Fatty Acid and Lipid Metabolism in Oil Palm: From Biochemistry to Molecular Mechanisms
Abstract
:1. Introduction
2. Fatty Acid Synthesis and Accumulation in Oil Palm
3. Regulation of Lipid Biosynthesis and Accumulation in Oil Palm
4. Molecular Mechanisms of Fatty Acid Synthesis and Oil Accumulation in Oil Palm
5. Advances in Oil Palm Breeding: Leveraging Biotechnology and Molecular Tools for Enhanced Yields
6. Key Challenges in the Future of Palm Oil Production
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Corley, R.H.V.; Tinker, P.B. The Oil Palm, 5th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2015; pp. 1–655. [Google Scholar]
- Murphy, D. The future of oil palm as a major global crop: Opportunities and challenges. J. Oil Palm Res. 2014, 26, 1–24. [Google Scholar]
- Singh, R.; Ong-Abdullah, M.; Low, E.T.; Manaf, M.A.; Rosli, R.; Nookiah, R.; Ooi, L.C.-L.; Ooi, S.-E.; Chan, K.-L.; Halim, M.A.; et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New Worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Sambanthamurthi, R.; Sundram, K.; Tan, Y.-A. Chemistry and biochemistry of palm oil. Prog. Lipid Res. 2000, 39, 507–558. [Google Scholar] [CrossRef] [PubMed]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef]
- Henderson, J.; Osborne, D.J. The oil palm in all our lives: How this came about. Endeavour 2000, 24, 63–68. [Google Scholar] [CrossRef]
- Tranbarger, T.J.; Dussert, S.; Joët, T.; Argout, X.; Summo, M.; Champion, A.; Cros, D.; Omore, A.; Nouy, B.; Morcillo, F. Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiol. 2011, 156, 564–584. [Google Scholar] [CrossRef]
- Dussert, S.; Guerin, C.; Andersson, M.; Joët, T.; Tranbarger, T.J.; Pizot, M.; Sarah, G.; Omore, A.; Durand-Gasselin, T.; Morcillo, F. Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol. 2013, 162, 1337–1358. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Linh Giang, V.N.; Waminal, N.E.; Park, H.-S.; Kim, N.-H.; Jang, W.; Lee, J.; Yang, T.-J. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J. Ginseng Res. 2020, 44, 135–144. [Google Scholar] [CrossRef]
- Yeap, W.C.; Lee, F.C.; Shabari Shan, D.K.; Musa, H.; Appleton, D.R.; Kulaveerasingam, H. WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 2017, 91, 97–113. [Google Scholar] [CrossRef]
- Murgianto, E.F.; Ardiyanto, A.; Astuti, E.J.; Ahmad, M.P. Preprocessing Factors Affected Free Fatty Acid Content in Crude Palm Oil Quality. J. Ilmu Pertan. Indones. 2022, 27, 177–181. [Google Scholar]
- Teh, H.F.; Neoh, B.K.; Hong, M.P.; Low, J.Y.; Ng, T.L.; Ithnin, N.; Thang, Y.M.; Mohamed, M.; Chew, F.T.; Yusof, H.M.; et al. Differential metabolite profiles during fruit development in high-yielding oil palm mesocarp. PLoS ONE 2013, 8, e61344. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Teh, H.F.; Mebus, K.; Ooi, T.E.K.; Kwong, O.P.; Koo, K.L.; Ong, C.k.; Mayes, S.; Chew, F.T.; David, R.A.; et al. Differential gene expression at different stages of mesocarp development in high- and low-yielding oil palm. BMC Genom. 2017, 18, 470. [Google Scholar] [CrossRef]
- Mirande-Ney, C.; Tcherkez, G.; Gilard, F.; Ghashghaie, J.; Lamade, E. Effects of Potassium Fertilization on Oil Palm Fruit Metabolism and Mesocarp Lipid Accumulation. J. Agric. Food Chem. 2019, 67, 33. [Google Scholar] [CrossRef]
- Wei, L.; Yang, C.; JohnMartin, J.J.; Li, R.; Zhou, L.; Cheng, S.; Cao, H.; Liu, X. Metabonomics and Transcriptomic Analysis of Free FattyAcid Synthesis in Seedless and TeneraOil Palm. Int. J. Mol. Sci. 2024, 25, 1686. [Google Scholar] [CrossRef]
- White, S.W.; Zheng, J.; Zhang, Y.M. Rock, The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Stahl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid: Diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef]
- Ting, N.C.; Sherbina, K.; Khoo, J.S.; Kamaruddin, K.; Chan, P.-L.; Chan, K.-L.; Halim, M.A.A.; Sritharan, K.; Yaakub, Z.; Mayes, S.; et al. Expression of Fatty Acid and Triacylglycerol Synthesis Genes in Interspecific Hybrids of Oil Palm. Sci. Rep. 2020, 10, 16296. [Google Scholar] [CrossRef]
- Chollet, R.; Vidal, J.; O’Leary, M.H. PHOSPHOENOLPYRUVATE CARBOXYLASE: A Ubiquitous, Highly Regulated Enzyme in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 273–298. [Google Scholar] [CrossRef]
- Xu, W.; John Martin, J.J.; Li, X.; Liu, X.; Zhang, R.; Hou, M.; Cao, H.; Cheng, S. Unveiling the Secrets of Oil Palm Genetics: A Look into Omics Research. Int. J. Mol. Sci. 2024, 25, 8625. [Google Scholar] [CrossRef]
- Loei, H.; Lim, J.; Tan, M.; Lim, T.K.; Lin, Q.S.; Chew, F.T.; Kulaveerasingam, H.; Chung, M.C. Proteomic analysis of the oil palm fruit mesocarp reveals elevated oxidative phosphorylation activity is critical for increased storage oil production. J. Proteome Res. 2013, 12, 5096–5109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, W.; Martin, J.J.J.; Qadri, R.; Fu, X.; Feng, M.; Wei, L.; Zhang, A.; Yang, C.; Cao, H. Differential analysis of transcriptomic and metabolomic of free fatty acidrancidity process in oil palm (Elaeis guineensis) fruits of different husk types. Front. Plant Sci. 2023, 14, 1132024. [Google Scholar]
- Gutbrod, P.; Reichert, S.; Gutbrod, K.; Hamai, A.; Brehelin, C.; Ngando-Ebongue, G.; Dormann, P. Fatty acid isoprenoid alcohol ester synthesis in fruits of the African Oil Palm (Elaeis guineensis). J. Phytochem. 2021, 185, 112684. [Google Scholar] [CrossRef] [PubMed]
- Morcillo, F.; Vaissayre, V.; Serret, J.; Avallone, S.; Domonh’edo, H.; Jacob, F.; Dussert, S. Natural diversity in the carotene, tocochromanol and fatty acid composition of crude palm oil. Food Chem. 2021, 365, 130638. [Google Scholar] [CrossRef]
- Martin, J.J.J.; Wu, Q.; Feng, M.; Li, R.; Zhou, L.; Zhang, S.; Yang, C.; Cao, H. Lipidomic Profiles of Lipid Biosynthesis in Oil Palm during Fruit Development. Metabolites 2023, 13, 727. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, C.; Liang, Y.; Sun, R.; Gao, L.; Liu, T.; Li, D. Genome-wide association analysis of the lipid and fatty acid metabolism regulatory network in the mesocarp of oil palm (Elaeis guineensis Jacq.) based on small noncoding RNA sequencing. J. Tree Physiol. 2018, 39, 356–371. [Google Scholar] [CrossRef]
- Jin, Y.; Li, M.; Zou, J.; Zheng, Y.; Li, D. EgbHLH63 negatively regulates palm fruit oil accumulation by repressing EgDGAT1 transcription. J. Ind. Crops Prod. 2024, 213, 118479. [Google Scholar] [CrossRef]
- Ramli, U.S.; Darren, S.B.; Patti, A.Q.; John, L.H. Control mechanisms operating for lipid biosynthesis differ in oil-palm (Elaeis guineensis Jacq.) and olive (Olea europaea L.) callus cultures. Biochem. Soc. J. 2002, 364, 385–391. [Google Scholar] [CrossRef]
- Yuan, Y.; Arondel, V.; Domergue, F. Characterization and heterologous expression of three DGATs from oil palm (Elaeis guineensis) mesocarp in Saccharomyces cerevisiae. Biochimie 2020, 169, 18–28. [Google Scholar] [CrossRef]
- Hassan, H.; Tahir, N.I.; Rozali, N.L.; Chung, B.L.Y.; Othman, A.; Weckwerth, W.; Ramli, U.S. Integrative tissue-resolved proteomics and metabolomics analysis of oil palm (Elaeis guineensis Jacq.) fruit provides insights into stilbenoid biosynthesis at the interface of primary and secondary metabolism. Biocatal. Agric. Biotechnol. 2024, 224, 103308. [Google Scholar] [CrossRef]
- Neoh, B.K.; Teh, H.F.; Ng, T.L.M.; Tiong, S.H.; Thang, Y.M.; Ersad, M.A.; Mohamed, M.; Chew, F.T.; Kulaveerasingam, H.; Appleton, D.R. Profiling of Metabolites in Oil Palm Mesocarp at Different Stages of Oil Biosynthesis. J. Agric. Food Chem. 2013, 61, 1920–1927. [Google Scholar] [CrossRef] [PubMed]
- Apriyanto, A.; Compart, J.; Fettke, J. Transcriptomic analysis of mesocarp tissue during fruit development of the oil palm revealed specific isozymes related to starch metabolism that control oil yield. Front Plant Sci. 2023, 14, 1220237. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.; Amiruddin, M.D.; Weckwerth, W.; Ramli, U.S. Deciphering key proteins of oil palm (Elaeis guineensis Jacq.) fruit mesocarp development by proteomics and chemometrics. Electrophoresis 2019, 40, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, W.; Xu, X.; Li, S.Y.; Zheng, Y.; Li, D.D. Identification and Analysis of MADS-Box Genes Expressed in the Mesocarp of Oil Palm Fruit (Elaeis guineensis Jacq.). Biochem. Genet. 2023, 61, 2382–2400. [Google Scholar] [CrossRef]
- Khan, F.S.; Goher, F.; Zhang, D.; Shi, P.; Li, Z.; Htwe, Y.M.; Wang, Y. Is CRISPR/Cas9 a way forward to fast-track genetic improvement in commercial palms? Prospects and limits. Front. Plant Sci. 2022, 12, 1042828. [Google Scholar] [CrossRef]
- Yang, Y.; Kong, Q.; Ma, Z.; Lim, P.K.; Singh, S.K.; Pattanaik, S.; Mutwil, M.; Miao, Y.; Yuan, L.; Ma, W. Phase separation of MYB73 regulates seed oil biosynthesis in Arabidopsis. Plant Physiol. 2024, 197, kiae674. [Google Scholar] [CrossRef]
- Na, G.; Mu, X.; Grabowski, P.; Schmutz, J.; Lu, C. Enhancing microRNA167A expression in seed decreases the α-linolenic acid content and increases seed size in Camelina sativa. Plant J. 2019, 98, 346–358. [Google Scholar] [CrossRef]
- Ding, T.; Tomes, S.; Gleave, A.P.; Zhang, H.; Dare, A.P.; Plunkett, B.; Espley, R.V.; Luo, Z.; Zhang, R.; Allan, A.C.; et al. microRNA172 targets APETALA2 to regulate flavonoid biosynthesis in apple (Malus domestica). Hortic. Res. 2022, 9, uhab007. [Google Scholar] [CrossRef]
- Wang, F.; Zheng, T.; Hu, Z.; Wu, G.; Lang, C.; Liu, R. Overexpression of miR319a Altered Oil Body Morphogenesis and Lipid Content in Arabidopsis Seeds. Plant Mol. Biol. Report. 2020, 38, 531–537. [Google Scholar] [CrossRef]
- Lu, S.; Li, Q.; Wei, H.; Chang, M.-J.; Tunlaya-Anukit, S.; Kim, H.; Liu, J.; Song, J.; Sun, Y.-H.; Yuan, L.; et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2013, 110, 10848–10853. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis Is Mediated by Downregulation of miR398 and Important for Oxidative Stress Tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Taier, G.; Hang, N.; Shi, T.; Liu, Y.; Ye, W.; Zhang, W.; Wang, K. Ectopic Expression of Os-miR408 Improves Thermo-Tolerance of Perennial Ryegrass. Agronomy 2021, 11, 1930. [Google Scholar] [CrossRef]
- Kong, X.; Peng, K.; Shan, Y.; Yun, Z.; Dalmay, T.; Duan, X.; Jiang, Y.; Qu, H.; Zhu, H. Transcriptional regulation of miR528-PPO module by miR156 targeted SPLs orchestrates chilling response in banana. Mol. Hortic. 2025, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhu, Z.; Wang, H.; Wang, L.; Cheng, L.; Yu, Y.; Zheng, Y.; Li, D. Characterization and functional analysis of a plastidial FAD6 gene and its promoter in the mesocarp of oil palm (Elaeis guineensis). J. Sci. Hortic. 2018, 239, 163–170. [Google Scholar] [CrossRef]
- Sun, R.; Gao, L.; Yu, X.; Zheng, Y.; Li, D.; Wang, X. Identification of a Δ12 fatty acid desaturase from oil palm (Elaeis guineensis Jacq.) involved in the biosynthesis of linoleic acid by heterologous expression in Saccharomyces cerevisiae. Gene 2016, 591, 21–26. [Google Scholar] [CrossRef]
- Zhang, Q.; Jin, Y.; Zou, J.; Zheng, Y.; Li, D. Characterization and functional analysis of the MADS-box EgAGL9 transcription factor from the mesocarp of oil palm (Elaeis guineensis Jacq.). J. Plant Sci. 2022, 321, 111317. [Google Scholar] [CrossRef]
- Luo, S.; Huang, J.; Jin, L.; Zou, J.; Zheng, Y.; Li, D. Transcription factor EgGRP2A regulates EgFATA expression and promotes oleic acid accumulation in oil palm (Elaeis guineensis). J. Plant Physiol. 2024, 299, 154263. [Google Scholar] [CrossRef]
- Xu, X.; Li, M.; Zou, J.X.; Zheng, Y.S.; Li, D.D. EgMYB108 regulates very long-chain fatty acid (VLCFA) anabolism in the mesocarp of oil palm. Plant Cell Rep. 2022, 41, 1449–1460. [Google Scholar] [CrossRef]
- Wang, Y.; Zou, J.; Zhao, J.; Zheng, Y.; Li, D. EgmiR5179 Regulates Lipid Metabolism by Targeting EgMADS16 in the Mesocarp of Oil Palm (Elaeis guineensis). Front. Plant Sci. 2021, 12, 722596. [Google Scholar] [CrossRef]
- Martin, J.J.J.; Song, Y.; Hou, M.; Zhou, L.; Liu, X.; Li, X.; Fu, D.; Li, Q.; Cao, H.; Li, R. Multi-Omics Approaches in Oil Palm Research: A Comprehensive Review of Metabolomics, Proteomics, and Transcriptomics Based on Low-Temperature Stress. Int. J. Mol. Sci. 2024, 25, 7695. [Google Scholar] [CrossRef]
- Yue, G.H.; Ye, B.Q.; Lee, M. Molecular approaches for improving oil palm for oil. Mol. Breed. 2021, 41, 22. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.B.; Mathur, R.K. Molecular breeding in oil palm (Elaeis guineensis): Status and Future perspectives. Horticulture 2016, 48, 123–131. [Google Scholar] [CrossRef]
- Ithain, M.; Wendy, T.V.; Shin, M.G.; Suryawanshi, V.; Sherbina, K.; Zolkafi, S.H.; Serdari, N.M.; Amiruddin, M.D.; Abdullah, N.; Mustaffs, S.; et al. Genomic diversity and genome-wide association analysis related to yield and fatty acid composition of wild American oil palm. J. Plant Sci. 2021, 304, 110731. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Masani, M.Y.; Izawati, A.M.D.; Abdul-Rasid, O.; Parveez, G.K.A. Biotechnology of oil palm: Current status of oil palm genetic transformation. Biocatal. Agric. Biotechnol. 2018, 15, 335–347. [Google Scholar] [CrossRef]
- Teh, H.F.; Neoh, B.K.; Ithnin, N.; Daim, L.D.J.; Ooi, T.E.K.; Appleton, D.R. Review: Omics and Strategic Yield Improvement in Oil Crops. J. Am. Oil Chem. Soc. 2017, 94, 1225–1244. [Google Scholar] [CrossRef]
- Montoya, C.; Cochard, B.; Flori, A.; Cros, D.; Lopes, R.; Cuellar, T.; Espeout, S.; Syaputra, I.; Villeneuve, P.; Pina, M.; et al. Genetic Architecture of Palm Oil Fatty Acid Composition in Cultivated Oil Palm (Elaeis guineensis Jacq.) Compared to Its Wild Relative E. oleifera (H.B.K) Corte’s. PLoS ONE 2014, 9, e95412. [Google Scholar] [CrossRef]
- Bai, B.; Wang, L.; Lee, M.; Zhang, Y.; Rahmadsyah Alfiko, Y.; Ye, B.Q.; Wan, Z.Y.; Lim, C.H.; Suwanto, A.; Chua, N.H.; et al. Genome-wide identification of markers for selecting higher oil content in oil palm. BMC Plant Biol. 2017, 17, 93. [Google Scholar] [CrossRef]
- Kwong, Q.B.; The, C.K.; Ong, A.L.; Chew, F.T.; Mayes, S.; Kulaveerasingam, H.; Tammi, M.; Yeoh, S.H.; Appleton, D.R.; Harikrishna, J.A. Evaluation of methods and marker Systems in Genomic Selection of oil palm (Elaeis guineensis Jacq.). BMC Genet. 2017, 18, 107. [Google Scholar] [CrossRef]
- Ebongue, G.F.N.; Koona, P.; Nouy, B.; Zok, S.; Carrière, F.; Zollo, P.A.; Aronde, V. Identification of oil palm breeding lines producing oils with low acid values. Eur. J. Lipid Sci. Technol. 2008, 110, 505–509. [Google Scholar] [CrossRef]
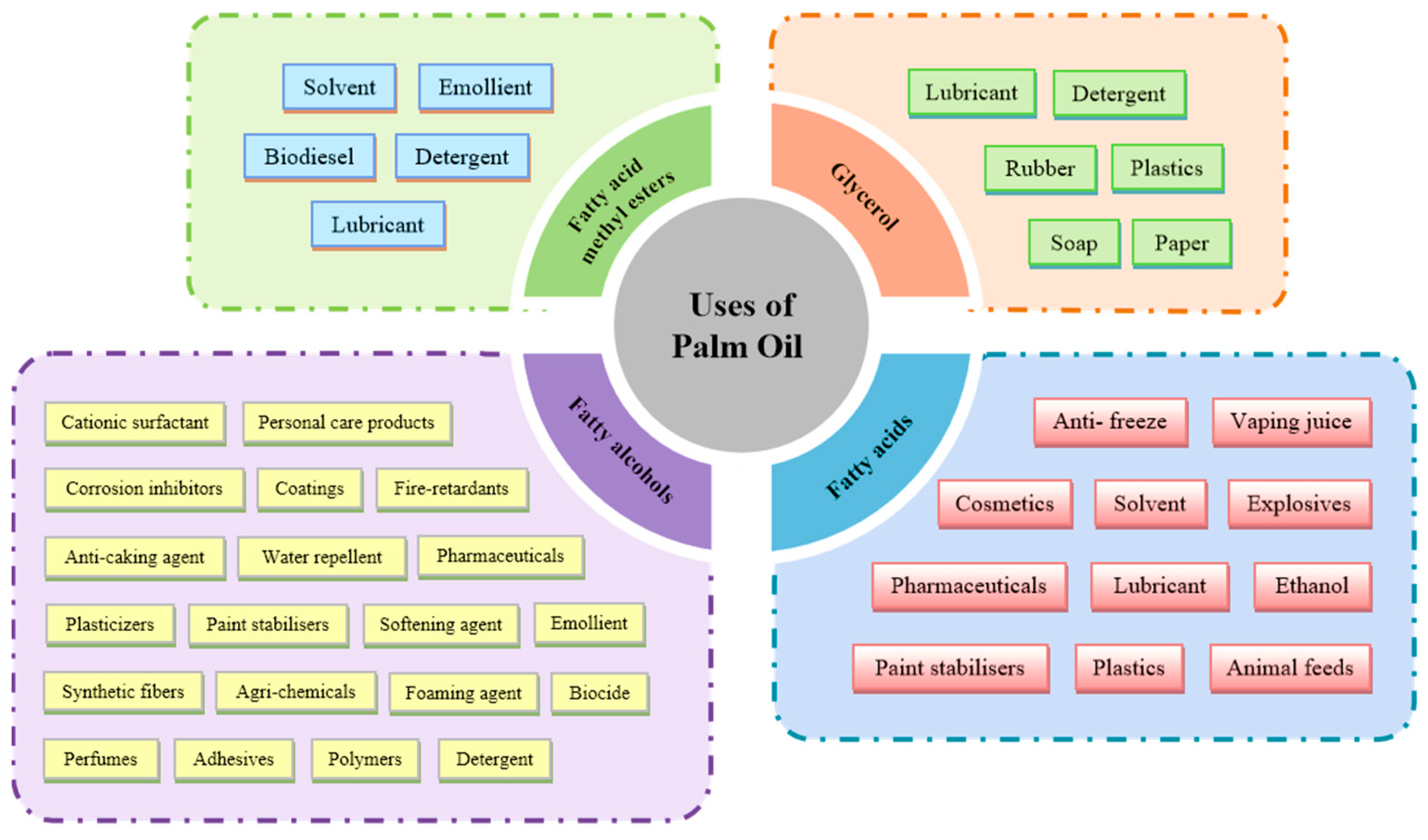
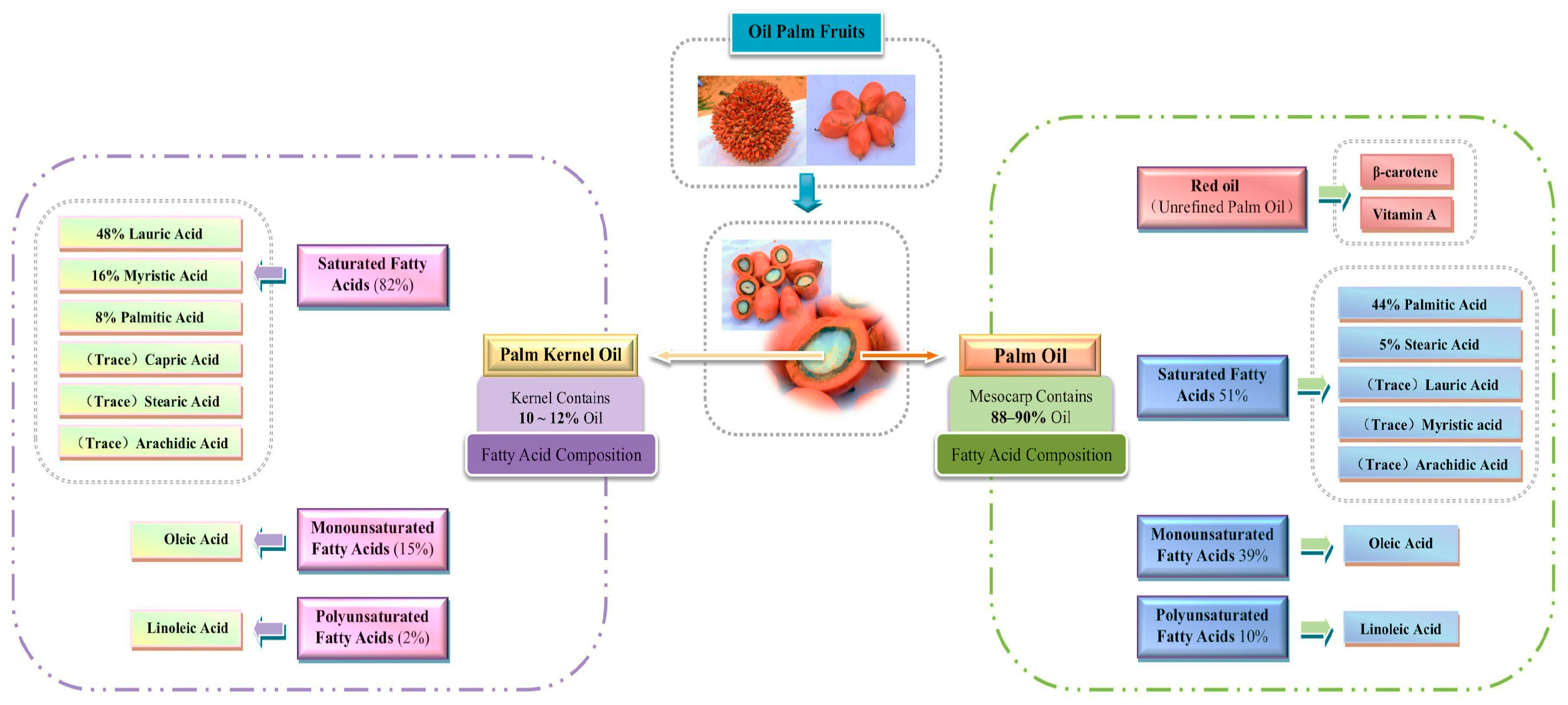

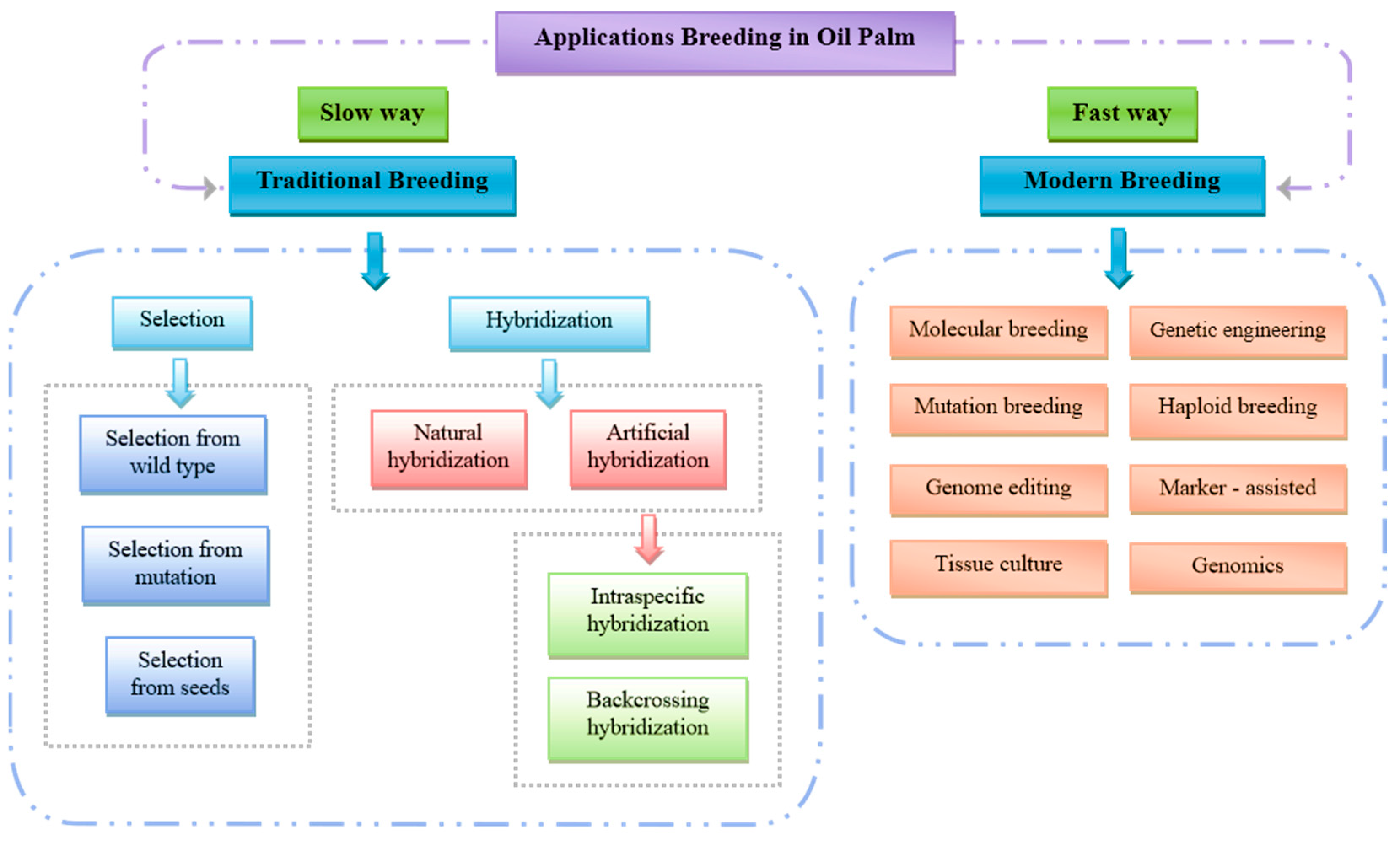
| Cultivar | Growth Period | Palmitic Acid (C16:0) | Oleic Acid (C18:1) | Linoleic Acid (C18:2) | Stearic Acid (C18:0) | Other Fatty Acids |
|---|---|---|---|---|---|---|
| Tenera (Dura × Pisifera) | 12 months | 44.5% | 39.2% | 10.1% | 4.5% | 1.7% |
| Dura | 12 months | 47.3% | 36.8% | 9.8% | 4.9% | 1.2% |
| Pisifera | 12 months | 42.1% | 41.5% | 11.2% | 3.8% | 1.4% |
| Tenera (Nigeria) | 18 months | 43.8% | 40.1% | 10.5% | 4.2% | 1.4% |
| Tenera (Côte d’Ivoire) | 18 months | 42.9% | 41.3% | 10.8% | 3.9% | 1.1% |
| Tenera (Ghana) | 24 months | 41.5% | 42.6% | 11.0% | 3.5% | 1.4% |
| Deli Dura | 24 months | 46.8% | 37.5% | 9.5% | 4.8% | 1.4% |
| Gene | Enzyme/Protein | Function | Role in Fatty Acid Metabolism | Reference |
|---|---|---|---|---|
| KAS I/II | 3-Ketoacyl-ACP Synthase I/II | Elongates fatty acid chains during biosynthesis | Modulates elongation steps in fatty acid synthesis | [4] |
| KAS III | 3-Ketoacyl-ACP Synthase III | Initiates fatty acid biosynthesis by condensing acetyl-CoA and malonyl-ACP | Regulates early stages of fatty acid biosynthesis by targeting KAS III transcripts | [17] |
| FATA/B | Acyl-ACP Thioesterase A/B | Terminates fatty acid synthesis by releasing free fatty acids | Influences the release of fatty acids from ACP | [8] |
| SAD | Stearoyl-ACP Desaturase | Converts stearic acid to oleic acid (introduces double bonds) | Regulates desaturation of fatty acids | [3] |
| FAD2 | Fatty Acid Desaturase 2 | Desaturates oleic acid to linoleic acid | Controls polyunsaturated fatty acid synthesis | [7] |
| DGAT1/2 | Diacylglycerol Acyltransferase 1/2 | Catalyzes the final step in triacylglycerol (TAG) biosynthesis | Regulates oil accumulation by targeting DGAT genes | [5] |
| PDAT | Phospholipid:DAG Acyltransferase | Alternative enzyme for TAG synthesis | Modulates TAG synthesis under stress conditions | [18] |
| ACP | Acyl Carrier Protein | Carries fatty acid chains during synthesis | Affects fatty acid chain elongation and transport | [19] |
| PEPC | Phosphoenolpyruvate Carboxylase | Provides carbon skeletons for fatty acid synthesis | Regulates carbon flux into fatty acid biosynthesis | [20] |
| WRI1 | WRINKLED1 Transcription Factor | Regulates genes involved in fatty acid biosynthesis | Modulates expression of WRI1, affecting oil accumulation | [5] |
| miRNA | Target Genes/Proteins | Function in Lipid Metabolism | Reference |
|---|---|---|---|
| miR156 | Squamosa Promoter-Binding Protein (SBP) | Regulates transcription factors, influencing lipid biosynthesis | [27] |
| miR159 | MYB Transcription Factors | Modulates gene expression linked to fatty acid biosynthesis | [37] |
| miR167 | Auxin Response Factor (ARF) | Influences fatty acid desaturation via hormonal regulation | [38] |
| miR172 | APETALA2 (AP2) | Indirectly affects fatty acid synthesis pathways | [39] |
| miR319 | TCP Transcription Factors | Regulates lipid metabolism and organ development | [40] |
| miR397 | Laccase Genes | Impacts secondary metabolism, indirectly affecting lipid pathways | [41] |
| miR398 | Cu/Zn Superoxide Dismutase (SOD) | Protects lipid membranes from oxidative damage | [42] |
| miR408 | Plastocyanin-Like Proteins | Affects lipid metabolism under stress conditions | [43] |
| miR528 | Acetyl-CoA Carboxylase (ACCase) | Regulates the first committed step in fatty acid biosynthesis | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afifi, E.H.; John Martin, J.J.; Wang, Q.; Li, X.; Liu, X.; Zhou, L.; Li, R.; Fu, D.; Li, Q.; Ye, J.; et al. Fatty Acid and Lipid Metabolism in Oil Palm: From Biochemistry to Molecular Mechanisms. Int. J. Mol. Sci. 2025, 26, 2531. https://doi.org/10.3390/ijms26062531
Afifi EH, John Martin JJ, Wang Q, Li X, Liu X, Zhou L, Li R, Fu D, Li Q, Ye J, et al. Fatty Acid and Lipid Metabolism in Oil Palm: From Biochemistry to Molecular Mechanisms. International Journal of Molecular Sciences. 2025; 26(6):2531. https://doi.org/10.3390/ijms26062531
Chicago/Turabian StyleAfifi, Eman H., Jerome Jeyakumar John Martin, Qi Wang, Xinyu Li, Xiaoyu Liu, Lixia Zhou, Rui Li, Dengqiang Fu, Qihong Li, Jianqiu Ye, and et al. 2025. "Fatty Acid and Lipid Metabolism in Oil Palm: From Biochemistry to Molecular Mechanisms" International Journal of Molecular Sciences 26, no. 6: 2531. https://doi.org/10.3390/ijms26062531
APA StyleAfifi, E. H., John Martin, J. J., Wang, Q., Li, X., Liu, X., Zhou, L., Li, R., Fu, D., Li, Q., Ye, J., & Cao, H. (2025). Fatty Acid and Lipid Metabolism in Oil Palm: From Biochemistry to Molecular Mechanisms. International Journal of Molecular Sciences, 26(6), 2531. https://doi.org/10.3390/ijms26062531





