Functional Study of GbSMXL8-Mediated Strigolactone Signaling Pathway in Regulating Cotton Fiber Elongation and Plant Growth
Abstract
1. Introduction
2. Results
2.1. Gene Member Identification of SMXL Family in Cotton
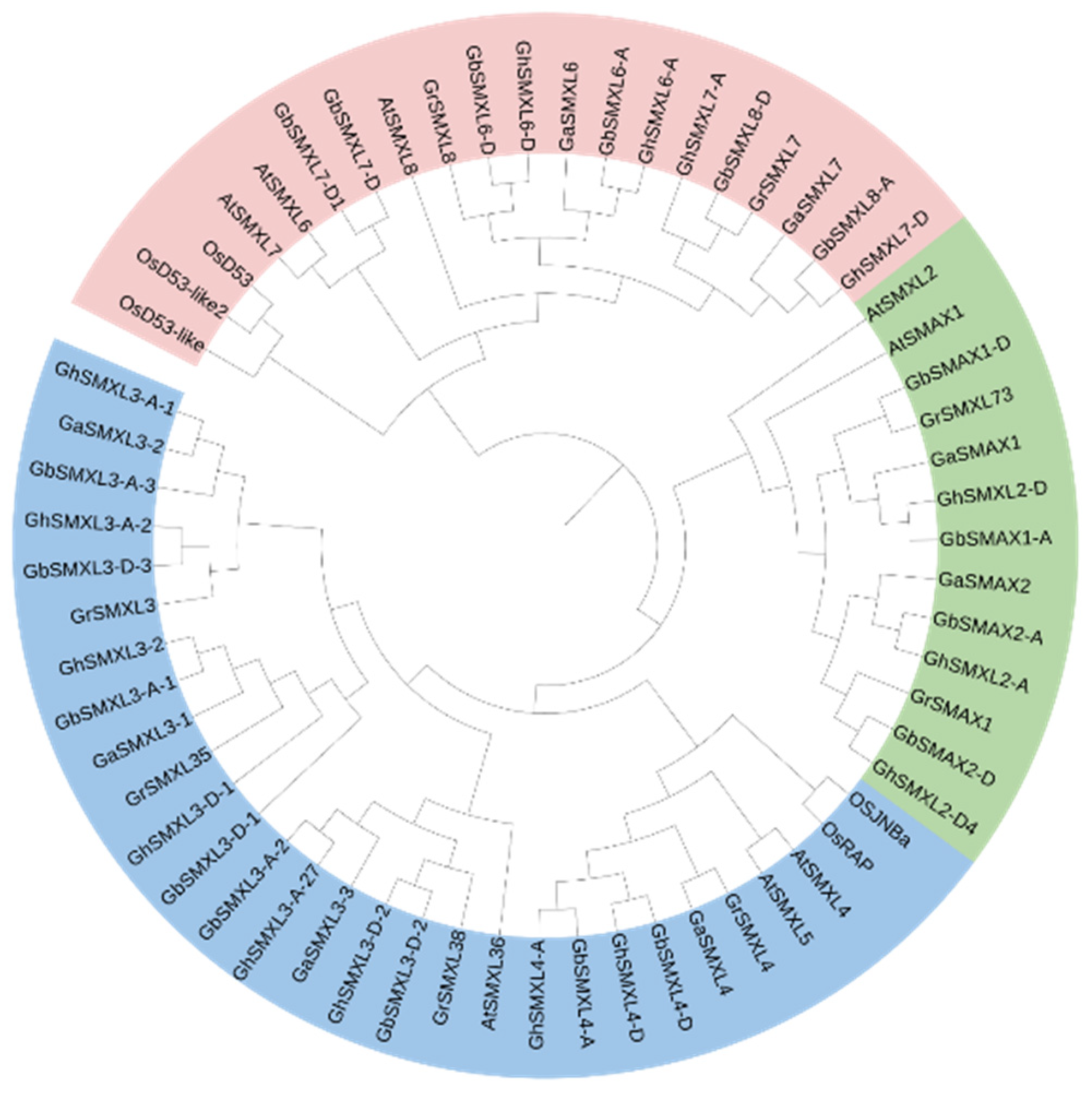
2.2. Phylogenetic Evolution and Gene Structure Analysis of SMXL Family
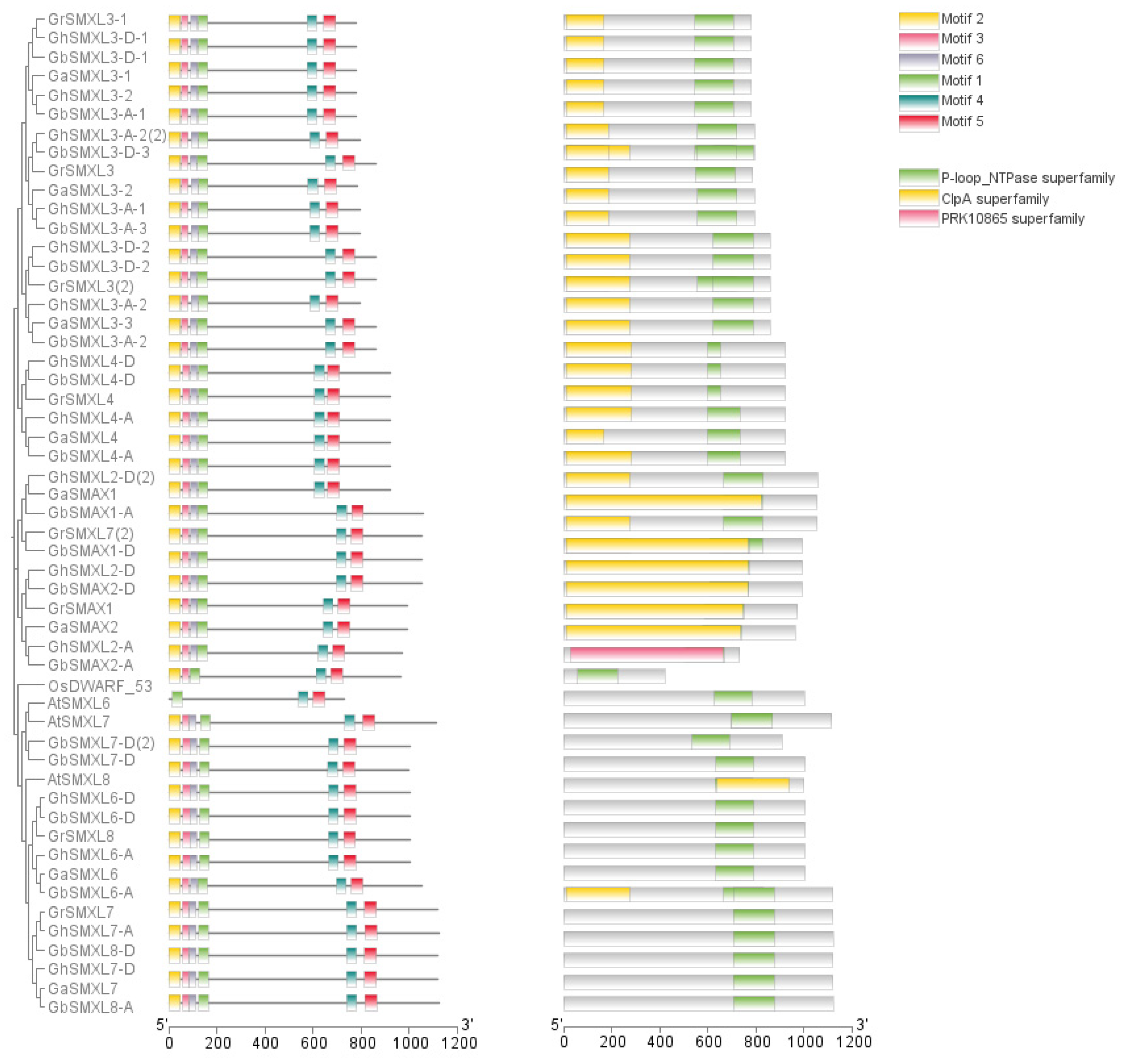
2.3. Analysis of Protein Domains of SMXL Family in Cotton
2.4. Cis-Acting Elements Analysis
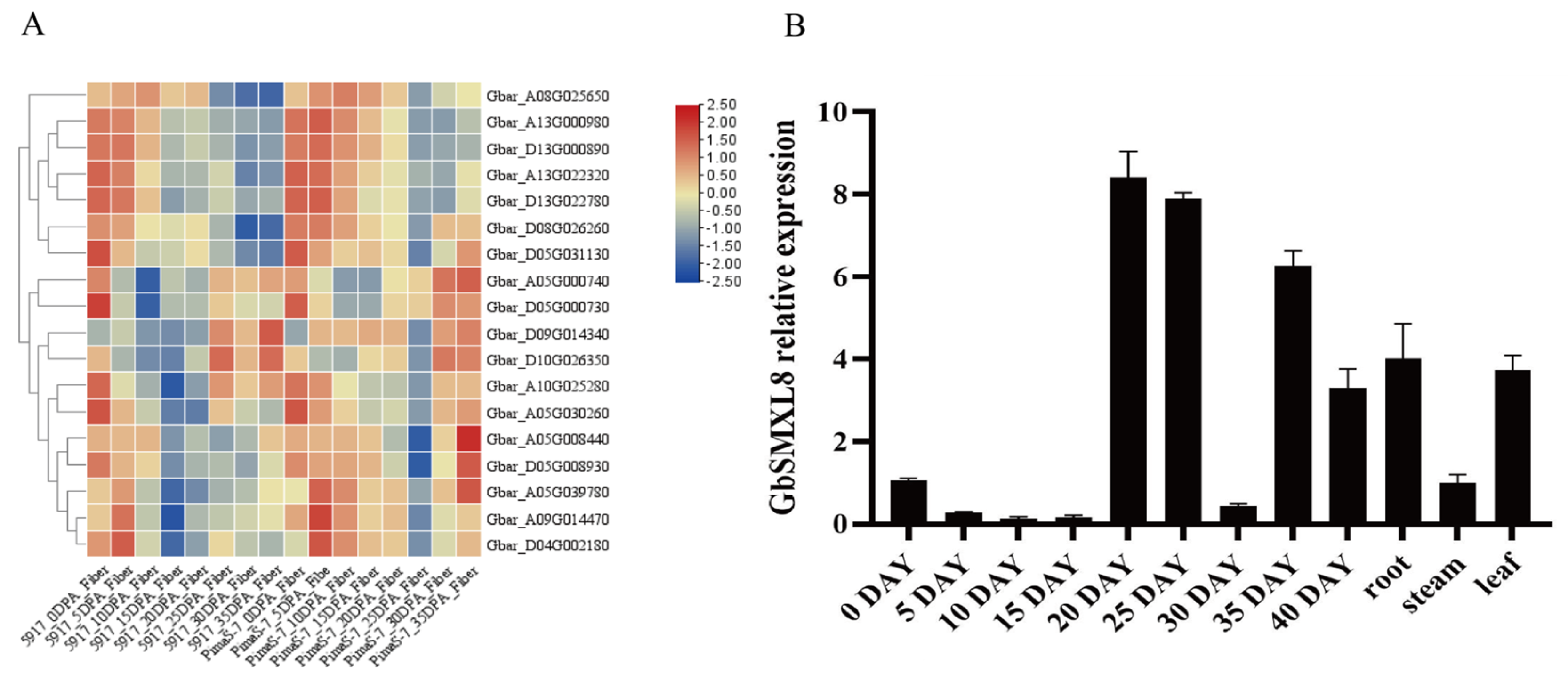
2.5. Analysis of SMXL8 Gene Expression Characteristics in Gossypium barbadense
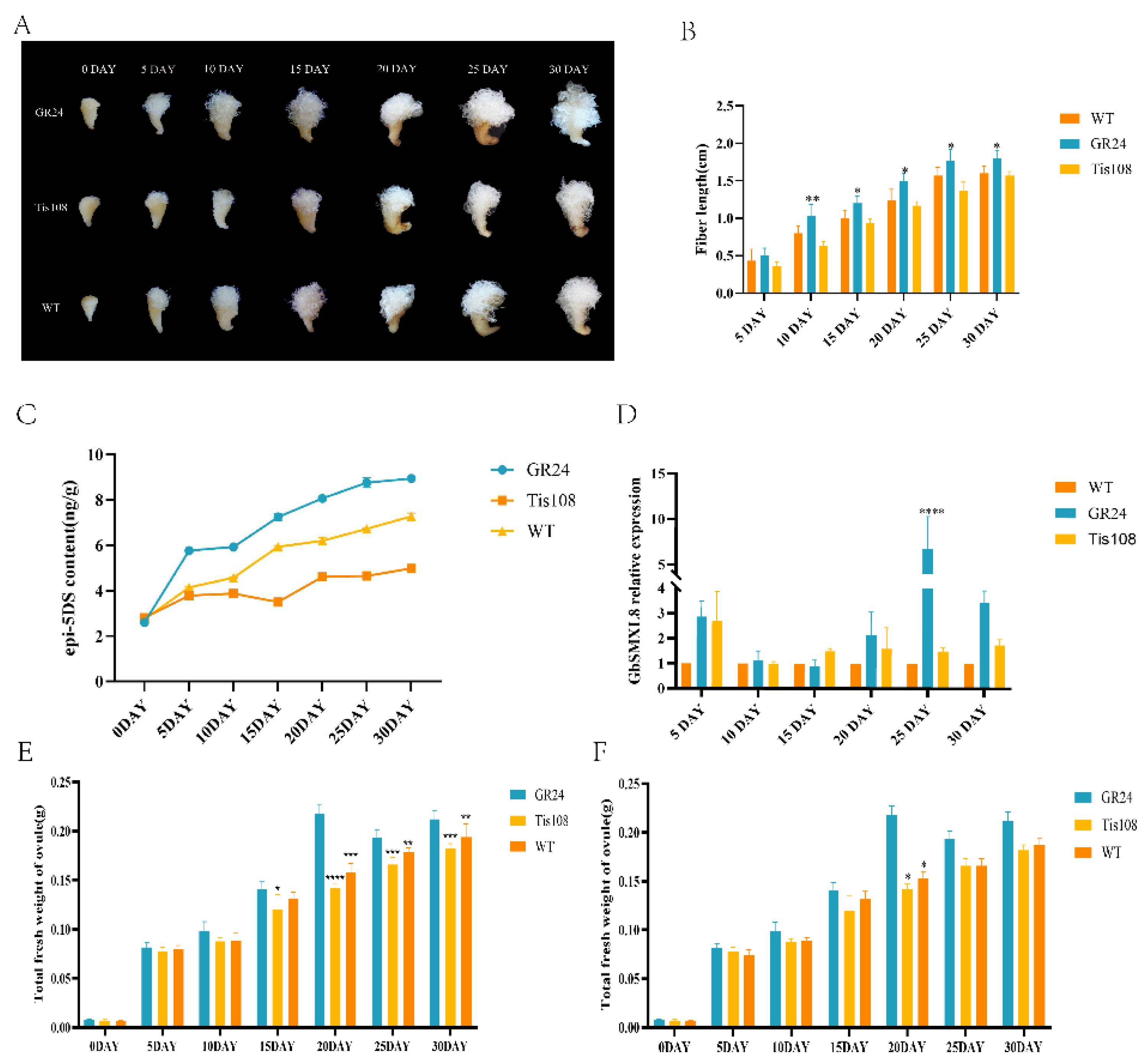
2.6. Effect of Auricolactone on Fiber Development
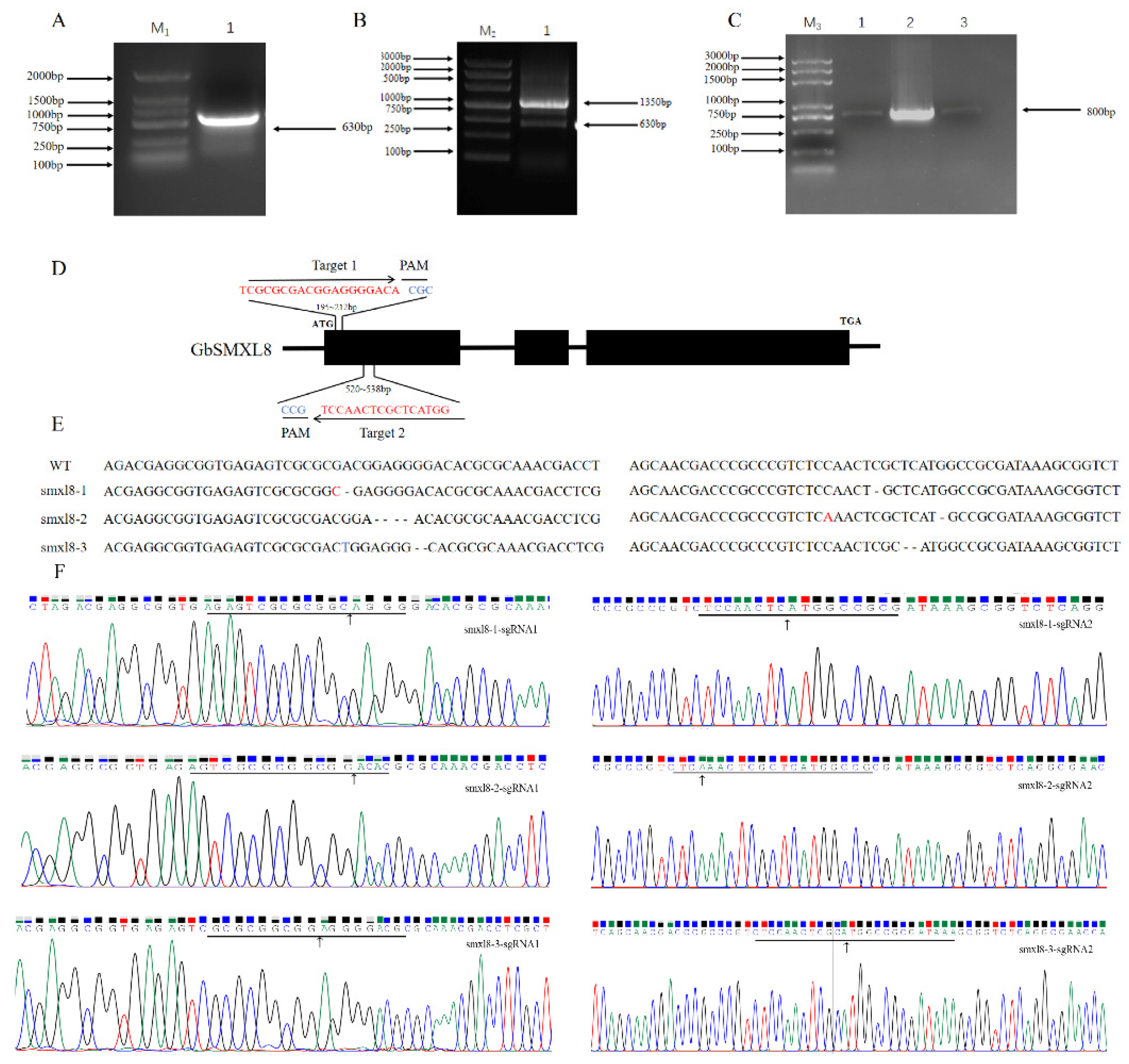
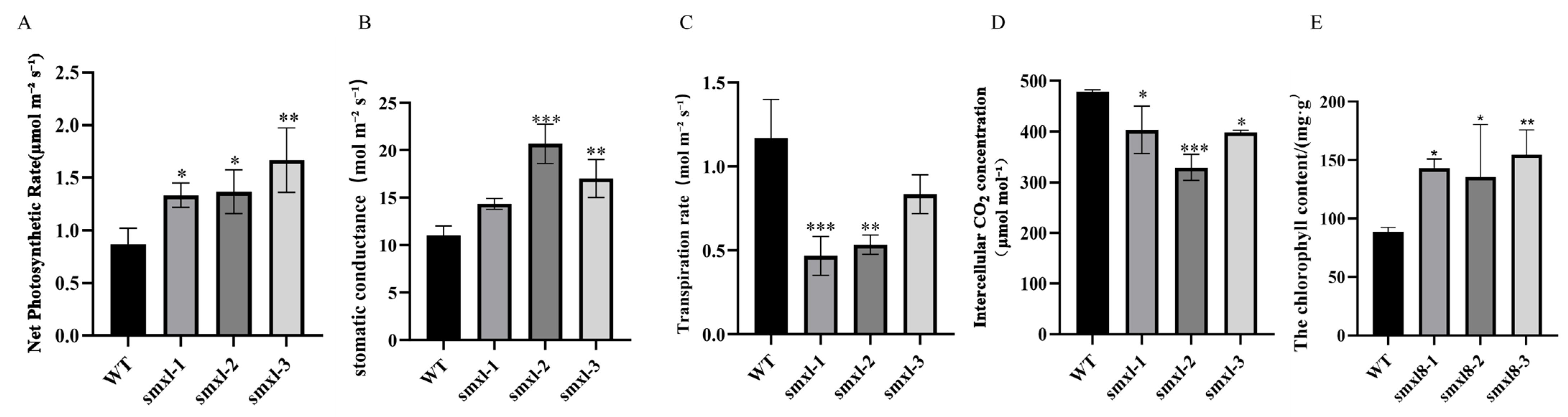
2.7. Impact of Editing GbSMXL8 on Fibers
3. Discussion
4. Materials and Methods
4.1. Structural Analysis of SMXL Gene Family in Cotton
4.2. Phylogenetic Analysis of Cotton SMXL Family
4.3. Gene Structure and Conserved Motif Analysis
4.4. Cotton SMXL Cis-Acting Element Prediction
4.5. Expression Characterization of the SMXL Family in Cotton
4.6. Ovule Culture and Observation
4.7. Expression Pattern Analysis
4.8. CRISPR/Cas9 System-Mediated Editing of Cotton
4.9. Statistics of Fiber and Epidermal Hair Length of Transgenic Cotton
4.10. Determination of Physiological and Biochemical Indexes
4.10.1. Measurement of Photosynthetic Rate
4.10.2. Chlorophyll Content Determination
4.11. Determination of Auricolactone Content
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Song, X.; Zhu, G.; Hou, S.; Ren, Y.; Amjid, M.W.; Li, W.; Guo, W. Genome-Wide Association Analysis Reveals Loci and Candidate Genes Involved in Fiber Quality Traits Under Multiple Field Environments in Cotton (Gossypium hirsutum). Front. Plant Sci. 2021, 12, 695503. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, V.; Koleva, M.; Valkova, N. Heterosis and combining ability in intraspecific (G. hirsutum L.) and interspecific (G. hirsutum L. × G. barbadense L.) cotton hybrids. Bulg. J. Crop Sci. 2023, 60, 37–47. [Google Scholar] [CrossRef]
- Malinga, L.N. The importance and economic status of cotton production. Acad. Lett. 2021. [Google Scholar] [CrossRef]
- Cao, J. C4H1 Gene Cloning and Expression Analysis between Sea-Island Cotton (Gossypium barbadense L.) Fiber and Upland Cotton (Gossypium hirsutum L.). Fiber Xinjiang Agric. Sci. 2015, 52, 1773–1781. [Google Scholar]
- Chen, G.; Liu, Z.; Li, S.; Liu, L.; Lu, L.; Wang, Z.; Mendu, V.; Li, F.; Yang, Z. Characterization of chromatin accessibility and gene expression reveal the key genes involved in cotton fiber elongation. Physiol. Plant. 2023, 175, e13972. [Google Scholar] [CrossRef]
- Xu, E.; Chai, L.; Zhang, S.; Yu, R.; Zhang, X.; Xu, C.; Hu, Y. Catabolism of strigolactones by a carboxylesterase. Nat. Plants 2021, 7, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Jiu, S.T.; Xu, Y.; Zhang, C.X.; Wang, L.; Ma, C.; Xu, W.P.; Wang, S.P. Research advancements on strigolactones and its regulatory effect on root growth and development in plants. Mol. Plant Breed. 2020, 18. [Google Scholar]
- Zheng, J.; Hong, K.; Zeng, L.; Wang, L.; Kang, S.; Qu, M.; Dai, J.; Zou, L.; Zhu, L.; Tang, Z.; et al. Karrikin Signaling Acts Parallel to and Additively with Strigolactone Signaling to Regulate Rice Mesocotyl Elongation in Darkness. Plant Cell 2020, 32, 2780–2805. [Google Scholar] [CrossRef]
- Xu, X.; Jibran, R.; Wang, Y.; Dong, L.; Flokova, K.; Esfandiari, A.; McLachlan, A.R.G.; Heiser, A.; Sutherland-Smith, A.J.; Brummell, D.A.; et al. Strigolactones regulate sepal senescence in Arabidopsis. J. Exp. Bot. 2021, 72, 5462–5477. [Google Scholar] [CrossRef]
- Wu, F.; Gao, Y.; Yang, W.; Sui, N.; Zhu, J. Biological Functions of Strigolactones and Their Crosstalk with Other Phytohormones. Front. Plant Sci. 2022, 13, 821563. [Google Scholar] [CrossRef]
- Zwanenburg, B.; Blanco-Ania, D. Strigolactones: New plant hormones in the spotlight. J. Exp. Bot. 2018, 69, 2205–2218. [Google Scholar] [CrossRef]
- Qu, B.; Qin, Y.; Bai, Y. From signaling to function: How strigolactones regulate plant development. Sci. China Life Sci. 2020, 63, 1768–1770. [Google Scholar] [CrossRef] [PubMed]
- Agusti, J.; Herold, S.; Schwarz, M.; Sanchez, P.; Ljung, K.; Dun, E.A.; Brewer, P.B.; Beveridge, C.A.; Sieberer, T.; Sehr, E.M.; et al. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 20242–20247. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Yu, H.; Yuan, K.; Liao, Z.; Meng, X.; Jing, Y.; Liu, G.; Chu, J.; Li, J. Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice. Proc. Natl. Acad. Sci. USA 2019, 116, 14319–14324. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef]
- Kee, Y.J.; Ogawa, S.; Ichihashi, Y.; Shirasu, K.; Yoshida, S. Strigolactones in Rhizosphere Communication: Multiple Molecules with Diverse Functions. Plant Cell Physiol. 2023, 64, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.-Y.; Chen, Y.-C.; Guo, S.-H.; Gan, X.-Y.; Tian, L.; Huang, L.-Q.; Yuan, Y. Research progress in strigolactones and application prospect in medicinal plants. Zhongguo Zhongyao Zazhi 2023, 48, 3132–3139. [Google Scholar]
- Hu, J.; Ji, Y.; Hu, X.; Sun, S.; Wang, X. BES1 functions as the co-regulator of D53-like SMXLs to inhibit BRC1 expression in strigolactone-regulated shoot branching in Arabidopsis. Plant Commun. 2020, 1, 100014. [Google Scholar] [CrossRef]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef]
- Song, X.; Lu, Z.; Yu, H.; Shao, G.; Xiong, J.; Meng, X.; Jing, Y.; Liu, G.; Xiong, G.; Duan, J.; et al. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017, 27, 1128–1141. [Google Scholar] [CrossRef]
- Ma, H.; Duan, J.; Ke, J.; He, Y.; Gu, X.; Xu, T.H.; Yu, H.; Wang, Y.; Brunzelle, J.S.; Jiang, Y.; et al. A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci. Adv. 2017, 3, e1601217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, L.; Gao, Y.; Guo, Y.; Zheng, N.; Xu, X.; Xu, M.; Wang, W.; Liu, C.; Liu, W.; et al. Genome-Wide Identification of SMXL Gene Family in Soybean and Expression Analysis of GmSMXLs under Shade Stress. Plants 2022, 11, 2410. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; An, J.P.; You, C.X.; Wang, X.F.; Hao, Y.J. Genome-wide analysis and identification of the SMXL gene family in apple (Malus × domestica). Tree Genet. Genomes 2018, 14, 61. [Google Scholar]
- Pan, Y.; Zhan, J.; Jiang, Y.; Xia, D.; Scheuring, S. A concerted ATPase cycle of the protein transporter AAA-ATPase Bcs1. Nat. Commun. 2023, 14, 6369. [Google Scholar] [CrossRef]
- Matias, P.M.; Baek, S.H.; Bandeiras, T.M.; Dutta, A.; Houry, W.A.; Llorca, O.; Rosenbaum, J. The AAA+ proteins Pontin and Reptin enter adult age: From understanding their basic biology to the identification of selective inhibitors. Front. Mol. Biosci. 2015, 2, 17. [Google Scholar] [CrossRef]
- Marand, A.P.; Eveland, A.L.; Kaufmann, K.; Springer, N.M. cis-Regulatory Elements in Plant Development, Adaptation, and Evolution. Annu. Rev. Plant Biol. 2023, 74, 111–137. [Google Scholar] [CrossRef]
- Sakagami, J.I.; Iwata, Y.; Nurrahma, A.H.I.; Siaga, E.; Junaedi, A.; Yabuta, S. Plant adaptations to anaerobic stress caused by flooding. IOP Conf. Ser. Earth Environ. Sci. 2020, 418, 012080. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Z.; Wu, H.; Xu, Z.; Zhang, H.; Qian, W.; Gao, W.; She, H. Genome-Wide Identification and Characterization of MYB Gene Family and Analysis of Its Sex-Biased Expression Pattern in Spinacia oleracea L. Int. J. Mol. Sci. 2024, 25, 795. [Google Scholar] [CrossRef]
- Krasylenko, Y.; Komis, G.; Hlynska, S.; Vavrdová, T.; Ovečka, M.; Pospíšil, T.; Šamaj, J. GR24, A Synthetic Strigolactone Analog, and Light Affect the Organization of Cortical Microtubules in Arabidopsis Hypocotyl Cells. Front. Plant Sci. 2021, 12, 675981. [Google Scholar] [CrossRef]
- Ito, S.; Umehara, M.; Hanada, A.; Yamaguchi, S.; Asami, T. Effects of strigolactone-biosynthesis inhibitor TIS108 on Arabidopsis. Plant Signal. Behav. 2013, 8, e24193. [Google Scholar] [CrossRef]
- Lee, J.A. Revision of the genetics of the hairiness-smoothness system of Gossypium. J. Hered. 1985, 76, 123–126. [Google Scholar] [CrossRef]
- Serna, L.; Martin, C. Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 2006, 11, 274–280. [Google Scholar] [CrossRef]
- Hu, H.; He, X.; Tu, L.; Zhu, L.; Zhu, S.; Ge, Z.; Zhang, X. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J. 2016, 88, 921–935. [Google Scholar] [CrossRef]
- Xu, B.; Zhang, J.; Shi, Y.; Dai, F.; Jiang, T.; Xuan, L.; He, Y.; Zhang, Z.; Deng, J.; Zhang, T.; et al. GoSTR, a negative modulator of stem trichome formation in cotton. Plant J. 2023, 116, 389–403. [Google Scholar] [CrossRef]
- Xiao, G.; Zhao, P.; Zhang, Y. A Pivotal Role of Hormones in Regulating Cotton Fiber Development. Front. Plant Sci. 2019, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Ramireddy, E.; Schmülling, T. Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. J. Exp. Bot. 2013, 64, 5021–5032. [Google Scholar] [CrossRef]
- Mayzlish-Gati, E.; De-Cuyper, C.; Goormachtig, S.; Beeckman, T.; Vuylsteke, M.; Brewer, P.B.; Beveridge, C.A.; Yermiyahu, U.; Kaplan, Y.; Enzer, Y.; et al. Strigolactones Are Involved in Root Response to Low Phosphate Conditions in Arabidopsis. Plant Physiol. 2012, 160, 1329–1341. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- T ian, Z.; Zhang, Y.; Zhu, L.; Jiang, B.; Wang, H.; Gao, R.; Friml, J.; Xiao, G. Strigolactones act downstream of gibberellins to regulate fiber cell elongation and cell wall thickness in cotton (Gossypium hirsutum). Plant Cell 2022, 34, 4816–4839. [Google Scholar]
- Ge, X.; Xu, J.; Yang, Z.; Yang, X.; Wang, Y.; Chen, Y.; Wang, P.; Li, F. Efficient genotype-independent cotton genetic transformation and genome editing. J. Integr. Plant Biol. 2023, 65, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Konstantinou, G.N. Enzyme-Linked Immunosorbent Assay (ELISA). In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2022; pp. 115–134. [Google Scholar]



| Gene Name | Gene ID | Chromosomal Location | Gene Length (bp) | Number of Amino Acids (aa) | Chromosomal Assignment | Isoelectric Point (IP) | Protein Molecular Weight (kD) |
|---|---|---|---|---|---|---|---|
| GaSMXL3-1 | Ga04G1902 | Chr04 | 2583 | 860 | 95242458–95245014 | 7.692 | 87.256 |
| GaSMAX1 | Ga05G0071 | Chr05 | 3174 | 1057 | 748814–752504 | 8.021 | 115.686 |
| GaSMXL7 | Ga05G3318 | Chr05 | 3294 | 1097 | 43831373–43835849 | 6.859 | 120.874 |
| GaSMXL2 | Ga08G2769 | Chr08 | 3366 | 1121 | 127820527–127828153 | 6.425 | 123.610 |
| GaSMXL3-2 | Ga09G1542 | Chr09 | 2583 | 860 | 72503416–72506219 | 6.833 | 95.317 |
| GaSMXL6 | Ga10G0058 | Chr10 | 3015 | 1004 | 617219–621650 | 8.049 | 111.242 |
| GaSMXL3-3 | Ga13G0098 | Chr13 | 2574 | 857 | 977357–979930 | 6.817 | 88.262 |
| GaSMXL4 | Ga13G2559 | Chr10 | 2769 | 922 | 617219–621650 | 7.633 | 103.048 |
| GbSMAX1-D | GB_D05G0073 | D05 | 3168 | 1055 | 702090–705771 | 8.024 | 115.392 |
| GbSMAX2-A | GB_A08G2797 | A08 | 2388 | 795 | 119163347–119165732 | 6.689 | 89.16 |
| GbSMAX2-D | GB_D08G2782 | D08 | 3168 | 1055 | 66175674–66178844 | 8.024 | 115.392 |
| GbSMAX1-A | GB_A05G0073 | A05 | 3168 | 1055 | 812909–816590 | 8.024 | 115.392 |
| GbSMXL3-1-A | GB_A05G4239 | A05 | 2583 | 860 | 104306914–104309470 | 6.833 | 95.365 |
| GbSMXL3-2-A | GB_A09G1634 | A09 | 2346 | 781 | 67281364–67284167 | 7.939 | 87.135 |
| GbSMXL3-3-A | GB_A13G0093 | A13 | 2583 | 795 | 888559–891131 | 6.817 | 88.262 |
| GbSMXL3-1-D | GB_D04G0224 | D04 | 2346 | 781 | 2897103–2899659 | 7.96 | 87.437 |
| GbSMXL3-2-D | GB_D09G1478 | D09 | 2583 | 860 | 42975932–42978737 | 6.962 | 95.336 |
| GbSMXL3-3-D | GB_D13G0095 | D13 | 2769 | 795 | 744946–747524 | 6.833 | 95.365 |
| GbSMXL4-A | GB_A13G2506 | A13 | 2769 | 922 | 110467899–110471049 | 8.124 | 103.066 |
| GbSMXL4-D | GB_D13G2442 | D13 | 2388 | 795 | 58526390–58529536 | 6.728 | 89.203 |
| GbSMXL6-A | GB_A10G2794 | A10 | 3018 | 1005 | 110924219–110928660 | 8.286 | 111.286 |
| GbSMXL6-D | GB_D10G2753 | D10 | 3006 | 1001 | 66148734–66153240 | 8.334 | 110.726 |
| GbSMXL7-A | GB_A05G3145 | A05 | 3351 | 1116 | 42056154–42060637 | 6.784 | 122.845 |
| GbSMXL7-D | GB_D05G3116 | D05 | 3351 | 1116 | 33847628–33852136 | 6.618 | 122.798 |
| GbSMXL8-A | GB_A05G0893 | A05 | 3369 | 1122 | 8339616–8343711 | 6.281 | 123.759 |
| GbSMXL8-D | GB_D05G0872 | D05 | 3369 | 1122 | 7368329–7372426 | 6.395 | 123.767 |
| GhSMAX1 | Gh_A05G005600 | A05 | 3168 | 1055 | 874949–879694 | 8.024 | 115.441 |
| GhSMXL2-A | Gh_A08G270100 | A08 | 2583 | 860 | 123972508–123980097 | 6.962 | 95.162 |
| GhSMXL2-D | Gh_D08G260600 | D08 | 2583 | 860 | 67120320–67123606 | 6.962 | 95.162 |
| GhSMXL3-1-A | Gh_A05G402300 | A05 | 2346 | 781 | 106202251–106205180 | 7.961 | 87.156 |
| GhSMXL3-2-A | Gh_A09G154300 | A09 | 2583 | 860 | 71317475–71320277 | 6.833 | 95.33 |
| GhSMXL3-3-A | Gh_A13G010300 | A13 | 2388 | 795 | 1013150–1016196 | 6.751 | 89.172 |
| GhSMXL3-1-D | Gh_D04G021100 | D04 | 2346 | 781 | 2840294–2843087 | 7.965 | 87.403 |
| GhSMXL3-2-D | Gh_D09G145500 | D09 | 2583 | 860 | 41594390–41597580 | 6.962 | 95.162 |
| GhSMXL3-3-D | Gh_D13G010500 | D13 | 2388 | 795 | 881251–884228 | 6.728 | 89.203 |
| GhSMXL4-A | Gh_A13G235100 | A13 | 2769 | 922 | 106078787–106082139 | 8.124 | 103.048 |
| GhSMXL6-A | Gh_A10G247300 | A10 | 3006 | 1001 | 114136161–114140859 | 8.334 | 110.726 |
| GhSMXL6-D | Gh_D10G279400 | D10 | 3018 | 1005 | 65267514–65272027 | 7.945 | 111.045 |
| GhSMXL7-A | Gh_A05G297200 | A05 | 3315 | 1116 | 43110739–43115841 | 6.784 | 122.859 |
| GhSMXL7-D | Gh_D05G306200 | D05 | 3351 | 1116 | 34015087–34020029 | 6.618 | 122.772 |
| GhSMXL4-D | Gh_D13G238200 | D13 | 2769 | 922 | 61213860–61217008 | 7.402 | 102.95 |
| GrSMXL3-1 | Grai_04G022220 | Chr04 | 2583 | 860 | 3077802–3080358 | 6.962 | 95.293 |
| GrSMXL6 | Grai_05G000730 | Chr05 | 3351 | 1116 | 113065942–113068498 | 6.618 | 122.716 |
| GrSMXL7 | Grai_05G033070 | Chr05 | 3366 | 1121 | 114074546–114076917 | 6.507 | 123.658 |
| GrSMAX1 | Grai_08G030620 | Chr08 | 3270 | 1089 | 2150749–2153436 | 7.65 | 111.174 |
| GrSMXL3 | Grai_09G016790 | Chr09 | 2346 | 781 | 71185776–71188579 | 7.574 | 87.25 |
| GrSMXL6 | Grai_10G031250 | Chr10 | 3270 | 1089 | 41926634–41929438 | 7.65 | 111.174 |
| GrSMXL3-2 | Grai_13G001050 | Chr13 | 2388 | 795 | 1013713–1016286 | 6.665 | 89.293 |
| GrSMXL4 | Grai_13G027690 | Chr13 | 2769 | 922 | 881673–884251 | 7.402 | 103.055 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Xu, W.; Zhang, L.; Chen, Q.; Cai, Y.; Chen, Q.; Zheng, K. Functional Study of GbSMXL8-Mediated Strigolactone Signaling Pathway in Regulating Cotton Fiber Elongation and Plant Growth. Int. J. Mol. Sci. 2025, 26, 2293. https://doi.org/10.3390/ijms26052293
Chen L, Xu W, Zhang L, Chen Q, Cai Y, Chen Q, Zheng K. Functional Study of GbSMXL8-Mediated Strigolactone Signaling Pathway in Regulating Cotton Fiber Elongation and Plant Growth. International Journal of Molecular Sciences. 2025; 26(5):2293. https://doi.org/10.3390/ijms26052293
Chicago/Turabian StyleChen, Lingyu, Wennuo Xu, Lingyu Zhang, Qin Chen, Yongsheng Cai, Quanjia Chen, and Kai Zheng. 2025. "Functional Study of GbSMXL8-Mediated Strigolactone Signaling Pathway in Regulating Cotton Fiber Elongation and Plant Growth" International Journal of Molecular Sciences 26, no. 5: 2293. https://doi.org/10.3390/ijms26052293
APA StyleChen, L., Xu, W., Zhang, L., Chen, Q., Cai, Y., Chen, Q., & Zheng, K. (2025). Functional Study of GbSMXL8-Mediated Strigolactone Signaling Pathway in Regulating Cotton Fiber Elongation and Plant Growth. International Journal of Molecular Sciences, 26(5), 2293. https://doi.org/10.3390/ijms26052293






