In Silico Characterization and Determination of Gene Expression Levels Under Saline Stress Conditions in the Zinc Finger Family of the C1-2i Subclass in Chenopodium quinoa Willd
Abstract
1. Introduction
2. Results
2.1. Identification of CqZAT Genes and Analysis of Conserved Domains, Motifs, and Gene Structure
2.2. Physicochemical Characterization of CqZAT Genes
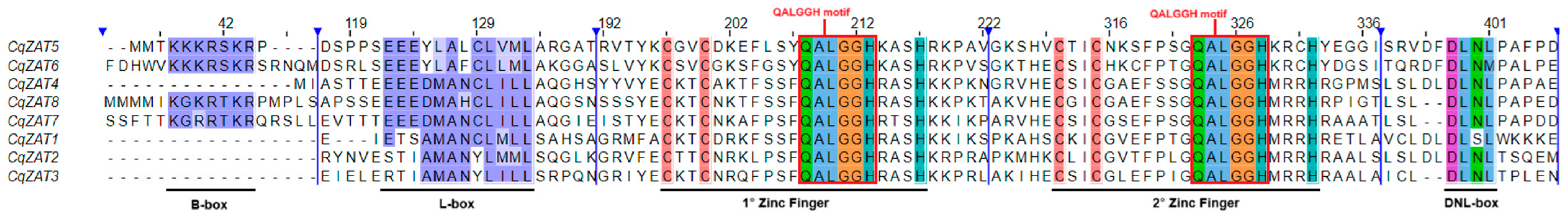
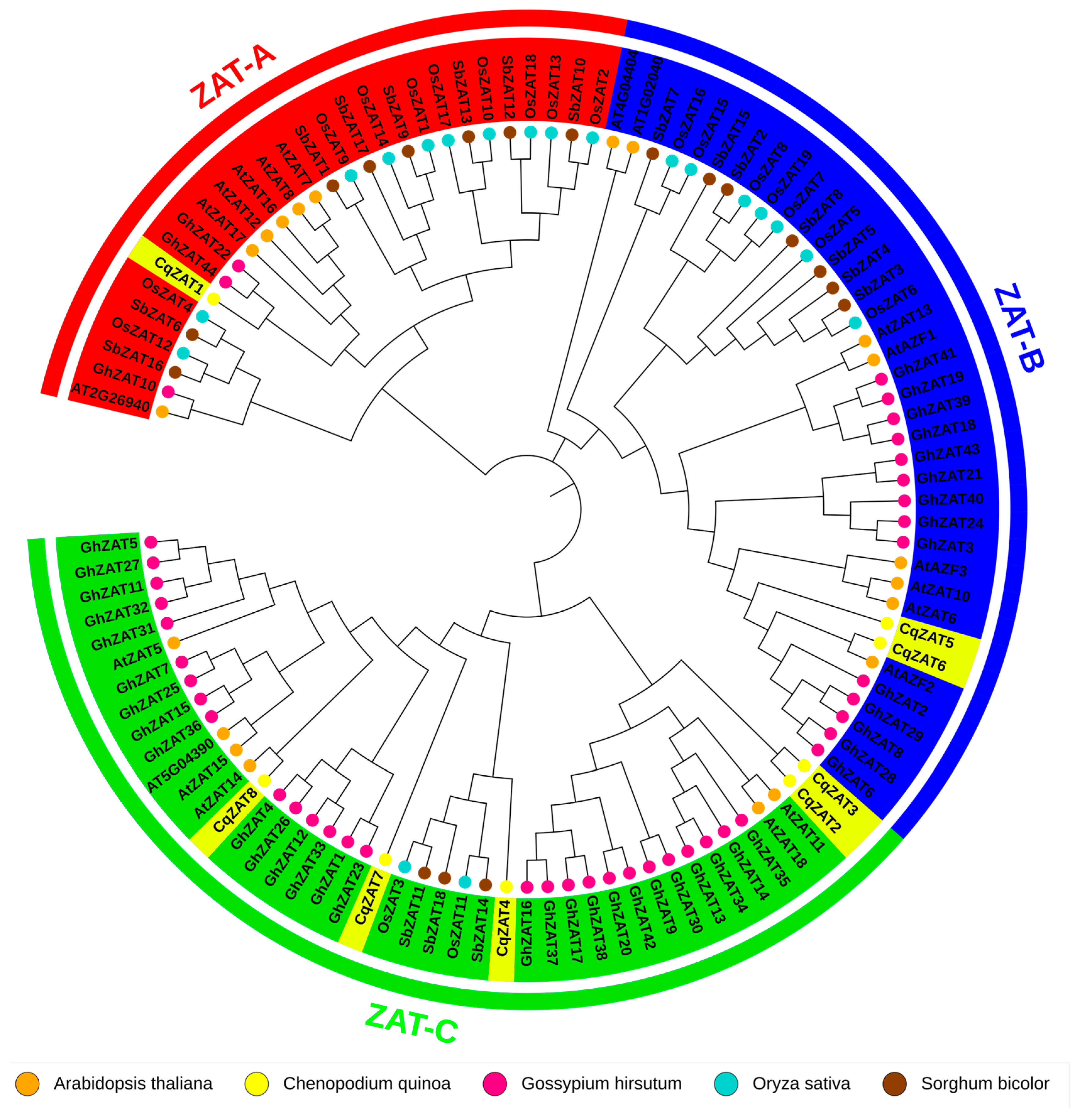
2.3. Sequence Alignment and Phylogenetic Analysis
2.4. Cis-Regulatory Element Analysis of CqZAT Promoter Regions
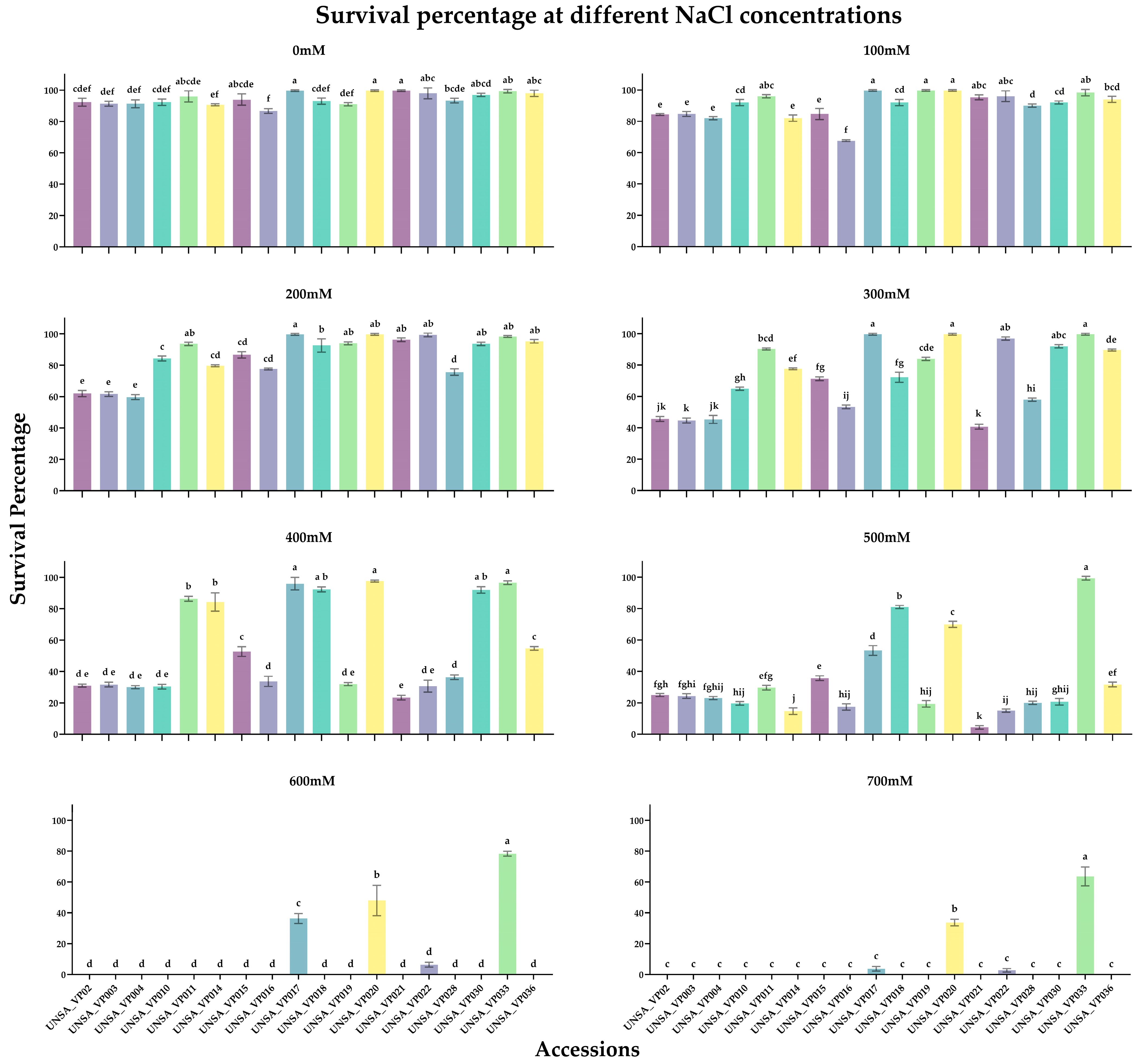
2.5. Germination Percentage and Identification of Halotolerant and Halosensitive Accessions
2.6. Determination of Halotolerant and Halosensitive Accessions in a Hydroponic System
2.7. Determination of Dry Matter Percentage
2.8. Determination of Relative Water Content (RWC)
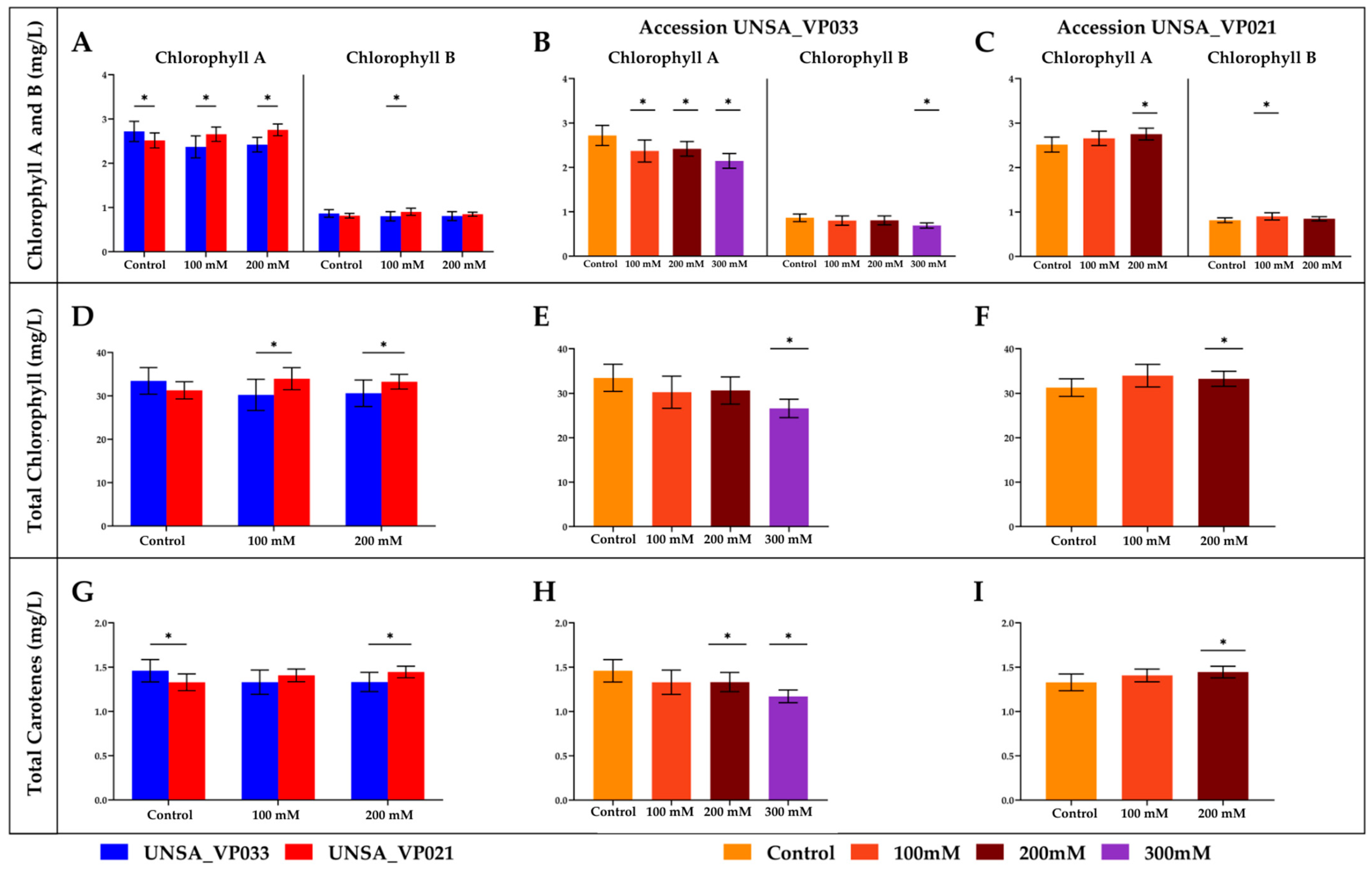
2.9. Chlorophyll and Carotenoid Content
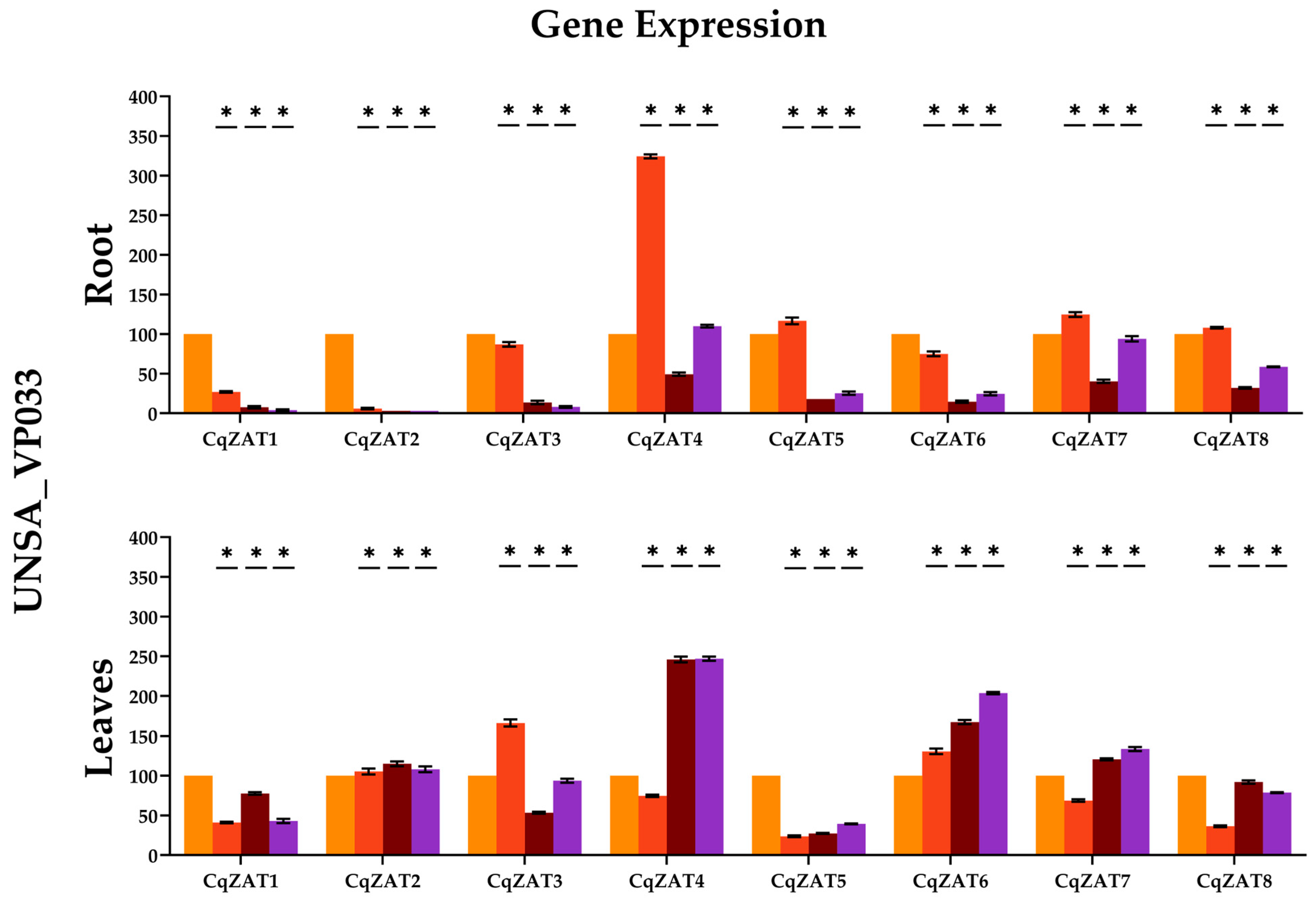
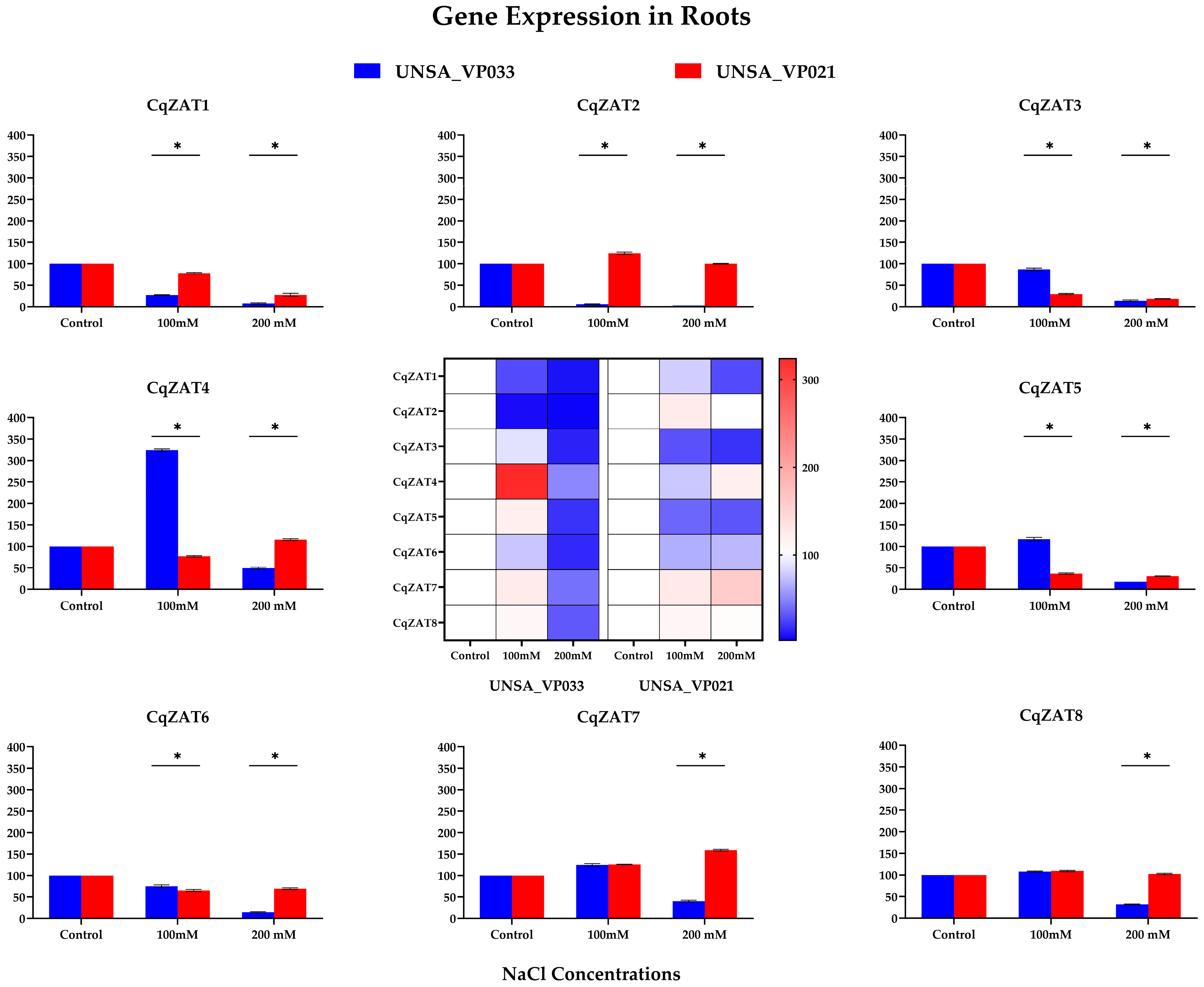
2.10. Relative Expression of ZAT Genes in C. quinoa
3. Discussion
3.1. In Silico Characterization of ZAT Proteins from the C2H2 Zinc Finger Subclass C1-2i in Chenopodium quinoa
3.2. Identification of a Halotolerant and a Halosensitive Accession in Chenopodium quinoa
3.3. Gene Expression Analysis in Tolerant and Sensitive Accessions of C. quinoa
4. Materials and Methods
4.1. Identification of ZAT Genes, Members of the C1-2i Subclass in Chenopodium quinoa
4.2. Domain Evaluation, Gene Structure Analysis, and Prediction of Conserved Motifs
4.3. Sequence Alignment and Phylogenetic Analysis
4.4. Analysis of Cis-Acting Promoter Elements in CqZAT Genes
4.5. Plant Material and Experimental Treatments
- a.
- Germination Percentage
- b.
- Selection of Halotolerant and Halosensitive Accessions
- c.
- Chenopodium quinoa Seedlings Subjected to Saline Stress in Hydroponic Systems
- d.
- Determination of Morphological and Physiological Parameters
- -
- Dry Matter and Plant Length: Dry matter percentage was determined using the formula:Dry Matter (%) = (Dry Sample Weight)/(Fresh Sample Weight) × 100Following the methodology of Ulloa and Valle (2021) [60], five samples were taken from each biological replicate for each treatment. The fresh weight of both the aerial and root parts was measured using a precision balance. The samples were then oven-dried at 80 °C for 24 h to determine their dry weight. Post-drying, the samples were weighed to calculate the dry mass of aerial parts (leaves + stems) and roots.The length of the aerial part of each sample was measured using graph paper, recording the measurements in millimeters. Root length was similarly measured from the base of the plant to the tip of the longest root. Fresh weight, dry weight, and length data were recorded for subsequent analysis.
- -
- Relative Water Content (RWC): RWC was determined following the methodology of Jensen et al. (2000) [61]. For each treatment, five leaves were collected from the middle third of 10 different plants, and their fresh weight (FW) was recorded immediately to avoid water loss due to transpiration. The leaves were then immersed in distilled water for 24 h at 24 °C under low light conditions. Afterward, the turgid weight (TW) was recorded, and leaves were oven-dried at 80 °C for 24 h to measure the dry weight (DW). The RWC was calculated using the following formula:RWC (%) = 100 × ((FW − DW)/(TW − DW))
- -
- Chlorophyll Content: Chlorophyll content was determined according to the methodology of Sumanta et al. (2014) [62]. Precisely 0.5 g of fresh leaf tissue was weighed and ground in a mortar with 10 mL of cold 95% ethanol. The mixture was centrifuged at 10,000 rpm for 15 min, and the supernatant was collected. A 0.5 mL aliquot of the supernatant was mixed with 4.5 mL of cold 95% ethanol. Chlorophyll-a, chlorophyll-b, and total carotenoid contents were quantified using an Epoch2 microplate spectrophotometer (BioTek, New Haven, CT, USA) at absorbances of 664 nm, 649 nm, and 470 nm, respectively. The equations used for quantification are as follows:
- e.
- Statistical Analysis
4.6. Primer Design
4.7. RNA Extraction and qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | Linear dichroism |
Appendix A
| Abbreviation | Name | Freq, |
|---|---|---|
| A | Alanine | 0.0808 |
| C | Cysteine | 0.0246 |
| D | Aspartic acid | 0.0477 |
| E | Glutamic acid | 0.0567 |
| F | Phenylalanine | 0.0269 |
| G | Glycine | 0.0704 |
| H | Histidine | 0.043 |
| I | Isoleucine | 0.0373 |
| K | Lysine | 0.0614 |
| L | Leucine | 0.0737 |
| M | Methionine | 0.026 |
| N | Asparagine | 0.0633 |
| P | Proline | 0.0435 |
| Q | Glutamine | 0.0288 |
| R | Arginine | 0.0463 |
| S | Serine | 0.121 |
| T | Threonine | 0.0831 |
| V | Valine | 0.0477 |
| W | Tryptophan | 0.00236 |
| Y | Tyrosine | 0.0156 |
| Cis-Element | Sequence | Function |
|---|---|---|
| Plant Hormone | ||
| ABRE | ACGTG | cis-acting element involved in the abscisic acid responsiveness |
| TGACG-motif | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
| CGTCA-motif | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| TATC-box | TATCCCA | cis-acting element involved in gibberellin-responsiveness |
| TCA-element | CCATCTTTTT | cis-acting element involved in salicylic acid responsiveness |
| Stress Responsiveness | ||
| GC-motif | CCCCCG | enhancer-like element involved in anoxic specific inducibility |
| LTR | CCGAAA | cis-acting element involved in low-temperature responsiveness |
| MBS | CAACTG | MYB binding site involved in drought-inducibility |
| TC-rich repeats | GTTTTCTTAC | cis-acting element involved in defense and stress responsiveness |
| WUN-motif | AAATTTCCT | wound-responsive element |
| Plant Growth | ||
| AACA_motif | TAACAAACTCCA | involved in endosperm-specific negative expression |
| ARE | AAACCA | cis-acting regulatory element essential for the anaerobic induction |
| CAT-box | GCCACT | cis-acting regulatory element related to meristem expression |
| circadian | CAAAGATATC | cis-acting regulatory element involved in circadian control |
| GCN4_motif | TGAGTCA | cis-regulatory element involved in endosperm expression |
| MBSI | aaaAaaC(G/C)GTTA | MYB binding site involved in flavonoid biosynthetic genes regulation |
| O2-site | GATGATGTGG | cis-acting regulatory element involved in zein metabolism regulation |
| Light Responsiveness | ||
| G-Box | CACGTT | cis-acting regulatory element involved in light responsiveness |
| AAAC-motif | CAATCAAAACCT | light responsive element |
| GT1-motif | GGTTAAT | light responsive element |
| Sp1 | GGGCGG | light responsive element |
| MRE | AACCTAA | MYB binding site involved in light responsiveness |
| ATC-motif | AGTAATCT | part of a conserved DNA module involved in light responsiveness |
| ATCT-motif | AATCTAATCC | part of a conserved DNA module involved in light responsiveness |
| Box 4 | ATTAAT | part of a conserved DNA module involved in light responsiveness |
| chs-CMA1a | TTACTTAA | part of a light responsive element |
| GA-motif | ATAGATAA | part of a light responsive element |
| Gap-box | CAAATGAA(A/G)A | part of a light responsive element |
| GATA-motif | AAGATAAGATT | part of a light responsive element |
| I-box | AGATAAGG | part of a light responsive element |
| TCCC-motif | TCTCCCT | part of a light responsive element |
| TCT-motif | TCTTAC | part of a light responsive element |
| AE-box | AGAAACAA | part of a module for light response |
| Symbol | Compound | Concentration | |
|---|---|---|---|
| Molarity | g/L | ||
| A | Ca(NO3)2.4H2O | 1.00 M | 236.00 |
| B | KNO3 | 1.00 M | 101.00 |
| C | MgSO4.7H2O | 1.00 M | 247.00 |
| D | KH2PO4 | 1.00M | 136.00 |
| E | Micronutrientes | ||
| F | Fe-EDTA | ||
| Micronutrients | Concentration g/L | Micronutrients | Concentration g/L |
| MnCl2.4H2O | 1.81 | H3BO3 | 2.86 |
| ZnSO4.7H2O | 0.22 | CuSO4.5H2O | 0.10 |
| H2MoO4.H2O | 0.10 | ||
| Code | Gen | Primers | |
|---|---|---|---|
| AUR62001834 | CqZAT1 | >CqZAT1_F | GTTGGAGGTGGAAGTTCTGAG |
| >CqZAT1_R | TTACACGAGTGAGCCTTCGG | ||
| AUR62038382 | CqZAT2 | >CqZAT2_F | TAAAGACCGCAACCGTGACA |
| >CqZAT2_R | CGGTTTTCCCTTCGTTGTATG | ||
| AUR62038383 | CqZAT3 | >CqZAT3_F | AGCGACGTAAGAAAGGCGAAA |
| >CqZAT3_R | TAAGCGAAAGCGAAAGCGAAAG | ||
| AUR62039327 | CqZAT4 | >CqZAT4_F | TCGTCAGATGATGAAGAACCAC |
| >CqZAT4_R | TGCTCGATTTGTCAGTTGAAGT | ||
| XP_021725368 | CqZAT5 | >CqZAT5_F | GTGGAGTATGTGACAAGGAGTTTCT |
| >CqZAT5_R | CAGTGGTTGCTATTGTGGTATGTAA | ||
| XP_021728442 | CqZAT6 | >CqZAT6_F | TGATTCCCGTCTATCCGAAG |
| >CqZAT6_R | TATTGCTCTGCTTGGTGTCG | ||
| XP_021766394 | CqZAT7 | >CqZAT7_F | CCGAGTTTCCGTCAGGACAA |
| >CqZAT7_R | GCAGTGGTGGTACTTATTGGAGA | ||
| XP_021774557 | CqZAT8 | >CqZAT8_F | AAAGCGAAGGTGCACGAATG |
| >CqZAT8_R | CGCTTCGTGATGGACTAGGG | ||

| Accession | Repetition | 0 mM | 100 mM | 200 mM | 300 mM | 400 mM | 500 mM | 600 mM | 700 mM |
|---|---|---|---|---|---|---|---|---|---|
| UNSA_VP002 | 1 | 90 | 84 | 60 | 46 | 30 | 24 | 0 | 0 |
| 2 | 95 | 84 | 64 | 47 | 32 | 26 | 0 | 0 | |
| 3 | 92 | 84 | 62 | 44 | 31 | 25 | 0 | 0 | |
| UNSA_VP003 | 1 | 91 | 86 | 62 | 46 | 33 | 23 | 0 | 0 |
| 2 | 93 | 83 | 60 | 43 | 30 | 24 | 0 | 0 | |
| 3 | 90 | 85 | 63 | 45 | 32 | 26 | 0 | 0 | |
| UNSA_VP004 | 1 | 94 | 82 | 58 | 45 | 29 | 22 | 0 | 0 |
| 2 | 91 | 81 | 60 | 48 | 30 | 23 | 0 | 0 | |
| 3 | 89 | 83 | 61 | 43 | 31 | 24 | 0 | 0 | |
| UNSA_VP010 | 1 | 93 | 92 | 86 | 64 | 32 | 19 | 0 | 0 |
| 2 | 90 | 90 | 84 | 65 | 29 | 19 | 0 | 0 | |
| 3 | 94 | 94 | 83 | 66 | 30 | 21 | 0 | 0 | |
| UNSA_VP011 | 1 | 92 | 96 | 93 | 90 | 88 | 30 | 0 | 0 |
| 2 | 97 | 95 | 93 | 90 | 86 | 28 | 0 | 0 | |
| 3 | 99 | 97 | 95 | 91 | 85 | 31 | 0 | 0 | |
| UNSA_VP014 | 1 | 91 | 84 | 80 | 78 | 82 | 14 | 0 | 0 |
| 2 | 90 | 80 | 80 | 78 | 91 | 17 | 0 | 0 | |
| 3 | 91 | 82 | 79 | 77 | 80 | 13 | 0 | 0 | |
| UNSA_VP015 | 1 | 90 | 88 | 74 | 70 | 50 | 36 | 0 | 0 |
| 2 | 95 | 85 | 78 | 72 | 56 | 37 | 0 | 0 | |
| 3 | 97 | 81 | 89 | 72 | 52 | 34 | 0 | 0 | |
| UNSA_VP016 | 1 | 88 | 68 | 78 | 54 | 36 | 18 | 0 | 0 |
| 2 | 85 | 68 | 78 | 54 | 30 | 19 | 0 | 0 | |
| 3 | 87 | 67 | 77 | 52 | 35 | 15 | 0 | 0 | |
| UNSA_VP017 | 1 | 100 | 100 | 100 | 100 | 100 | 72 | 40 | 2 |
| 2 | 100 | 100 | 100 | 100 | 96 | 56 | 34 | 5 | |
| 3 | 100 | 100 | 100 | 100 | 92 | 50 | 35 | 4 | |
| UNSA_VP018 | 1 | 95 | 90 | 88 | 70 | 91 | 81 | 0 | 0 |
| 2 | 91 | 92 | 94 | 76 | 92 | 80 | 0 | 0 | |
| 3 | 93 | 94 | 96 | 71 | 94 | 82 | 0 | 0 | |
| UNSA_VP019 | 1 | 90 | 100 | 94 | 84 | 32 | 20 | 0 | 0 |
| 2 | 91 | 99 | 93 | 83 | 31 | 21 | 0 | 0 | |
| 3 | 92 | 100 | 95 | 85 | 33 | 17 | 0 | 0 | |
| UNSA_VP020 | 1 | 100 | 100 | 100 | 100 | 98 | 70 | 59 | 36 |
| 2 | 99 | 99 | 99 | 99 | 97 | 68 | 40 | 33 | |
| 3 | 100 | 100 | 100 | 100 | 98 | 72 | 45 | 32 | |
| UNSA_VP021 | 1 | 100 | 95 | 97 | 39 | 22 | 5 | 0 | 0 |
| 2 | 100 | 97 | 95 | 41 | 25 | 3 | 0 | 0 | |
| 3 | 100 | 94 | 97 | 42 | 23 | 5 | 0 | 0 | |
| UNSA_VP022 | 1 | 100 | 94 | 98 | 98 | 29 | 16 | 5 | 2 |
| 2 | 94 | 94 | 100 | 96 | 28 | 14 | 8 | 4 | |
| 3 | 100 | 100 | 100 | 97 | 35 | 15 | 6 | 2 | |
| UNSA_VP028 | 1 | 92 | 90 | 74 | 58 | 36 | 20 | 0 | 0 |
| 2 | 95 | 89 | 75 | 57 | 35 | 19 | 0 | 0 | |
| 3 | 93 | 91 | 78 | 59 | 38 | 21 | 0 | 0 | |
| UNSA_VP030 | 1 | 97 | 92 | 93 | 92 | 90 | 19 | 0 | 0 |
| 2 | 96 | 91 | 93 | 91 | 92 | 20 | 0 | 0 | |
| 3 | 98 | 93 | 95 | 93 | 94 | 23 | 0 | 0 | |
| UNSA_VP033 | 1 | 100 | 100 | 99 | 100 | 98 | 100 | 78 | 69 |
| 2 | 98 | 96 | 98 | 100 | 96 | 98 | 80 | 65 | |
| 3 | 100 | 99 | 98 | 100 | 96 | 100 | 77 | 57 | |
| UNSA_VP036 | 1 | 96 | 94 | 96 | 90 | 56 | 33 | 0 | 0 |
| 2 | 100 | 96 | 94 | 70 | 54 | 30 | 0 | 0 | |
| 3 | 98 | 92 | 96 | 86 | 44 | 32 | 0 | 0 |
| Accession | Repetition | Concentration | |||||
|---|---|---|---|---|---|---|---|
| 0 mM | 100 mM | 200 mM | 300 mM | 400 mM | 500 mM | ||
| UNSA_VP021 | 1 | 100 | 100 | 100 | 100 | 10 | 0 |
| 2 | 100 | 100 | 100 | 100 | 11 | 0 | |
| 3 | 100 | 100 | 100 | 100 | 10 | 0 | |
| UNSA_VP033 | 1 | 100 | 100 | 100 | 32 | 0 | 0 |
| 2 | 100 | 100 | 100 | 34 | 0 | 0 | |
| 3 | 100 | 100 | 100 | 36 | 0 | 0 | |
| Accession | Repet | Control | 100 mM | 200 mM | 300 mM | ||||
|---|---|---|---|---|---|---|---|---|---|
| Leave | Root | Leave | Root | Leave | Root | Leave | Root | ||
| UNSA_VP021 | 1 | 8.29 | 8.55 | 9.15 | 9.10 | 11.14 | 10.32 | - | - |
| 2 | 8.72 | 6.92 | 8.97 | 8.08 | 10.98 | 10.08 | - | - | |
| 3 | 7.67 | 7.30 | 8.99 | 9.08 | 11.02 | 10.41 | - | - | |
| UNSA_VP031 | 1 | 8.72 | 10.92 | 10.68 | 8.82 | 10.56 | 10.69 | 11.48 | 10.46 |
| 2 | 8.68 | 10.80 | 9.96 | 8.62 | 10.73 | 10.80 | 12.03 | 10.41 | |
| 3 | 7.98 | 11.22 | 9.42 | 9.19 | 10.57 | 10.70 | 11.68 | 10.42 | |
| Accession | Repetition | Control | 100 mM | 200 mM | 300 mM |
|---|---|---|---|---|---|
| UNSA_VP021 | 1 | 261.4 | 234.4 | 254 | - |
| 2 | 213.6 | 275.6 | 274 | - | |
| 3 | 248 | 238 | 266 | - | |
| UNSA_VP031 | 1 | 146 | 217.4 | 241.8 | 234.6 |
| 2 | 193.6 | 179 | 183.8 | 228.6 | |
| 3 | 157.4 | 223.2 | 196.1 | 242 |
| Accession | Repetition | Control | 100 mM | 200 mM | 300 mM |
|---|---|---|---|---|---|
| UNSA_VP021 | 1 | 69.55 | 59.05 | 47.66 | - |
| 2 | 73.87 | 55.73 | 47.22 | - | |
| 3 | 74 | 58 | 47 | - | |
| UNSA_VP031 | 1 | 79.71 | 65.71 | 57.10 | 64.93 |
| 2 | 79.44 | 68.48 | 59.62 | 65.08 | |
| 3 | 76.87 | 64.46 | 58.28 | 65.32 |
| UNSA_VP021 | UNSA_VP033 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Repetition | Control | 100 mM | 200 mM | Control | 100 mM | 200 mM | 300 mM | ||
| Chlorophyll | A | 1 | 2.33 | 2.58 | 2.61 | 2.63 | 2.59 | 2.57 | 2.00 |
| 2 | 2.51 | 2.52 | 2.77 | 2.51 | 2.05 | 2.48 | 2.08 | ||
| 3 | 2.71 | 2.86 | 2.89 | 3.01 | 2.47 | 2.21 | 2.35 | ||
| B | 1 | 0.77 | 0.89 | 0.79 | 0.84 | 0.89 | 0.91 | 0.66 | |
| 2 | 0.81 | 0.83 | 0.86 | 0.79 | 0.67 | 0.82 | 0.67 | ||
| 3 | 0.87 | 0.98 | 0.89 | 0.96 | 0.84 | 0.70 | 0.74 | ||
| Total | 1 | 29.25 | 33.27 | 31.26 | 32.38 | 33.30 | 33.64 | 25.11 | |
| 2 | 31.13 | 31.76 | 33.68 | 30.84 | 25.70 | 31.14 | 25.85 | ||
| 3 | 33.45 | 36.85 | 34.87 | 37.17 | 31.72 | 27.06 | 28.83 | ||
| Total Carotens | 1 | 1.23 | 1.40 | 1.37 | 1.41 | 1.44 | 1.44 | 1.11 | |
| 2 | 1.32 | 1.34 | 1.45 | 1.35 | 1.15 | 1.35 | 1.16 | ||
| 3 | 1.44 | 1.49 | 1.51 | 1.61 | 1.39 | 1.20 | 1.24 | ||
| Organ | Treat | Repetition | CqZAT1 | CqZAT2 | CqZAT3 | CqZAT4 | CqZAT5 | CqZAT6 | CqZAT7 | CqZAT8 |
|---|---|---|---|---|---|---|---|---|---|---|
| Roots | 0 mM | 1 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| 2 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||
| 3 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||
| 100 mM | 1 | 76.4 | 127.8 | 29.6 | 76.2 | 37.5 | 62.8 | 125.5 | 107.5 | |
| 2 | 78.6 | 122.5 | 30.0 | 75.9 | 36.8 | 65.4 | 125.8 | 109.0 | ||
| 3 | 79.1 | 123.0 | 27.6 | 79.3 | 35.4 | 67.8 | 124.7 | 110.8 | ||
| 200 mM | 1 | 31.49 | 100.20 | 18.64 | 117.01 | 30.52 | 67.00 | 159.00 | 103.13 | |
| 2 | 26.84 | 101.38 | 18.41 | 112.50 | 29.95 | 70.95 | 160.88 | 101.26 | ||
| 3 | 24.47 | 99.32 | 18.11 | 116.72 | 30.72 | 69.52 | 157.23 | 103.87 | ||
| Leaves | 0 mM | 1 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| 2 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||
| 3 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||
| 100 mM | 1 | 18.28 | 36.18 | 17.76 | 332.44 | 7.44 | 6.68 | 62.21 | 138.13 | |
| 2 | 16.24 | 28.78 | 13.35 | 335.34 | 7.21 | 4.19 | 53.99 | 139.35 | ||
| 3 | 19.68 | 29.19 | 11.69 | 339.64 | 6.83 | 4.10 | 56.29 | 133.70 | ||
| 200 mM | 1 | 11.94 | 23.46 | 28.76 | 203.16 | 21.45 | 7.60 | 55.84 | 100.61 | |
| 2 | 11.40 | 26.58 | 23.72 | 204.56 | 17.48 | 8.40 | 48.70 | 103.00 | ||
| 3 | 12.34 | 29.08 | 24.81 | 207.27 | 20.87 | 7.49 | 48.13 | 101.91 |
| Organ | Treat. | Repetition | CqZAT1 | CqZAT2 | CqZAT3 | CqZAT4 | CqZAT5 | CqZAT6 | CqZAT7 | CqZAT8 |
|---|---|---|---|---|---|---|---|---|---|---|
| Roots | 0 mM | 1 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| 2 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||
| 3 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||
| 100 mM | 1 | 28.06 | 6.79 | 89.57 | 323.68 | 119.83 | 75.40 | 128.43 | 109.30 | |
| 2 | 26.50 | 6.16 | 87.29 | 321.65 | 118.08 | 78.36 | 121.78 | 106.51 | ||
| 3 | 26.76 | 4.75 | 84.42 | 326.98 | 112.49 | 72.16 | 124.02 | 107.96 | ||
| 200 mM | 1 | 5.52 | 3.07 | 12.10 | 49.79 | 17.63 | 14.52 | 41.35 | 32.40 | |
| 2 | 7.59 | 3.10 | 13.03 | 51.06 | 17.91 | 12.86 | 42.03 | 31.05 | ||
| 3 | 8.58 | 3.14 | 15.53 | 47.40 | 17.55 | 16.02 | 37.56 | 32.82 | ||
| 300 mM | 1 | 3.31 | 2.53 | 7.58 | 107.58 | 25.92 | 27.32 | 90.15 | 57.53 | |
| 2 | 4.98 | 3.49 | 7.49 | 110.64 | 23.22 | 24.08 | 95.51 | 59.27 | ||
| 3 | 4.33 | 3.07 | 8.77 | 110.84 | 27.30 | 23.25 | 95.53 | 59.42 | ||
| Leaves | 0 mM | 1 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| 2 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||
| 3 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||
| 100 mM | 1 | 41.17 | 107.05 | 161.22 | 73.15 | 24.56 | 134.03 | 69.56 | 37.09 | |
| 2 | 39.50 | 107.80 | 170.03 | 76.03 | 22.59 | 127.35 | 66.99 | 34.56 | ||
| 3 | 41.69 | 101.28 | 167.20 | 74.79 | 22.51 | 130.55 | 68.55 | 36.66 | ||
| 200 mM | 1 | 78.73 | 118.37 | 51.92 | 249.03 | 26.93 | 164.66 | 122.46 | 94.37 | |
| 2 | 76.32 | 115.14 | 53.60 | 246.66 | 27.49 | 169.63 | 120.19 | 90.15 | ||
| 3 | 77.77 | 111.60 | 53.79 | 241.64 | 28.32 | 165.64 | 119.70 | 91.60 | ||
| 300 mM | 1 | 40.83 | 107.05 | 91.48 | 250.32 | 39.41 | 205.18 | 130.86 | 78.79 | |
| 2 | 42.44 | 111.60 | 96.31 | 246.28 | 40.04 | 202.34 | 134.29 | 77.52 | ||
| 3 | 45.77 | 104.85 | 94.11 | 244.97 | 38.54 | 203.66 | 136.48 | 79.15 |
References
- Otterbach, S.; Wellman, G.; Schmöckel, S.M. (Eds.) Compendium of Plant Genomes. In The Quinoa Genome; Springer International Publishing: Cham, Switzerland, 2021; ISBN 978-3-030-65236-4. [Google Scholar]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The Genome of Chenopodium Quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef]
- Alandia, G.; Odone, A.; Rodriguez, J.P.; Bazile, D.; Condori, B. Quinoa—Evolution and Future Perspectives. In The Quinoa Genome; Schmöckel, S.M., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 179–195. ISBN 978-3-030-65237-1. [Google Scholar]
- Shi, P.; Gu, M. Transcriptome Analysis and Differential Gene Expression Profiling of Two Contrasting Quinoa Genotypes in Response to Salt Stress. BMC Plant Biol. 2020, 20, 568. [Google Scholar] [CrossRef] [PubMed]
- Grenfell-Shaw, L.; Tester, M. Abiotic Stress Tolerance in Quinoa. In The Quinoa Genome; Schmöckel, S.M., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 139–167. ISBN 978-3-030-65237-1. [Google Scholar]
- Lamz Piedra, A.; González Cepero, M.C. La Salinidad Como Problema En La Agricultura: La Mejora Vegetal Una Solución Inmediata. Cultiv. Trop. 2013, 34, 31–42. [Google Scholar]
- Hao, S.; Wang, Y.; Yan, Y.; Liu, Y.; Wang, J.; Chen, S. A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance. Horticulturae 2021, 7, 132. [Google Scholar] [CrossRef]
- Schmöckel, S.M.; Lightfoot, D.J.; Razali, R.; Tester, M.; Jarvis, D.E. Identification of Putative Transmembrane Proteins Involved in Salinity Tolerance in Chenopodium Quinoa by Integrating Physiological Data, RNAseq, and SNP Analyses. Front. Plant Sci. 2017, 8, 1023. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ding, Y.; Cai, C.; Chen, Z.; Zhu, C. The Role of C2H2 Zinc Finger Proteins in Plant Responses to Abiotic Stresses. Physiol. Plant. 2019, 165, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Wang, N.; Peng, Z.; He, S.; Zhao, Z.; Gao, Q.; Wang, Z.; Li, H.; Du, X. Identification of C2H2 Subfamily ZAT Genes in Gossypium Species Reveals GhZAT34 and GhZAT79 Enhanced Salt Tolerance in Arabidopsis and Cotton. Int. J. Biol. Macromol. 2021, 184, 967–980. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, Z.; Li, S.; Qanmber, G.; Liu, L.; Guo, M.; Lu, L.; Ma, S.; Li, F.; Yang, Z. Genome-Wide Analysis of ZAT Gene Family Revealed GhZAT6 Regulates Salt Stress Tolerance in G. Hirsutum. Plant Sci. 2021, 312, 111055. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yue, M.; Jiang, L.; Liu, Y.; Zhang, N.; Liu, X.; Ye, Y.; Lin, X.; Zhang, Y.; Lin, Y.; et al. Genome-Wide Identification of Strawberry C2H2-ZFP C1-2i Subclass and the Potential Function of FaZAT10 in Abiotic Stress. Int. J. Mol. Sci. 2022, 23, 13079. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Y.; Zeng, L.; Liu, F.; Xu, Y.; Jin, Q.; Wang, Y. Genome-Wide Identification of the Q-Type C2H2 Zinc Finger Protein Gene Family and Expression Analysis under Abiotic Stress in Lotus (Nelumbo nucifera). BMC Genom. 2024, 25, 648. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-R.; Yang, K.; Wang, X.; You, C.-X. A C2H2-Type Zinc Finger Transcription Factor, MdZAT17, Acts as a Positive Regulator in Response to Salt Stress. J. Plant Physiol. 2022, 275, 153737. [Google Scholar] [CrossRef] [PubMed]
- Ciftci-Yilmaz, S.; Morsy, M.R.; Song, L.; Coutu, A.; Krizek, B.A.; Lewis, M.W.; Warren, D.; Cushman, J.; Connolly, E.L.; Mittler, R. The EAR-Motif of the Cys2/His2-Type Zinc Finger Protein Zat7 Plays a Key Role in the Defense Response of Arabidopsis to Salinity Stress *. J. Biol. Chem. 2007, 282, 9260–9268. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, Z.; Xu, X.; Zhang, H.; Li, C. Genome-Wide Analysis of C2H2 Zinc-Finger Family Transcription Factors and Their Responses to Abiotic Stresses in Poplar (Populus trichocarpa). PLoS ONE 2015, 10, e0134753. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Zhou, Y.; Qin, Z.; Li, A.; Wang, Q.; Li, Z.; Hou, F.; Zhang, L. Genome-Wide Identification of the C2H2 Zinc Finger Gene Family and Expression Analysis under Salt Stress in Sweetpotato. Front. Plant Sci. 2023, 14, 1301848. [Google Scholar] [CrossRef]
- Sun, S.-J.; Guo, S.-Q.; Yang, X.; Bao, Y.-M.; Tang, H.-J.; Sun, H.; Huang, J.; Zhang, H.-S. Functional Analysis of a Novel Cys2/His2-Type Zinc Finger Protein Involved in Salt Tolerance in Rice. J. Exp. Bot. 2010, 61, 2807–2818. [Google Scholar] [CrossRef]
- Tang, W.; Luo, C. Overexpression of Zinc Finger Transcription Factor ZAT6 Enhances Salt Tolerance. Open Life Sci. 2018, 13, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Liu, R.; Guo, B.; Huang, K.; Wang, L.; Han, Y.; Li, H.; Hou, S. Ectopic Expression of GmZAT4, a Putative C2H2-Type Zinc Finger Protein, Enhances PEG and NaCl Stress Tolerances in Arabidopsis thaliana. 3 Biotech 2019, 9, 166. [Google Scholar] [CrossRef]
- Sakamoto, H.; Maruyama, K.; Sakuma, Y.; Meshi, T.; Iwabuchi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2-Type Zinc-Finger Proteins Function as Transcription Repressors under Drought, Cold, and High-Salinity Stress Conditions. Plant Physiol. 2004, 136, 2734–2746. [Google Scholar] [CrossRef]
- Englbrecht, C.C.; Schoof, H.; Böhm, S. Conservation, Diversification and Expansion of C2H2 Zinc Finger Proteins in the Arabidopsis thaliana Genome. BMC Genom. 2004, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Sun, J.; Gong, D.; Kong, Y. The Roles of Arabidopsis C1-2i Subclass of C2H2-Type Zinc-Finger Transcription Factors. Genes 2019, 10, 653. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; Li, M.; Mao, P.; Zhou, Q.; Liu, W.; Liu, Z. Genome-Wide Identification of the Q-Type C2H2 Transcription Factor Family in Alfalfa (Medicago sativa) and Expression Analysis under Different Abiotic Stresses. Genes 2021, 12, 1906. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, X.; Wang, M.-M.; Tang, H.-J.; Ding, L.-Y.; Shen, Y.; Zhang, H.-S. A Novel Rice C2H2-Type Zinc Finger Protein Lacking DLN-Box/EAR-Motif Plays a Role in Salt Tolerance. Biochim. Biophys. Acta BBA—Gene Struct. Expr. 2007, 1769, 220–227. [Google Scholar] [CrossRef]
- Chow, V.; Kirzinger, M.W.; Kagale, S. Lend Me Your EARs: A Systematic Review of the Broad Functions of EAR Motif-Containing Transcriptional Repressors in Plants. Genes 2023, 14, 270. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-M.; Nguyen, X.C.; Kim, K.E.; Han, H.J.; Yoo, J.; Lee, K.; Kim, M.C.; Yun, D.-J.; Chung, W.S. Phosphorylation of the Zinc Finger Transcriptional Regulator ZAT6 by MPK6 Regulates Arabidopsis Seed Germination under Salt and Osmotic Stress. Biochem. Biophys. Res. Commun. 2013, 430, 1054–1059. [Google Scholar] [CrossRef]
- Zhao, T.; Wu, T.; Zhang, J.; Wang, Z.; Pei, T.; Yang, H.; Li, J.; Xu, X. Genome-Wide Analyses of the Genetic Screening of C2H2-Type Zinc Finger Transcription Factors and Abiotic and Biotic Stress Responses in Tomato (Solanum lycopersicum) Based on RNA-Seq Data. Front. Genet. 2020, 11, 540. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yu, A.; Sun, R.; Liu, A. Function and Evolution of C1-2i Subclass of C2H2-Type Zinc Finger Transcription Factors in POPLAR. Genes 2022, 13, 1843. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Chen, J.; Liu, M.; Zhang, H.; Zhang, S.; Liu, D.; Chen, S. Genome-Wide Analysis of C2H2 Zinc Finger Gene Family and Its Response to Cold and Drought Stress in Sorghum [Sorghum bicolor (L.) Moench]. Int. J. Mol. Sci. 2022, 23, 5571. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Jiang, C.; Ye, L.; Gao, Y.; Hou, X. Genome-Wide Identification of C2H2-ZFPs and Functional Analysis of BcZAT12 and BcZAT10 under Cold Stress in Non-Heading Chinese Cabbage. Environ. Exp. Bot. 2024, 217, 105574. [Google Scholar] [CrossRef]
- Liu, K.; Hou, Q.; Yu, R.; Deng, H.; Shen, L.; Wang, Q.; Wen, X. Genome-Wide Analysis of C2H2 Zinc Finger Family and Their Response to Abiotic Stresses in Apple. Gene 2024, 904, 148164. [Google Scholar] [CrossRef] [PubMed]
- Shariatipour, N.; Heidari, B. Meta-Analysis of Expression of the Stress Tolerance Associated Genes and Uncover Their Cis-Regulatory Elements in Rice (Oryza sativa L.). Open Bioinform. J. 2020, 13, 39–49. [Google Scholar] [CrossRef]
- Luo, J.; Yu, C.M.; Yan, M.; Chen, Y.H. Molecular Characterization of the Promoter of the Stress-Inducible ZmMYB30 Gene in Maize. Biol. Plant. 2020, 64, 200–210. [Google Scholar] [CrossRef]
- Li, Q.; Liu, J.; Tan, D.; Allan, A.C.; Jiang, Y.; Xu, X.; Han, Z.; Kong, J. A Genome-Wide Expression Profile of Salt-Responsive Genes in the Apple Rootstock Malus Zumi. Int. J. Mol. Sci. 2013, 14, 21053–21070. [Google Scholar] [CrossRef]
- Li, P.; Zheng, T.; Li, L.; Zhuo, X.; Jiang, L.; Wang, J.; Cheng, T.; Zhang, Q. Identification and Comparative Analysis of the CIPK Gene Family and Characterization of the Cold Stress Response in the Woody Plant Prunus Mume. PeerJ 2019, 7, e6847. [Google Scholar] [CrossRef] [PubMed]
- Jahandar, S.Z. The HKT2;3 Gene Expression Analysis in Bread Wheat Genotypes under Salt Stress Imposition. Inst. Mol. Biol. Biotechnol. Trans. 2022, 6, 12–16. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, Z.; Yang, L.; Chan, Z. Transcriptional Variation Analysis of Arabidopsis Ecotypes in Response to Drought and Salt Stresses Dissects Commonly Regulated Networks. Physiol. Plant. 2021, 172, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, B.; Sudiro, C.; Jabir, R.; Schiavo, F.; Hyder, M.; Yasmin, T. Adaptive Behaviour of Roots under Salt Stress Correlates with Morpho—Physiological Changes and Salinity Tolerance in Rice. Int. J. Agric. Biol. 2019, 21, 667–674. [Google Scholar] [CrossRef]
- Muhammad, N. Effects of Salt Stress on Cotton Growth and Germination at Various Stages. Heliyon 2023, 57. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Y.; Sun, L.; Dai, J.; Dong, H. Mitigating Salinity Stress and Improving Cotton Productivity with Agronomic Practices. Agronomy 2023, 13, 2486. [Google Scholar] [CrossRef]
- Robin, A.H.K.; Matthew, C.; Uddin, M.J.; Bayazid, K.N. Salinity-Induced Reduction in Root Surface Area and Changes in Major Root and Shoot Traits at the Phytomer Level in Wheat. J. Exp. Bot. 2016, 67, 3719–3729. [Google Scholar] [CrossRef] [PubMed]
- Patwa, N.; Pandey, V.; Gupta, O.P.; Yadav, A.; Meena, M.R.; Ram, S.; Singh, G. Unravelling Wheat Genotypic Responses: Insights into Salinity Stress Tolerance in Relation to Oxidative Stress, Antioxidant Mechanisms, Osmolyte Accumulation and Grain Quality Parameters. BMC Plant Biol. 2024, 24, 875. [Google Scholar] [CrossRef] [PubMed]
- Saddiq, M.S.; Iqbal, S.; Hafeez, M.B.; Ibrahim, A.M.H.; Raza, A.; Fatima, E.M.; Baloch, H.; Jahanzaib; Woodrow, P.; Ciarmiello, L.F. Effect of Salinity Stress on Physiological Changes in Winter and Spring Wheat. Agronomy 2021, 11, 1193. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, H. Plant Salt Response: Perception, Signaling, and Tolerance. Front. Plant Sci. 2023, 13, 1053699. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Z.; Sui, N. Sensitivity and Responses of Chloroplasts to Salt Stress in Plants. Front. Plant Sci. 2024, 15, 1374086. [Google Scholar] [CrossRef]
- Hussin, S.A.; Ali, S.H.; Lotfy, M.E.; El-Samad, E.H.A.; Eid, M.A.; Abd-Elkader, A.M.; Eisa, S.S. Morpho-Physiological Mechanisms of Two Different Quinoa Ecotypes to Resist Salt Stress. BMC Plant Biol. 2023, 23, 374. [Google Scholar] [CrossRef]
- Kumar, S.; Li, G.; Yang, J.; Huang, X.; Ji, Q.; Liu, Z.; Ke, W.; Hou, H. Effect of Salt Stress on Growth, Physiological Parameters, and Ionic Concentration of Water Dropwort (Oenanthe javanica) Cultivars. Front. Plant Sci. 2021, 12, 660409. [Google Scholar] [CrossRef] [PubMed]
- Moradi, F.; Ismail, A.M. Responses of Photosynthesis, Chlorophyll Fluorescence and ROS-Scavenging Systems to Salt Stress During Seedling and Reproductive Stages in Rice. Ann. Bot. 2007, 99, 1161–1173. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiang, L.; Hai-Xing, S.; Xiang-Min, R.; Ismail, A. Responses of Different Rice (Oryza sativa L.) Genotypes to Salt Stress and Relation to Carbohydrate Metabolism and Chlorophyll Content. Afr. J. Agric. Res. 2012, 7, 19–27. [Google Scholar] [CrossRef]
- Shah, S.H.; Houborg, R.; McCabe, M.F. Response of Chlorophyll, Carotenoid and SPAD-502 Measurement to Salinity and Nutrient Stress in Wheat (Triticum aestivum L.). Agronomy 2017, 7, 61. [Google Scholar] [CrossRef]
- Razzaghi, F.; Ahmadi, S.H.; Adolf, V.; Jensen, C.; Jacobsen, S.-E.; Andersen, M. Water Relations and Transpiration of Quinoa (Chenopodium quinoa Willd.) Under Salinity and Soil Drying. J. Agron. Crop Sci. 2011, 197, 348–360. [Google Scholar] [CrossRef]
- Panuccio, M.R.; Jacobsen, S.E.; Akhtar, S.S.; Muscolo, A. Effect of Saline Water on Seed Germination and Early Seedling Growth of the Halophyte Quinoa. AoB PLANTS 2014, 6, plu047. [Google Scholar] [CrossRef] [PubMed]
- Hariadi, Y.; Marandon, K.; Tian, Y.; Jacobsen, S.-E.; Shabala, S. Ionic and Osmotic Relations in Quinoa (Chenopodium quinoa Willd.) Plants Grown at Various Salinity Levels. J. Exp. Bot. 2011, 62, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Alanoca Quispe, C.; Guzman, D.; Lutino, S.; Iquize Villca, E. Evaluación de La Germinación de Accesiones Del Banco de Germoplasma de Quinua (Chenopodium quinoa Willd.) Del Estado Plurinacional de Bolivia. Rev. Científica Investig. INFO-INIAF 2013, 1, 23. [Google Scholar]
- Yang, H.W.; Liu, W.Y.; Shen, B.Y.; Li, C.Z. Seed Germination and Physiological Characteristics of Chenopodium Quinoa under Salt Stress. Acta Prataculturae Sin. 2017, 26, 146. [Google Scholar] [CrossRef]
- Prado, F.E.; Boero, C.; Gallardo, M.R.A.; González, J.A. Effect of NaCl on Growth Germination and Soluble Sugars Content in Chenopodium quinoa Willd. Seeds; ACAD SINICA: Taipei, Taiwan, 2000. [Google Scholar]
- Cole, D.L.; Woolley, R.K.; Tyler, A.; Buck, R.L.; Hopkins, B.G. Mineral Nutrient Deficiencies in Quinoa Grown in Hydroponics with Single Nutrient Salt/Acid/Chelate Sources. J. Plant Nutr. 2020, 43, 1661–1673. [Google Scholar] [CrossRef]
- Ulloa, M.C.; Valle, L. Índice de cosecha con macro-nutrientes en grano de quinua (Chenopodium quinoa Willd). Rev. Alfa 2021, 5, 15–28. [Google Scholar] [CrossRef]
- Jensen, C.R.; Jacobsen, S.-E.; Andersen, M.N.; Núñez, N.; Andersen, S.D.; Rasmussen, L.; Mogensen, V.O. Leaf Gas Exchange and Water Relation Characteristics of Field Quinoa (Chenopodium quinoa Willd.) during Soil Drying. Eur. J. Agron. 2000, 13, 11–25. [Google Scholar] [CrossRef]
- Nayek, S.; Choudhury, I.; Haque; Jaishee, N.; Roy, S. Spectrophotometric Analysis of Chlorophylls and Carotenoids from Commonly Grown Fern Species by Using Various Extracting Solvents. Res. J. Chem. Sci. 2014, 4, 2231–2606. [Google Scholar] [CrossRef]
- López, T.d.R.; Lazo, H.E. Concentración de clorofila total en el extrato etanólico, procedente de Chlorella sp. cultivada en efluentes de la industria pesquera. Bol. Cent. Investig. Biol. 1998, 32, 179–193. [Google Scholar]
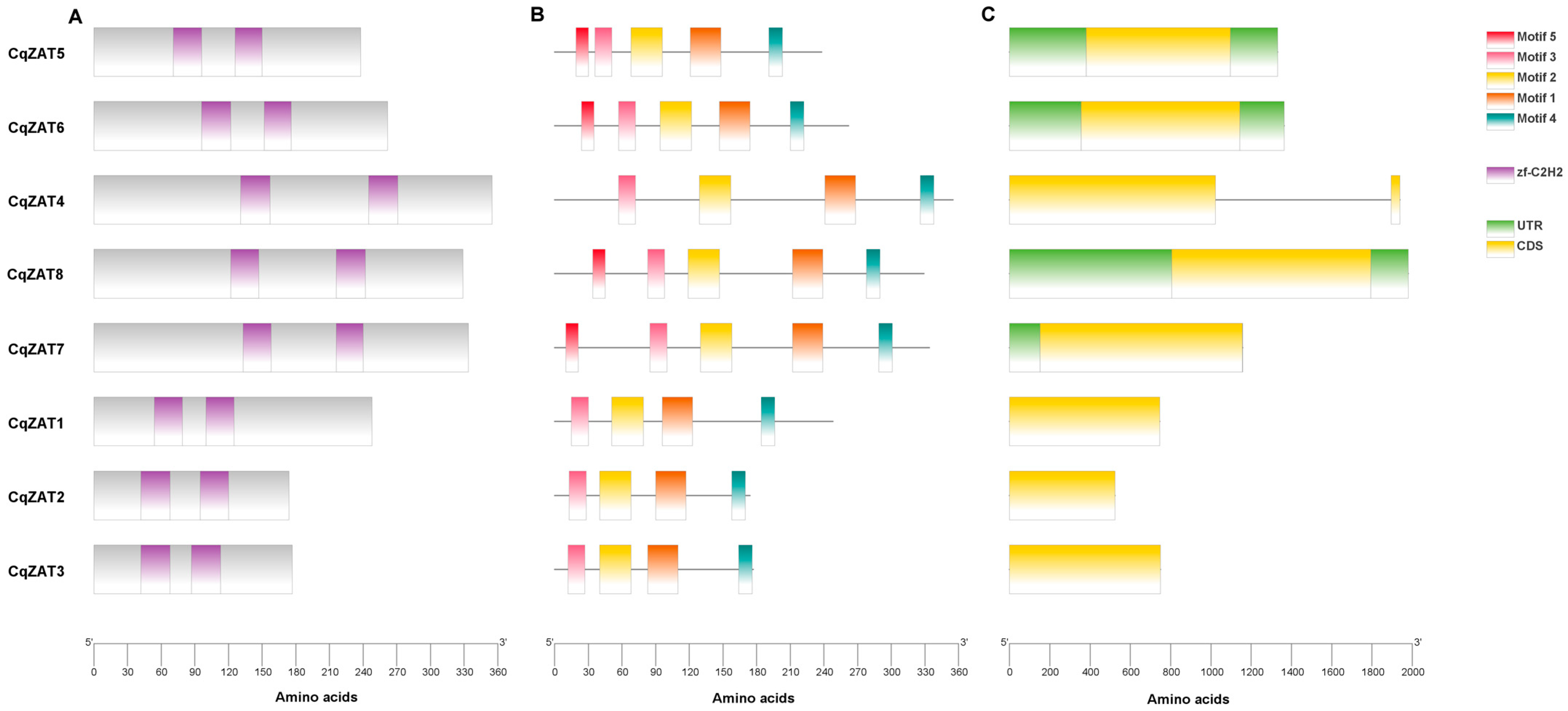



| Phylogenetic Group | Gen | Gene Report | Location | Gene (pb *) | CDS * (pb *) | PP * (aa *) | MW * (kDa *) | Aliphatic Index | pI * | Cellular Location * |
|---|---|---|---|---|---|---|---|---|---|---|
| ZAT-A | CqZAT1 | AUR62001834 | C_Quinoa_Scaffold_2716:9075033..9075780 reverse | 747 | 747 | 248 | 26.32 | 67.66 | 6.66 | Nu 1,3,4, Cy 1, Ec 2, Pe 2 |
| ZAT-C | CqZAT2 | AUR62038382 | C_Quinoa_Scaffold_1034:1966184..1966709 reverse | 525 | 525 | 174 | 19.38 | 72.30 | 9.48 | Nu 1,2,3,4, Cp 1, Ec 2 |
| ZAT-C | CqZAT3 | AUR62038383 | C_Quinoa_Scaffold_2493:1671380..1672130 reverse | 614 | 534 | 177 | 19.71 | 82.15 | 9.10 | Nu 1,3,4, Cy 2, Ec 2 |
| ZAT-C | CqZAT4 | AUR62039327 | C_Quinoa_Scaffold_1932:1438123..1440062 reverse | 1939 | 1068 | 355 | 38.12 | 64.39 | 5.84 | Nu 1,2,3,4 |
| ZAT-B | CqZAT5 | XP_021725368 | NW_018744124.1 (330074..331406) | 1333 | 717 | 238 | 25.58 | 61.55 | 8.79 | Nu 1,2,3,4 |
| ZAT-B | CqZAT6 | XP_021728442 | NW_018744301.1 (4034356..4035720) | 1365 | 789 | 262 | 28.14 | 58.13 | 9.13 | Nu 1,2,3,4 |
| ZAT-C | CqZAT7 | XP_021766394 | NW_018743050.1 (1137896..1139053, complement) | 1158 | 1005 | 334 | 35.34 | 64.34 | 5.94 | Nu 1,3,4, Cy 1, Ec 2 |
| ZAT-C | CqZAT8 | XP_021774557 | NW_018743389.1 (4219259..4221239, complement) | 1981 | 990 | 329 | 35.79 | 60.52 | 6.69 | Nu 1,2,3,4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Vasquez, A.; Lima-Huanca, L.; Bardales-Álvarez, R.; Valderrama-Valencia, M.; Condori-Pacsi, S. In Silico Characterization and Determination of Gene Expression Levels Under Saline Stress Conditions in the Zinc Finger Family of the C1-2i Subclass in Chenopodium quinoa Willd. Int. J. Mol. Sci. 2025, 26, 2570. https://doi.org/10.3390/ijms26062570
Alvarez-Vasquez A, Lima-Huanca L, Bardales-Álvarez R, Valderrama-Valencia M, Condori-Pacsi S. In Silico Characterization and Determination of Gene Expression Levels Under Saline Stress Conditions in the Zinc Finger Family of the C1-2i Subclass in Chenopodium quinoa Willd. International Journal of Molecular Sciences. 2025; 26(6):2570. https://doi.org/10.3390/ijms26062570
Chicago/Turabian StyleAlvarez-Vasquez, Andrea, Luz Lima-Huanca, Roxana Bardales-Álvarez, Maria Valderrama-Valencia, and Sandro Condori-Pacsi. 2025. "In Silico Characterization and Determination of Gene Expression Levels Under Saline Stress Conditions in the Zinc Finger Family of the C1-2i Subclass in Chenopodium quinoa Willd" International Journal of Molecular Sciences 26, no. 6: 2570. https://doi.org/10.3390/ijms26062570
APA StyleAlvarez-Vasquez, A., Lima-Huanca, L., Bardales-Álvarez, R., Valderrama-Valencia, M., & Condori-Pacsi, S. (2025). In Silico Characterization and Determination of Gene Expression Levels Under Saline Stress Conditions in the Zinc Finger Family of the C1-2i Subclass in Chenopodium quinoa Willd. International Journal of Molecular Sciences, 26(6), 2570. https://doi.org/10.3390/ijms26062570







