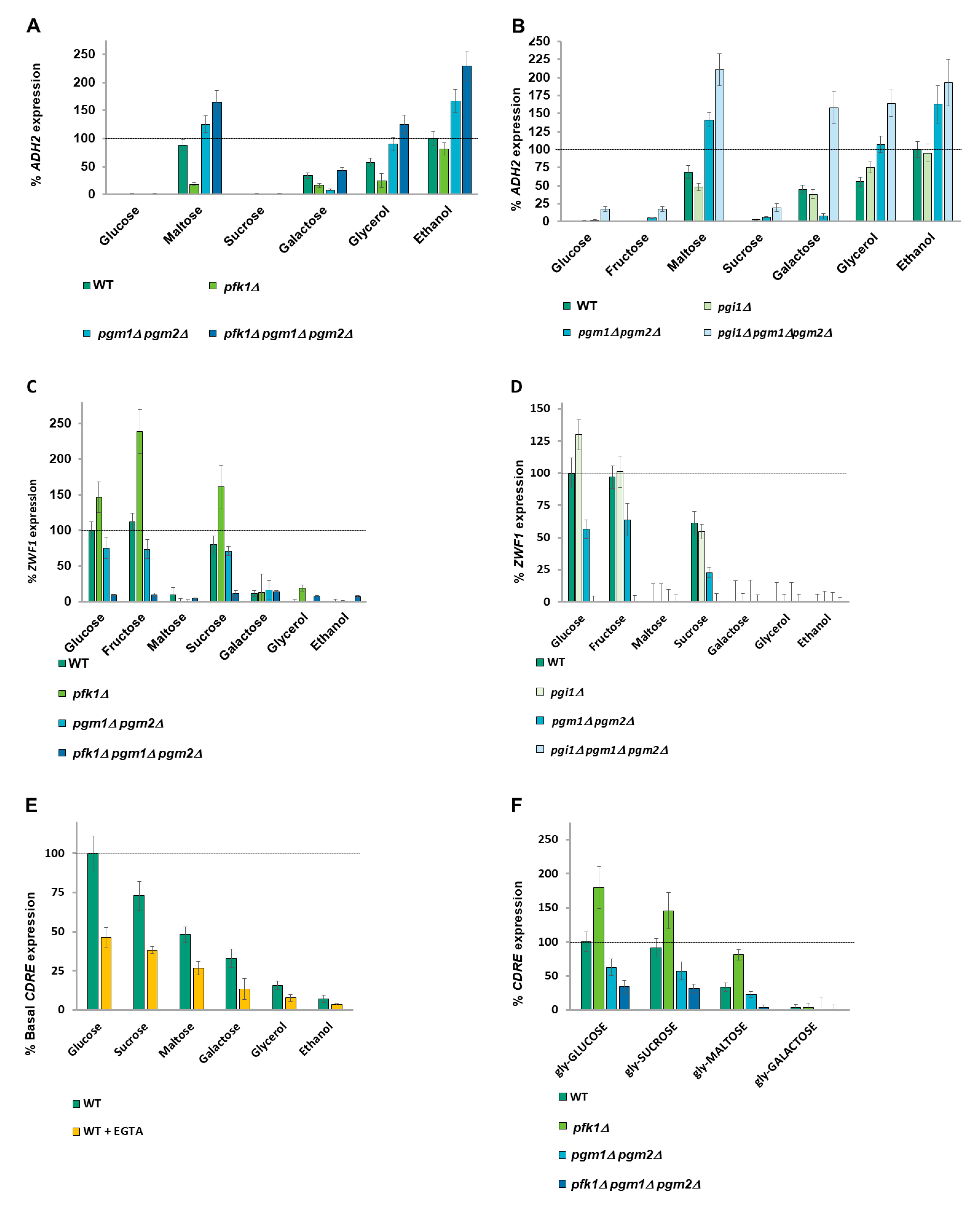
2.4. A Balance of Hexose Metabolism Regulates Gene Expression
2.5. ZWF1 Expression Is Also Regulated by Glucose-6-Phosphate and Glucose-1-Phosphate
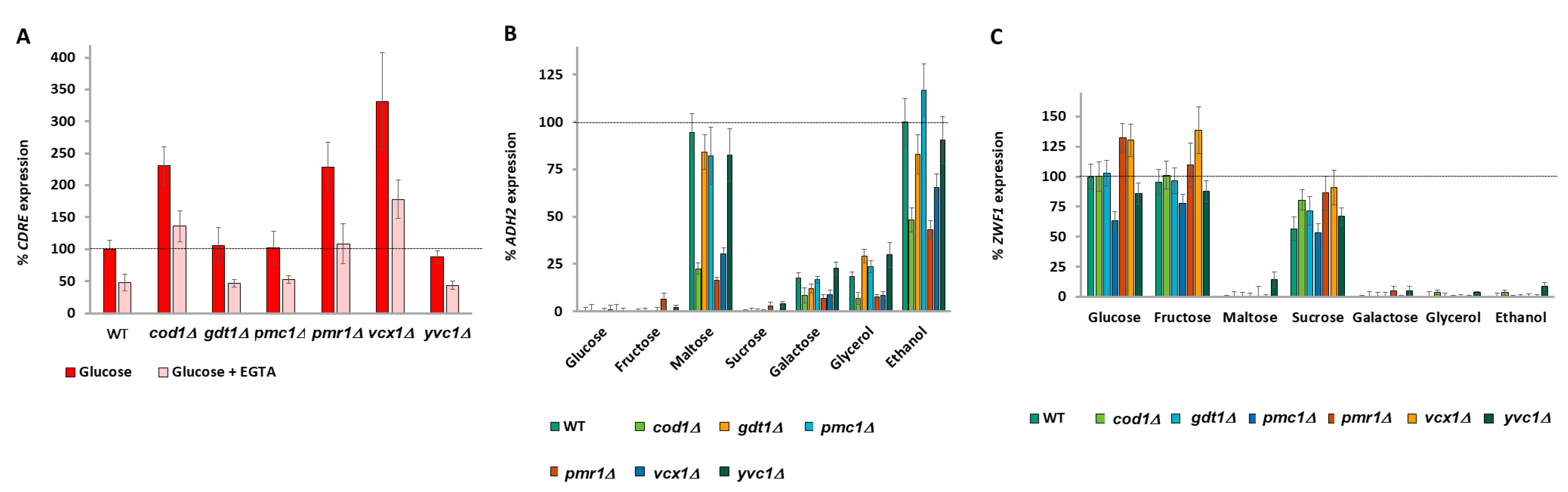
2.6. Calcium Signaling Is Regulated by All Carbon Sources
2.7. Glucose Metabolites Signal to Calcium
2.8. Nucleo-Cytoplasmic Calcium Levels Both Positively and Negatively Regulate Carbon Source-Dependent Gene Expression
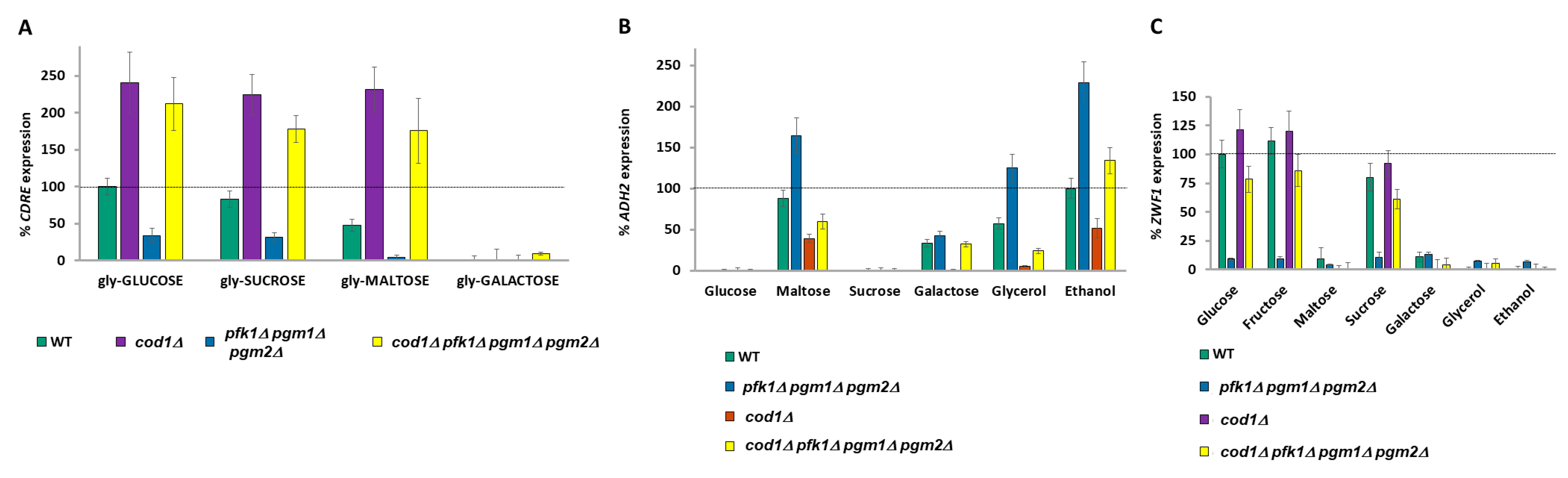
2.9. Raising Nucleo-Cytoplasmic Calcium Levels Suppresses Phenotypes Caused by Low G1P
2.10. The G1P/G6P Ratio Signals to Pma1 Proton Pumping
2.11. Signaling Downstream of Calcium Bifurcates
2.12. Calcium Storage Provides a Memory Mechanism for Previous Meals
3. Discussion
4. Materials and Methods
4.1. Contact for Reagent and Resource Sharing
4.2. Experimental Model and Subject Details
4.3. Figure 7C,D Experiments
4.4. Western Blot Experimental Details
4.5. Serial Dilution Experiment Details
4.6. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simpson-Lavy, K.; Kupiec, M. Carbon catabolite repression: Not only for glucose. Curr. Genet. 2019, 65, 1321–1323. [Google Scholar] [CrossRef] [PubMed]
- Gadura, N.; Robinson, L.C.; Michels, C.A. Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics 2006, 172, 1427–1439. [Google Scholar] [CrossRef] [PubMed]
- Simpson-Lavy, K.J.; Johnston, M. SUMOylation regulates the SNF1 protein kinase. Proc. Natl. Acad. Sci. USA 2013, 110, 17432–17437. [Google Scholar] [CrossRef] [PubMed]
- Moriya, H.; Johnston, M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 2004, 101, 1572–1577. [Google Scholar] [CrossRef]
- Kim, J.-H.; Brachet, V.; Moriya, H.; Johnston, M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot. Cell 2006, 5, 167–173. [Google Scholar] [CrossRef]
- Kim, J.-H.; Johnston, M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 26144–26149. [Google Scholar] [CrossRef]
- Goossens, A.; de La Fuente, N.; Forment, J.; Serrano, R.; Portillo, F. Regulation of yeast H+-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol. Cell. Biol. 2000, 20, 7654–7661. [Google Scholar] [CrossRef]
- Gancedo, J.M. The early steps of glucose signalling in yeast. FEMS Microbiol. Rev. 2008, 32, 673–704. [Google Scholar] [CrossRef]
- Tamaki, H. Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae. J. Biosci. Bioeng. 2007, 104, 245–250. [Google Scholar] [CrossRef]
- Coccetti, P.; Nicastro, R.; Tripodi, F. Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae. Microb. Cell 2018, 5, 482–494. [Google Scholar] [CrossRef]
- Rubenstein, E.M.; McCartney, R.R.; Zhang, C.; Shokat, K.M.; Shirra, M.K.; Arndt, K.M.; Schmidt, M.C. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 2008, 283, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Shashkova, S.; Wollman, A.J.M.; Leake, M.C.; Hohmann, S. The yeast Mig1 transcriptional repressor is dephosphorylated by glucose-dependent and -independent mechanisms. FEMS Microbiol. Lett. 2017, 364, fnx133. [Google Scholar] [CrossRef] [PubMed]
- Snowdon, C.; Schierholtz, R.; Poliszczuk, P.; Hughes, S.; van der Merwe, G. ETP1/YHL010c is a novel gene needed for the adaptation of Saccharomyces cerevisiae to ethanol. FEMS Yeast Res. 2009, 9, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, N.; Jacques, P.-E.; Klimova, N.; Guo, X.; Ricciardi, A.; Robert, F.; Turcotte, B. The switch from fermentation to respiration in Saccharomyces cerevisiae is regulated by the Ert1 transcriptional activator/repressor. Genetics 2014, 198, 547–560. [Google Scholar] [CrossRef]
- Kim, M.S.; Cho, K.H.; Park, K.H.; Jang, J.; Hahn, J.-S. Activation of Haa1 and War1 transcription factors by differential binding of weak acid anions in Saccharomyces cerevisiae. Nucleic Acids Res. 2019, 47, 1211–1224. [Google Scholar] [CrossRef]
- Simpson-Lavy, K.; Kupiec, M. Carbon Catabolite Repression in Yeast is Not Limited to Glucose. Sci. Rep. 2019, 9, 6491. [Google Scholar] [CrossRef]
- Smits, H.; Smits, G.J.; Postma, P.W.; Walsh, M.C.; van Dam, K. High-affinity glucose uptake in Saccharomyces cerevisiae is not dependent on the presence of glucose-phosphorylating enzymes. Yeast 1996, 12, 439–447. [Google Scholar] [CrossRef]
- Nogae, I.; Johnston, M. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene 1990, 96, 161–169. [Google Scholar] [CrossRef]
- Tökés-Füzesi, M.; Bedwell, D.M.; Repa, I.; Sipos, K.; Sümegi, B.; Rab, A.; Miseta, A. Hexose phosphorylation and the putative calcium channel component Mid1p are required for the hexose-induced transient elevation of cytosolic calcium response in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 44, 1299–1308. [Google Scholar] [CrossRef]
- Hong, M.-P.; Vu, K.; Bautos, J.; Gelli, A. Cch1 restores intracellular Ca2+ in fungal cells during endoplasmic reticulum stress. J. Biol. Chem. 2010, 285, 10951–10958. [Google Scholar] [CrossRef]
- Locke, E.G.; Bonilla, M.; Liang, L.; Takita, Y.; Cunningham, K.W. A homolog of voltage-gated Ca2+ channels stimulated by depletion of secretory Ca2+ in yeast. Mol. Cell. Biol. 2000, 20, 6686–6694. [Google Scholar] [CrossRef]
- Batiza, A.F.; Schulz, T.; Masson, P.H. Yeast respond to hypotonic shock with a calcium pulse. J. Biol. Chem. 1996, 271, 23357–23362. [Google Scholar] [CrossRef] [PubMed]
- Paidhungat, M.; Garrett, S. A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol. Cell. Biol. 1997, 17, 6339–6347. [Google Scholar] [CrossRef] [PubMed]
- Miseta, A.; Kellermayer, R.; Aiello, D.P.; Fu, L.; Bedwell, D.M. The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett. 1999, 451, 132–136. [Google Scholar] [CrossRef]
- Miseta, A.; Fu, L.; Kellermayer, R.; Buckley, J.; Bedwell, D.M. The Golgi apparatus plays a significant role in the maintenance of Ca2+ homeostasis in the vps33Delta vacuolar biogenesis mutant of Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 5939–5947. [Google Scholar] [CrossRef]
- Halachmi, D.; Eilam, Y. Calcium homeostasis in yeast cells exposed to high concentrations of calcium. Roles of vacuolar H+-ATPase and cellular ATP. FEBS Lett. 1993, 316, 73–78. [Google Scholar] [CrossRef]
- Groppi, S.; Belotti, F.; Brandão, R.L.; Martegani, E.; Tisi, R. Glucose-induced calcium influx in budding yeast involves a novel calcium transport system and can activate calcineurin. Cell Calcium 2011, 49, 376–386. [Google Scholar] [CrossRef]
- Peiter, E.; Fischer, M.; Sidaway, K.; Roberts, S.K.; Sanders, D. The Saccharomyces cerevisiae Ca2+ channel Cch1pMid1p is essential for tolerance to cold stress and iron toxicity. FEBS Lett. 2005, 579, 5697–5703. [Google Scholar] [CrossRef]
- Viladevall, L.; Serrano, R.; Ruiz, A.; Domenech, G.; Giraldo, J.; Barceló, A.; Ariño, J. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 43614–43624. [Google Scholar] [CrossRef]
- Denis, V.; Cyert, M.S. Internal Ca2+ release in yeast is triggered by hypertonic shock and mediated by a TRP channel homologue. J. Cell Biol. 2002, 156, 29–34. [Google Scholar] [CrossRef]
- Li, X.; Qian, J.; Wang, C.; Zheng, K.; Ye, L.; Fu, Y.; Han, N.; Bian, H.; Pan, J.; Wang, J.; et al. Regulating cytoplasmic calcium homeostasis can reduce aluminum toxicity in yeast. PLoS ONE 2011, 6, e21148. [Google Scholar] [CrossRef] [PubMed]
- Ruta, L.L.; Popa, V.C.; Nicolau, I.; Danet, A.F.; Iordache, V.; Neagoe, A.D.; Farcasanu, I.C. Calcium signaling mediates the response to cadmium toxicity in Saccharomyces cerevisiae cells. FEBS Lett. 2014, 588, 3202–3212. [Google Scholar] [CrossRef]
- Muller, E.M.; Locke, E.G.; Cunningham, K.W. Differential regulation of two Ca2+ influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics 2001, 159, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Nakajima-Shimada, J.; Iida, H.; Tsuji, F.I.; Anraku, Y. Monitoring of intracellular calcium in Saccharomyces cerevisiae with an apoaequorin cDNA expression system. Proc. Natl. Acad. Sci. USA 1991, 88, 6878–6882. [Google Scholar] [CrossRef]
- Kellermayer, R.; Szigeti, R.; Kellermayer, M.; Miseta, A. The intracellular dissipation of cytosolic calcium following glucose re-addition to carbohydrate depleted Saccharomyces cerevisiae. FEBS Lett. 2004, 571, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Pittman, J.K. Vacuolar Ca2+ uptake. Cell Calcium 2011, 50, 139–146. [Google Scholar] [CrossRef]
- Yao, W.; Chen, Y.; Zhang, Y.; Zhong, S.; Ye, M.; Chen, Y.; Fan, S.; Ye, M.; Yang, H.; Li, Y.; et al. Ca2+-triggered Atg11-Bmh1/2-Snf1 complex assembly initiates autophagy upon glucose starvation. J. Cell Biol. 2024, 223, e202310049. [Google Scholar] [CrossRef]
- Thomas, D.; Cherest, H.; Surdin-Kerjan, Y. Identification of the structural gene for glucose-6-phosphate dehydrogenase in yeast. Inactivation leads to a nutritional requirement for organic sulfur. EMBO J. 1991, 10, 547–553. [Google Scholar] [CrossRef]
- Aiello, D.P.; Fu, L.; Miseta, A.; Bedwell, D.M. Intracellular glucose 1-phosphate and glucose 6-phosphate levels modulate Ca2+ homeostasis in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 45751–45758. [Google Scholar] [CrossRef]
- Csutora, P.; Strassz, A.; Boldizsár, F.; Németh, P.; Sipos, K.; Aiello, D.P.; Bedwell, D.M.; Miseta, A. Inhibition of phosphoglucomutase activity by lithium alters cellular calcium homeostasis and signaling in Saccharomyces cerevisiae. Am. J. Physiol. Cell Physiol. 2005, 289, C58–C67. [Google Scholar] [CrossRef]
- Espeso, E.A. The CRaZy Calcium Cycle. Adv. Exp. Med. Biol. 2016, 892, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Locke, T.M.; Fields, R.; Gizinski, H.; Otto, G.M.; MacEwen, M.J.S.; Rusnac, D.-V.; He, P.; Shechner, D.M.; McGann, C.D.; Berg, M.D.; et al. High-throughput identification of calcium-regulated proteins across diverse proteomes. Cell Rep. 2024, 43, 114879. [Google Scholar] [CrossRef] [PubMed]
- Heux, S.; Cadiere, A.; Dequin, S. Glucose utilization of strains lacking PGI1 and expressing a transhydrogenase suggests differences in the pentose phosphate capacity among Saccharomyces cerevisiae strains. FEMS Yeast Res. 2008, 8, 217–224. [Google Scholar] [CrossRef]
- Persson, S.; Welkenhuysen, N.; Shashkova, S.; Cvijovic, M. Fine-Tuning of Energy Levels Regulates SUC2 via a SNF1-Dependent Feedback Loop. Front. Physiol. 2020, 11, 954. [Google Scholar] [CrossRef] [PubMed]
- Meurer, M.; Chevyreva, V.; Cerulus, B.; Knop, M. The regulatable MAL32 promoter in Saccharomyces cerevisiae: Characteristics and tools to facilitate its use. Yeast 2017, 34, 39–49. [Google Scholar] [CrossRef]
- Stockwell, S.R.; Landry, C.R.; Rifkin, S.A. The yeast galactose network as a quantitative model for cellular memory. Mol. Biosyst. 2015, 11, 28–37. [Google Scholar] [CrossRef]
- Schüller, H.J.; Entian, K.D. Isolation and expression analysis of two yeast regulatory genes involved in the derepression of glucose-repressible enzymes. Mol. Gen. Genet. 1987, 209, 366–373. [Google Scholar] [CrossRef]
- Ratnakumar, S.; Kacherovsky, N.; Arms, E.; Young, E.T. Snf1 controls the activity of adr1 through dephosphorylation of Ser230. Genetics 2009, 182, 735–745. [Google Scholar] [CrossRef]
- Spasskaya, D.S.; Karpov, D.S.; Mironov, A.S.; Karpov, V.L. Transcription factor Rpn4 promotes a complex antistress response in Saccharomyces cerevisiae cells exposed to methyl methanesulfonate. Mol. Biol. 2014, 48, 141–149. [Google Scholar] [CrossRef]
- Wang, Y.; Pierce, M.; Schneper, L.; Güldal, C.G.; Zhang, X.; Tavazoie, S.; Broach, J.R. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004, 2, E128. [Google Scholar] [CrossRef]
- Cannon, J.F.; Tatchell, K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 1987, 7, 2653–2663. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, T.; Powers, S.; McGill, C.; Fasano, O.; Strathern, J.; Broach, J.; Wigler, M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell 1984, 37, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Montllor-Albalate, C.; Kim, H.; Thompson, A.E.; Jonke, A.P.; Torres, M.P.; Reddi, A.R. Sod1 integrates oxygen availability to redox regulate NADPH production and the thiol redoxome. Proc. Natl. Acad. Sci. USA 2022, 119, e2023328119. [Google Scholar] [CrossRef]
- Fu, L.; Miseta, A.; Hunton, D.; Marchase, R.B.; Bedwell, D.M. Loss of the major isoform of phosphoglucomutase results in altered calcium homeostasis in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 5431–5440. [Google Scholar] [CrossRef]
- Heinisch, J. Isolation and characterization of the two structural genes coding for phosphofructokinase in yeast. Mol. Gen. Genet. 1986, 202, 75–82. [Google Scholar] [CrossRef]
- Breitenbach-Schmitt, I.; Schmitt, H.D.; Heinisch, J.; Zimmermann, F.K. Genetic and physiological evidence for the existence of a second glycolytic pathway in yeast parallel to the phosphofructokinase-aldolase reaction sequence. Mol. Gen. Genet. 1984, 195, 536–540. [Google Scholar] [CrossRef]
- Klinder, A.; Kirchberger, J.; Edelmann, A.; Kopperschläger, G. Assembly of phosphofructokinase-1 fromSaccharomyces cerevisiae in extracts of single-deletion mutants. Yeast 1998, 14, 323–334. [Google Scholar] [CrossRef]
- Chan, C.-Y.; Parra, K.J. Yeast phosphofructokinase-1 subunit Pfk2p is necessary for pH homeostasis and glucose-dependent vacuolar ATPase reassembly. J. Biol. Chem. 2014, 289, 19448–19457. [Google Scholar] [CrossRef]
- Boles, E.; Liebetrau, W.; Hofmann, M.; Zimmermann, F.K. A family of hexosephosphate mutases in Saccharomyces cerevisiae. Eur. J. Biochem. 1994, 220, 83–96. [Google Scholar] [CrossRef]
- Stathopoulos, A.M.; Cyert, M.S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997, 11, 3432–3444. [Google Scholar] [CrossRef]
- Puigpinós, J.; Casas, C.; Herrero, E. Altered intracellular calcium homeostasis and endoplasmic reticulum redox state in Saccharomyces cerevisiae cells lacking Grx6 glutaredoxin. Mol. Biol. Cell 2015, 26, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, K.W.; Fink, G.R. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 1994, 124, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Colinet, A.-S.; Sengottaiyan, P.; Deschamps, A.; Colsoul, M.-L.; Thines, L.; Demaegd, D.; Duchêne, M.-C.; Foulquier, F.; Hols, P.; Morsomme, P. Yeast Gdt1 is a Golgi-localized calcium transporter required for stress-induced calcium signaling and protein glycosylation. Sci. Rep. 2016, 6, 24282. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.; Frank, C.G.; Jakob, C.A.; Ng, D.T.W. Two distinctly localized p-type ATPases collaborate to maintain organelle homeostasis required for glycoprotein processing and quality control. Mol. Biol. Cell 2002, 13, 3955–3966. [Google Scholar] [CrossRef]
- Zhao, Y.; Du, J.; Zhao, G.; Jiang, L. Activation of calcineurin is mainly responsible for the calcium sensitivity of gene deletion mutations in the genome of budding yeast. Genomics 2013, 101, 49–56. [Google Scholar] [CrossRef]
- D’hooge, P.; Coun, C.; van Eyck, V.; Faes, L.; Ghillebert, R.; Mariën, L.; Winderickx, J.; Callewaert, G. Ca2+ homeostasis in the budding yeast Saccharomyces cerevisiae: Impact of ER/Golgi Ca2+ storage. Cell Calcium 2015, 58, 226–235. [Google Scholar] [CrossRef]
- Ma, T.-Y.; Deprez, M.-A.; Callewaert, G.; Winderickx, J. Coordinated glucose-induced Ca2+ and pH responses in yeast Saccharomyces cerevisiae. Cell Calcium 2021, 100, 102479. [Google Scholar] [CrossRef]
- Simpson-Lavy, K.J.; Kupiec, M. Regulation of yeast Snf1 (AMPK) by a polyhistidine containing pH sensing module. iScience 2022, 25, 105083. [Google Scholar] [CrossRef]
- Geva, Y.; Crissman, J.; Arakel, E.C.; Gómez-Navarro, N.; Chuartzman, S.G.; Stahmer, K.R.; Schwappach, B.; Miller, E.A.; Schuldiner, M. Two novel effectors of trafficking and maturation of the yeast plasma membrane H+-ATPase. Traffic 2017, 18, 672–682. [Google Scholar] [CrossRef]
- Cunningham, K.W.; Fink, G.R. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996, 16, 2226–2237. [Google Scholar] [CrossRef]
- Masuda, C.A.; Xavier, M.A.; Mattos, K.A.; Galina, A.; Montero-Lomeli, M. Phosphoglucomutase is an in vivo lithium target in yeast. J. Biol. Chem. 2001, 276, 37794–37801. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, L.L.; Diderich, J.A.; Slattery, M.G.; Heideman, W. Glucose regulation of Saccharomyces cerevisiae cell cycle genes. Eukaryot. Cell 2003, 2, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.; Serrano, R.; Ariño, J. Direct regulation of genes involved in glucose utilization by the calcium/calcineurin pathway. J. Biol. Chem. 2008, 283, 13923–13933. [Google Scholar] [CrossRef]
- Simpson-Lavy, K.; Xu, T.; Johnston, M.; Kupiec, M. The Std1 Activator of the Snf1/AMPK Kinase Controls Glucose Response in Yeast by a Regulated Protein Aggregation. Mol. Cell 2017, 68, 1120–1133.e3. [Google Scholar] [CrossRef]
- Qu, Y.; Jiang, J.; Liu, X.; Wei, P.; Yang, X.; Tang, C. Cell Cycle Inhibitor Whi5 Records Environmental Information to Coordinate Growth and Division in Yeast. Cell Rep. 2019, 29, 987–994.e5. [Google Scholar] [CrossRef]
- Velivela, S.D.; Kane, P.M. Compensatory Internalization of Pma1 in V-ATPase Mutants in Saccharomyces cerevisiae Requires Calcium- and Glucose-Sensitive Phosphatases. Genetics 2018, 208, 655–672. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, M.K.; Kim, U.; Lee, J.-M.; Hsieh, Y.S.; Seol, G.H. Lavandula angustifolia Mill. inhibits high glucose and nicotine-induced Ca2+ influx in microglia and neuron-like cells via two distinct mechanisms. Biomed. Pharmacother. 2024, 177, 117062. [Google Scholar] [CrossRef]
- Delgadillo-Silva, L.F.; Tasöz, E.; Singh, S.P.; Chawla, P.; Georgiadou, E.; Gompf, A.; Rutter, G.A.; Ninov, N. Optogenetic β cell interrogation in vivo reveals a functional hierarchy directing the Ca2+ response to glucose supported by vitamin B6. Sci. Adv. 2024, 10, eado4513. [Google Scholar] [CrossRef]
- Babenko, V.A.; Varlamova, E.G.; Saidova, A.A.; Turovsky, E.A.; Plotnikov, E.Y. Lactate protects neurons and astrocytes against ischemic injury by modulating Ca2+ homeostasis and inflammatory response. FEBS J. 2024, 291, 1684–1698. [Google Scholar] [CrossRef]
- Zaborska, K.E.; Dadi, P.K.; Dickerson, M.T.; Nakhe, A.Y.; Thorson, A.S.; Schaub, C.M.; Graff, S.M.; Stanley, J.E.; Kondapavuluru, R.S.; Denton, J.S.; et al. Lactate activation of α-cell KATP channels inhibits glucagon secretion by hyperpolarizing the membrane potential and reducing Ca2+ entry. Mol. Metab. 2020, 42, 101056. [Google Scholar] [CrossRef]
- Raghav, D.; Shukla, S.; Jadiya, P. Mitochondrial calcium signaling in non-neuronal cells: Implications for Alzheimer’s disease pathogenesis. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 167169. [Google Scholar] [CrossRef] [PubMed]
- Mitaishvili, E.; Feinsod, H.; David, Z.; Shpigel, J.; Fernandez, C.; Sauane, M.; La Parra, C.D. The Molecular Mechanisms behind Advanced Breast Cancer Metabolism: Warburg Effect, OXPHOS, and Calcium. Front. Biosci. (Landmark Ed.) 2024, 29, 99. [Google Scholar] [CrossRef] [PubMed]
- Saint-Prix, F.; Bönquist, L.; Dequin, S. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: The NADP+-dependent Ald6p and Ald5p isoforms play a major role in acetate formation. Microbiology 2004, 150, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Baker Brachmann, C.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Ma, H.; Kunes, S.; Schatz, P.J.; Botstein, D. Plasmid construction by homologous recombination in yeast. Gene 1987, 58, 201–216. [Google Scholar] [CrossRef]
- Nayak, V.; Zhao, K.; Wyce, A.; Schwartz, M.F.; Lo, W.-S.; Berger, S.L.; Marmorstein, R. Structure and dimerization of the kinase domain from yeast Snf1, a member of the Snf1/AMPK protein family. Structure 2006, 14, 477–485. [Google Scholar] [CrossRef]
- Orlova, M.; Barrett, L.; Kuchin, S. Detection of endogenous Snf1 and its activation state: Application to Saccharomyces and Candida species. Yeast 2008, 25, 745–754. [Google Scholar] [CrossRef]

| Number | Name | Genotype | Source |
|---|---|---|---|
| V5 145 | V5 WT | ho ura3Δ | Dequin lab [83] |
| V5 2 | V5 leu2 | V5 leu2::hisG | This project |
| V5 3 | V5 snf1 | V5 leu2::hisG snf1::NATMX | This project |
| V5 4 | V5 pp | V5 pgm1::NAT pgm2::HygBMX | This project |
| V5 5 | V5 pgi1 | V5 pgi1::KANMX | Dequin lab [43] |
| V5 6 | V5 pp pgi | V5 pgi1::KANMX pgm1::NATMX pgm2::HygBMX | This project |
| V5 7 | V5 pfk1 | V5 pfk1::NATMX (maltose) | This project |
| V5 8 | V5 ppp | V5 pfk1::NATMX pgm1::KANMX pgm2::HygBMX (maltose) | This project |
| V5 9 | V5 pp l2 | V5 leu2::hisG pgm1::NATMX pgm2::HygBMX | This project |
| V5 10 | V5 pp pgi1(hisG) | V5 pgi1::hisG pgm1::NATMX pgm2::HygBMX | This project |
| V5 11 | V5 ppp (hisG) | V5 pfk1::hisG pgm1::NATMXMX pgm2::HygB | This project |
| V5 12 | V5 cod1 | V5 cod1::KANMX | This project |
| V5 13 | V5 gdt2 | V5 gdt2::KANMX | This project |
| V5 14 | V5 pmc1 | V5 pmc1::KANMX | This project |
| V5 15 | V5 pmr1 | V5 pmr1::NATMX | This project |
| V5 16 | V5 vcx1 | V5 vcx1::KANMX | This project |
| V5 17 | V5 yvc1 | V5 ycv1::NATMX | This project |
| V5 18 | V5 pp cod1 | V5 pgm1::NATMX pgm2::HygBMX cod1::KANMX | This project |
| V5 19 | V5 pp vcx1 | V5 pgm1::NATMX pgm2::HygBMX vcx1::KANMX | This project |
| V5 20 | V5 pp pgi1 cod1 | V5 pgi1::hisG pgm1::NATMX pgm2::HygBMX cod1::KANMX | This project |
| V5 21 | V5 ppp cod1 | V5 pfk1::hisG pgm1::NATMX pgm2::HygBMX cod1::KANMX | This project |
| V5 22 | V5 ptk2 hrk1 | V5 hrk1::HygBMX ptk2::NATMX | This project |
| V5 23 | V5 psg1 | V5 psg1::KANMXMX | This project |
| V5 24 | V5 h30 | V5 leu2::hisG hsp30::KANMX | This project |
| V5 25 | V5 cod1 ph | V5 hrk1::HygBMX ptk2::NATMX cod1::KANMX | This project |
| V5 26 | V5 vcx1 ph | V5 hrk1::HygBMX ptk2::NATMX vcx1::KANMX | This project |
| V5 27 | V5 h30pp | V5 leu2::hisG hsp30::KANMX pgm1::NATMX pgm2::HygBMX | This project |
| V5 28 | V5 cmk1 | V5 leu2::hisG cmk1:NATMX (maltose) | This project |
| V5 29 | V5 gal80 | V5 gal80::KANMX | This project |
| V5 30 | V5 pp gal80 | V5 pgm1::KANMX pgm2::HygBMX gal80::NATMX | This project |
| BY | BY4741a his3D0 leu2D0 ura3D0 met15D0 | Lab stock | |
| BY 1 | BY snf1 | BY4741a snf1:HygBMX | P. Cocetti [10] |
| BY2 | BY rgt2snf3 | BY4741a rgt2::KANMX snf3::NATMX | This project |
| BY 2 | BY pp | BY4741a pgm1::KANMX pgm2::HygBMX | This project |
| BY 3 | BY gal80 | BY4741a gal80::KANMX | This project |
| BY 4 | BY pp gal80 | BY4741a pgm1::KANMX pgm2::HygBMX gal80::NATMX | This project |
| Number | Backbone | Contains | Original Backbone | Source |
|---|---|---|---|---|
| 85 | pRS 315 | prSNF1::SNF1-2HA | Berger lab [86] | |
| 319 | pRS 316 | 4x prCDRE::LacZ | pAMS366 | Arino lab [60] |
| 411 | pRS 316 | prZWF1::LacZ | Karpov lab [49] | |
| 467 | pRS 315 | prRAS2::Ras2G19V | pRS 316 | Cannon lab [51] |
| 488 | pRS 316 | prADH1:ADR1S230A | pRS 314 | Young lab [48] |
| 919 | pRS 316 | prADH2::LacZ | Young lab [48] | |
| 1367 | pRS 315 | prPMA1::Pma1D901 | Kupiec lab [68] |
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| mouse anti-HA | Santa Cruz | F7 Sc7392 |
| rabbit anti-phospho T172 AMPK | Cell Signaling | 40H9 2535 |
| Mouse anti-Pgk1 | Abcam | 113687 |
| Mouse anti-polyHIStidine | Abcam | 18184 |
| Goat anti-mouse HRP | Jackson | 115035003 |
| Goat anti-rabbit HRP | Jackson | 111035144 |
| Bacterial and Virus Strains | ||
| DH5a | Lab stock | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| YPER | Pierce | 78990 |
| ONPG | Sigma | N1127 |
| Na2CO3 | Sigma | 791768 |
| Experimental Models: Organisms/Strains | ||
| Please see Table 1 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simpson-Lavy, K.; Kupiec, M. Calcium Signaling Is a Universal Carbon Source Signal Transducer and Effects an Ionic Memory of Past Carbon Sources. Int. J. Mol. Sci. 2025, 26, 2198. https://doi.org/10.3390/ijms26052198
Simpson-Lavy K, Kupiec M. Calcium Signaling Is a Universal Carbon Source Signal Transducer and Effects an Ionic Memory of Past Carbon Sources. International Journal of Molecular Sciences. 2025; 26(5):2198. https://doi.org/10.3390/ijms26052198
Chicago/Turabian StyleSimpson-Lavy, Kobi, and Martin Kupiec. 2025. "Calcium Signaling Is a Universal Carbon Source Signal Transducer and Effects an Ionic Memory of Past Carbon Sources" International Journal of Molecular Sciences 26, no. 5: 2198. https://doi.org/10.3390/ijms26052198
APA StyleSimpson-Lavy, K., & Kupiec, M. (2025). Calcium Signaling Is a Universal Carbon Source Signal Transducer and Effects an Ionic Memory of Past Carbon Sources. International Journal of Molecular Sciences, 26(5), 2198. https://doi.org/10.3390/ijms26052198







