Characterization of the Complete Mitochondrial Genome of Three Satyrid Butterfly Species (Satyrinae:Amathusiini) and Reconstructed Phylogeny of Satyrinae
Abstract
:1. Introduction
2. Results
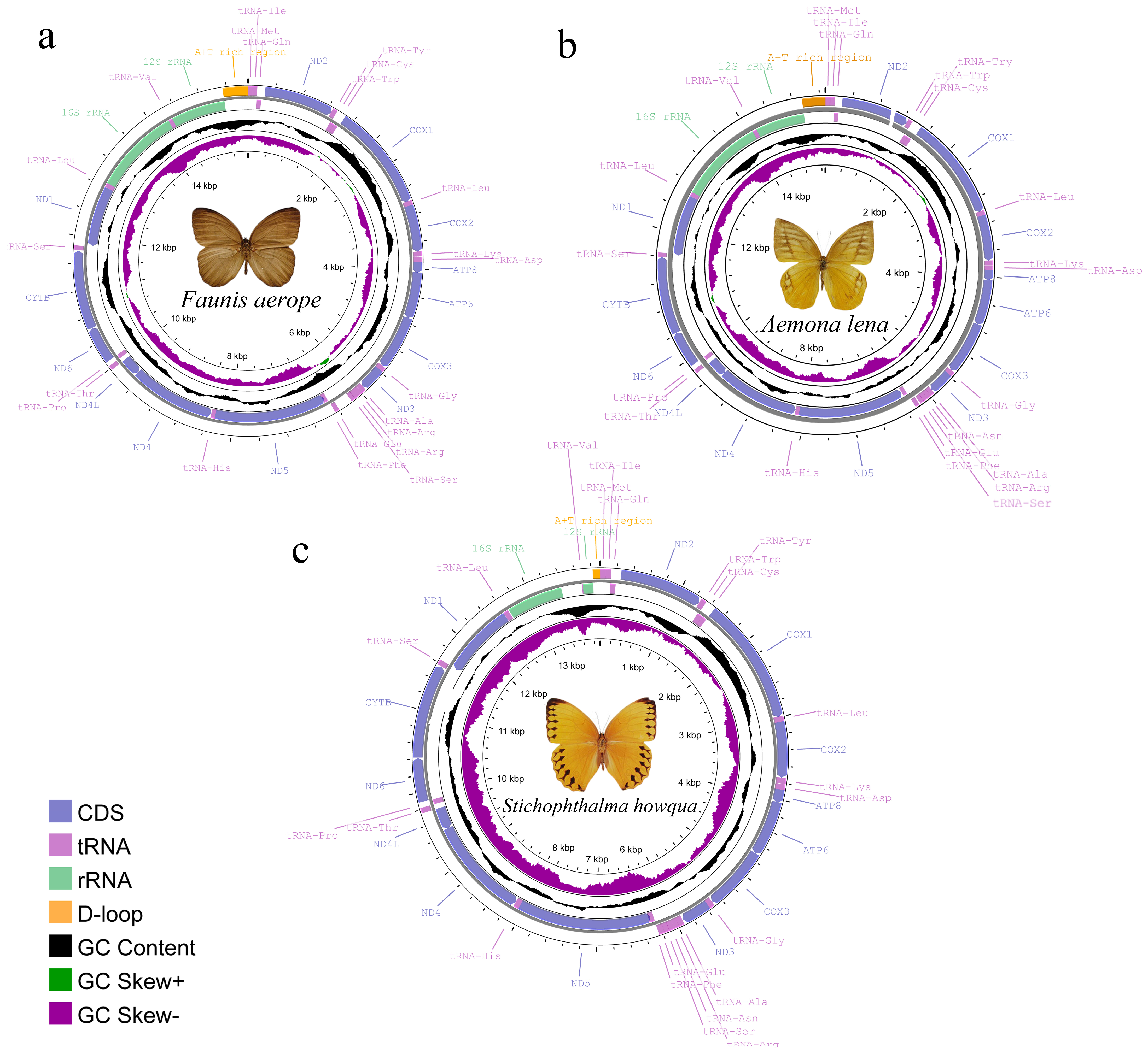
2.1. Mitochondrial Genome Assembly and Annotation
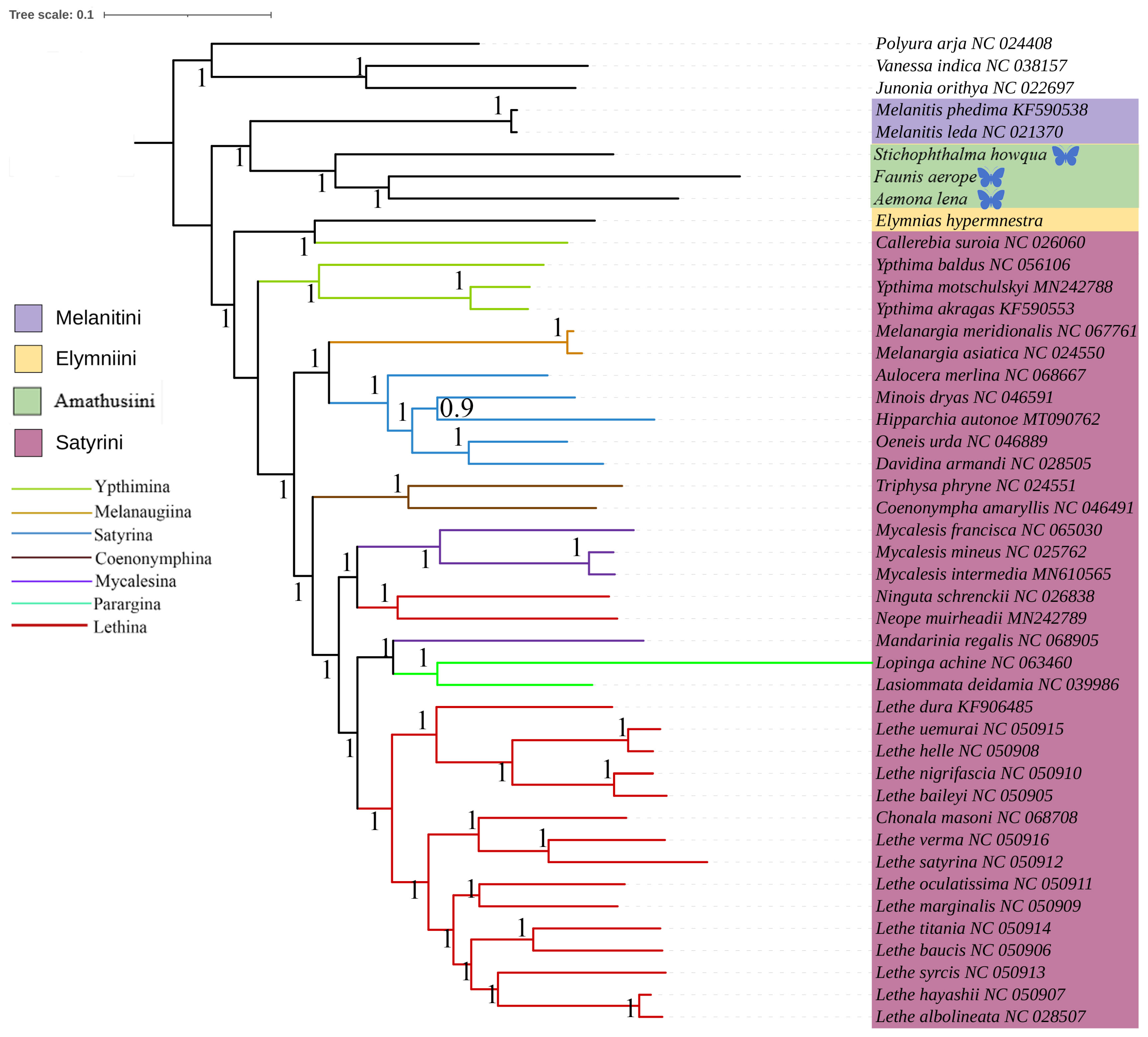
2.2. Phylogenetic Analyses
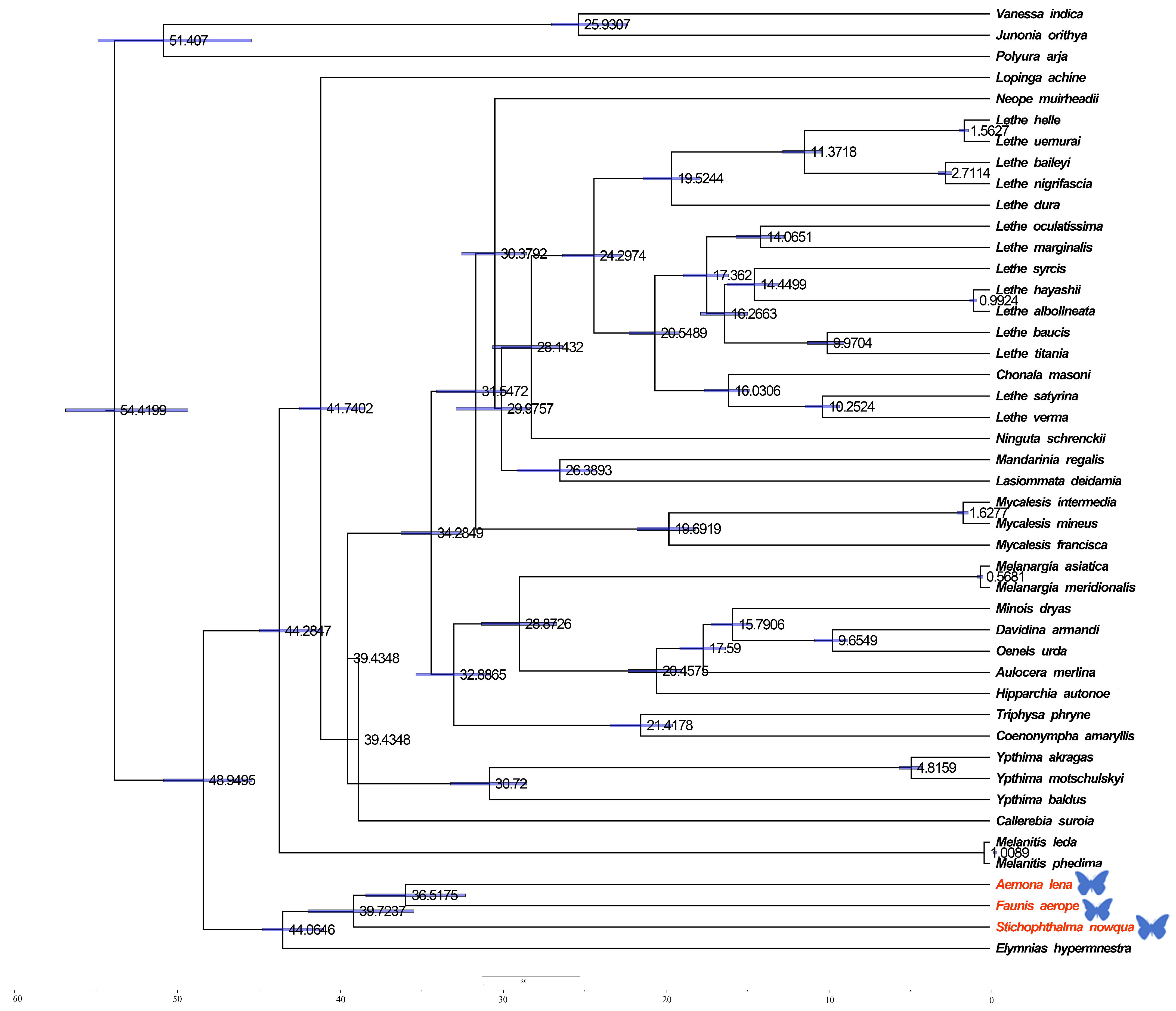
2.3. Divergence Time Estimation of Satyrinae Species
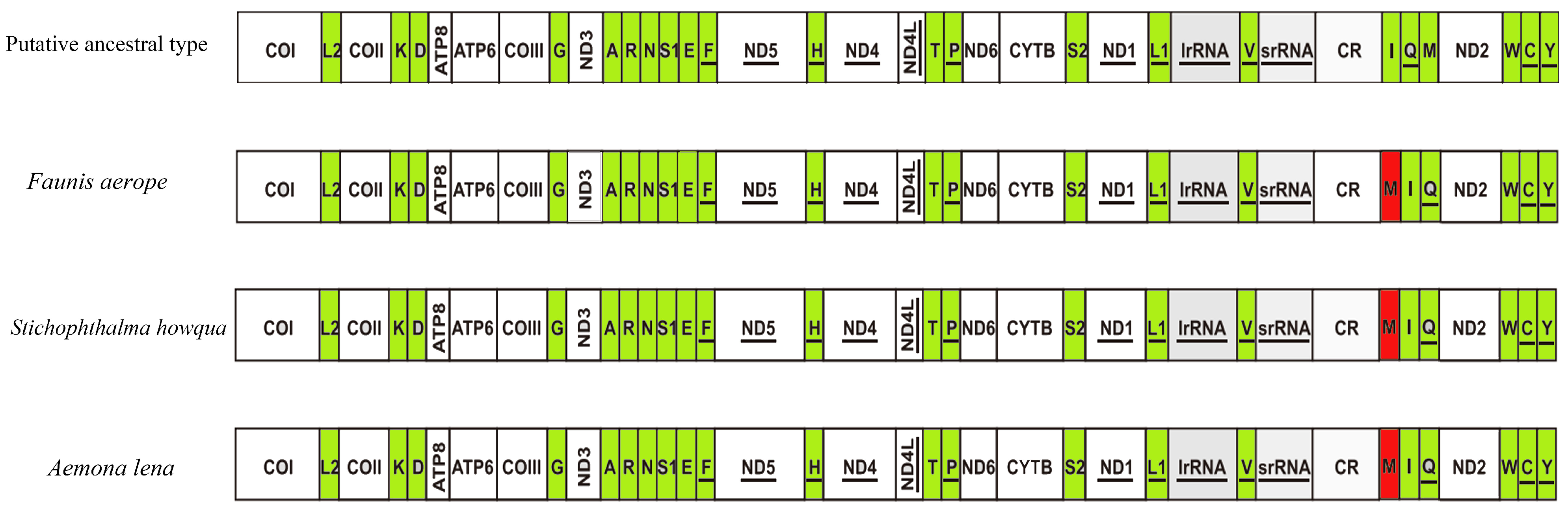
2.4. Mitogenomic Gene Rearrangements
3. Discussion
3.1. Three Satyrid Mitochondrial Genome Structure
3.2. Phylogenetic Analysis of Satyrinae
3.3. Mitogenomic Gene Rearrangements in Satyrinae
4. Materials and Methods
4.1. Sample Collection and DNA Extraction
4.2. Mitogenome Assembly, Annotation, and Analysis
4.3. Phylogenetic Analyses and Estimation of Divergence Time
| Taxon | Mitogenome Size (bp) | GenBank Accession No. | References | |
|---|---|---|---|---|
| Satyrinae | ||||
| Elymniini | ||||
| Elymnias hypermnestra | 15,167 | KF906484 | ||
| Melanitini | ||||
| Melanitis phedima | 15,142 | KF590538 | [30] | |
| Melanitis leda | 15,122 | JF905446 | [14] | |
| Satyrini | ||||
| Callerebia suroia | 15,208 | NC026060 | [31] | |
| Coenonympha amaryllis | 15,125 | NC046491 | [32] | |
| Davidina armandi | 15,214 | KF881046 | ||
| Hipparchia autonoe | 15,489 | OK094488 | [33] | |
| Lasiommata deidamia | 15,244 | MG880214 | [34] | |
| Lethe albolineata | 15,248 | NC028507 | [35] | |
| Lethe baileyi | 15,225 | NC050905 | [7] | |
| Lethe baucis | 15,251 | NC050906 | [7] | |
| Lethe dura | 15,259 | KF906485 | ||
| Lethe hayashii | 15,246 | NC050907 | [7] | |
| Lethe helle | 15,253 | NC050908 | [7] | |
| Lethe marginalis | 15,229 | NC050909 | [7] | |
| Lethe nigrifascia | 15,239 | NC050910 | [7] | |
| Lethe oculatissima | 15,243 | NC050911 | [7] | |
| Lethe satyrina | 15,271 | NC050912 | [7] | |
| Lethe syrcis | 15,252 | NC050913 | [7] | |
| Lethe titania | 15,257 | NC050914 | [7] | |
| Lethe uemurai | 15,272 | NC050915 | [7] | |
| Lethe verma | 15,239 | NC050916 | [7] | |
| Minois dryas | 15,194 | NC046591 | [36] | |
| Mycalesis francisca | 15,279 | MN242790 | [36] | |
| Mycalesis intermedia | 15,386 | MN610565 | [13] | |
| Mycalesis mineus | 15,267 | KM244676 | [37] | |
| Neope muirheadii | 15,217 | MN242789 | [36] | |
| Oeneis urda | 15,248 | NC046889 | [38] | |
| Triphysa phryne | 15,143 | KF906487 | [39] | |
| Ypthima akragas | 15,227 | KF590553 | [30] | |
| Ypthima motschulskyi | 15,232 | MN242788 | [36] | |
| Ypthima baldus | 15,304 | MN708051 | [6] | |
| Aulocera merlina | 15,259 | NC068667 | [40] | |
| Lopinga achine | 15,284 | NC063460 | [41] | |
| Mandarinia regalis | 15,267 | NC068905 | ||
| Melanargia asiatica | 15,142 | NC024550 | [42] | |
| Melanargia meridionalis | 15,442 | NC067761 | ||
| Ninguta schrenckii | 15,261 | NC026838 | [43] | |
| Chonala masoni | 15,278 | NC068708 | ||
| Aemona lena | 15,288 | This study | ||
| Faunis aerope | 15,512 | This study | ||
| Stichophthalma howqua | 13,914 | This study | ||
| Outgroup | Polyura arja | 15,363 | NC024408 | [44] |
| Junonia orithya | 15,241 | NC022697 | [45] | |
| Vanessa indica | 15,191 | NC038157 | [46] |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peña, C.; Wahlberg, N.; Weingartner, E.; Kodandaramaiah, U.; Nylin, S.; Freitas, A.V.; Brower, A.V. Higher level phylogeny of Satyrinae butterflies (Lepidoptera: Nymphalidae) based on DNA sequence data—ScienceDirect. Mol. Phylogenet. Evol. 2006, 40, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Marín, M.; Peña, C.; Freitas, A.; Wahlberg, N.; Uribe, S. From the Phylogeny of the Satyrinae Butterflies to the Systematics of Euptychiina (Lepidoptera: Nymphalidae): History, Progress and Prospects. Neotrop. Entomol. 2011, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wahlberg, N.; Leneveu, J.; Kodandaramaiah, U.; Pena, C.; Nylin, S.; Freitas, A.V.L.; Brower, A.V. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc. R. Soc. B Biol. Sci. 2009, 276, 4295–4302. [Google Scholar] [CrossRef]
- Peña, C.; Wahlberg, N. Prehistorical climate change increased diversification of a group of butterflies. Biol. Lett. 2008, 4, 274–278. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Li, X.D.; Hu, H.W.; Zhang, S.L.; Wang, J.W.; Li, R. Characterization of the complete mitochondrial genome of Ypthima baldus (Lepidoptera: Satyrinae) with phylogenetic analysis. Mitochondrial Dna Part B-Resour. 2020, 5, 1019–1020. [Google Scholar] [CrossRef]
- Chen, L.; Wahlberg, N.; Liao, C.Q.; Wang, C.B.; Ma, F.Z.; Huang, G.H. Fourteen complete mitochondrial genomes of butterflies from the genus Lethe (Lepidoptera, Nymphalidae, Satyrinae) with mitogenome-based phylogenetic analysis. Genomics 2020, 112, 4435–4441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, R.; Zhou, C. Complete mitochondrial genomes of Epeorus carinatus and E. dayongensis (Ephemeroptera: Heptageniidae): Genomic comparison and phylogenetic inference. Gene 2021, 777, 145467. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Su, T.; He, B.; Zhao, F.; Huang, Z. Complete mitochondrial genome of Casmara patrona (Lepidoptera: Oecophoridae). Mitochondrial DNA Part B 2021, 6, 325–326. [Google Scholar] [CrossRef]
- Yang, J.; Ren, Q.; Huang, Y. Complete mitochondrial genomes of three crickets (Orthoptera: Gryllidae) and comparative analyses within Ensifera mitogenomes. Zootaxa 2016, 4092, 529. [Google Scholar] [CrossRef]
- Liu, B.; Sun, H.; Zhan, Q.; Gai, Y. The complete mitochondrial genome of Orthaga achatina (Lepidoptera: Pyralidae). Mitochondrial DNA Part B 2021, 6, 821–823. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Su, X.; Li, C.; Zhou, Y.; Li, S.; Guo, J.; Fan, Q.; Lü, S.; Zhang, Y. Draft genome of the blister beetle, Epicauta chinensis. Int. J. Biol. Macromol. 2021, 193 Pt B, 1694–1706. [Google Scholar] [CrossRef]
- Wu, Y.P.; Lu, J.J.; Yang, J.; Wang, J.P.; Cao, T.W.; Fan, R.J. Complete mitochondrial genome of Mycalesis intermedia (Lepidoptera: Nymphalidae). Mitochondrial DNA Part B Resour. 2020, 5, 703–704. [Google Scholar] [CrossRef]
- Shi, Q.H.; Zhao, F.; Hao, J.S.; Yang, Q. Complete mitochondrial genome of the Common Evening Brown, Melanitis leda Linnaeus (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA 2013, 24, 492–494. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chang, Z.; Chen, L.; He, J.; Li, X. Genome size variation in butterflies (Insecta, Lepidotera, Papilionoidea): A thorough phylogenetic comparison. Syst. Entomol. 2020, 45, 571–582. [Google Scholar] [CrossRef]
- Xu, C.W.Y. Butterflies of China (Complete 4 Volumes); Straits Publishing House: Fuzhou, China, 2017. [Google Scholar]
- Zhou, Y. Chinese Butterfly Fauna; Henan Science and Technology Press: Zhengzhou, China, 1999. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juehling, F.; Stadler, P. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772. [Google Scholar]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Wiemers, M.; Chazot, N.; Wheat, C.W.; Schweiger, O.; Wahlberg, N. A complete time-calibrated multi-gene phylogeny of the European butterflies. ZooKeys 2020, 938, 97–124. [Google Scholar] [CrossRef]
- Chazot, N.; Wahlberg, N.; Freitas, A.; Eacute Lucci, V.; Mitter, C.; Labandeira, C.; Sohn, J.C.; Sahoo, R.K.; Seraphim, N.; de Jong, R.; et al. Priors and Posteriors in Bayesian Timing of Divergence Analyses: The Age of Butterflies Revisited. Syst. Biol. 2019, 68, 797–813. [Google Scholar] [CrossRef]
- Wu, L.W.; Lin, L.H.; Lees, D.C.; Hsu, Y.F. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genom. 2014, 15, 468. [Google Scholar] [CrossRef]
- Shi, Q.; Zhang, W.; Hao, J. The complete mitochondrial genome of Callerebia suroia (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 1463–1465. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, C.; Zhai, Q.; Zhang, Y. The complete mitochondrial genome sequence of Coenonympha amaryllis and monophyly of Satyrinae (Lepidoptera: Nymphalidae). Mitochondrial DNA Part B Resour. 2020, 5, 1223–1224. [Google Scholar] [CrossRef]
- Dan, Z.; Lei_Duan Chen, Z.; Xu, S. Mitogenomes of Three Satyrid Butterfly Species (Nymphalidae: Lepidoptera) and Reconstructed Phylogeny of Satyrinae. Diversity 2021, 13, 468. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, C.; Geng, X.; Li, J. Complete mitochondrial genome of Lasiommata deidamia and its phylogenetic implication to subfamily Satyrinae (Lepidoptera: Nymphalidae). Mitochondrial DNA Part B Resour. 2021, 6, 2943–2945. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, C.; Lei, Y.; Fan, C.; Gao, Y.; Xu, C.; Wang, R. Complete mitochondrial genome of a satyrid butterfly, Lethe albolineata (Lepidoptera: Nymphalidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 4195–4196. [Google Scholar] [CrossRef]
- Yang, M.; Song, L.; Zhou, L.; Shi, Y.; Song, N.; Zhang, Y. Mitochondrial genomes of four satyrine butterflies and phylogenetic relationships of the family Nymphalidae (Lepidoptera: Papilionoidea). Int. J. Biol. Macromol. 2020, 145, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.F.; Bonebrake, T.C. Consistent heat tolerance under starvation across seasonal morphs in Mycalesis mineus (Lepidoptera: Nymphalidae). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2022, 271, 111261. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Z.; Wang, S.; Zhong, H.; Wang, N.; Liang, B.J. A mitogenomic phylogeny of satyrid butterflies and complete mitochondrial genome of Oeneis urda (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA Part B 2020, 5, 1344–1345. [Google Scholar] [CrossRef]
- Zhang, W.; Gan, S.; Zuo, N.; Chen, C.; Wang, Y.; Hao, J. The complete mitochondrial genome of Triphysa phryne (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 474–475. [Google Scholar] [CrossRef]
- Shi, Q.; Xie, J.; Wu, J.; Chen, S.; Sun, G.; Zhang, J. Characterization of the complete mitochondrial genome of an endemic species in China, Aulocera merlina (Lepidoptera: Nymphalidae: Satyrinae) and phylogenetic analysis within Satyrinae. Ecol. Evol. 2024, 14, e11355. [Google Scholar] [CrossRef]
- Wu, J.L.; Bao, T.; Sun, G.; Xiao, Y.; Fang, Y.; Shi, Q.H. Complete mitochondrial genome of the Woodland Brown, Lopinga achine Scopoli, 1763 (Nymphalidae: Satyrinae) and its phylogenetic analysis. Mitochondrial DNA B Resour 2022, 7, 747–749. [Google Scholar] [CrossRef]
- Huang, D.; Hao, J.; Zhang, W.; Su, T.; Wang, Y.; Xu, X. The complete mitochondrial genome of Melanargia asiatica (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA Part A 2016, 27, 806–808. [Google Scholar] [CrossRef]
- Fan, C.; Xu, C.; Li, J.; Lei, Y.; Gao, Y.; Xu, C.; Wang, R. Complete mitochondrial genome of a satyrid butterfly, Ninguta schrenkii (Lepidoptera: Nymphalidae). Mitochondrial DNA Part A 2016, 27, 80–81. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Duan, L.; Hao, X.; Xiao, J.; Yuan, X. Characterization of complete mitochondrial genome of Polyura narcaeus (Lepidoptera: Nymphalidae: Charaxinae). Mitochondrial DNA Part B Resour. 2021, 6, 1654–1655. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Huang, D.; Wang, Y.; Hao, J. The complete mitochondrial genome of Blue Pansy, Junonia orithya (Lepidoptera: Nymphalidae: Nymphalinae). Mitochondrial DNA 2015, 26, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, N.; Xu, L.; Fang, J.; Wang, S. The complete mitochondrial genome of Vanessa indica and phylogenetic analyses of the family Nymphalidae. Genes Genom. 2018, 40, 1011–1022. [Google Scholar] [CrossRef]



| Feature | Size | A + T% | AT-Skew | GC-Skew | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fa | Ae | St | Fa | Ae | St | Fa | Ae | St | Fa | Ae | St | |
| Whole genome | 15,512 | 15,288 | 13,914 | 80 | 80 | 77.9 | −0.050 | −0.025 | −0.024 | −0.224 | −0.211 | 0.304 |
| Protein-coding genes | 11,210 | 11,246 | 11,170 | 78.4 | 78.6 | 77.7 | −0.159 | −0.155 | −0.161 | −0.163 | −0.134 | −0.217 |
| 1st codon position | 3736 | 3748 | 3722 | 74.4 | 73.2 | 72.1 | −0.012 | −0.055 | −0.026 | 0.438 | 0.456 | 0.464 |
| 2nd codon position | 3736 | 3748 | 3722 | 70.2 | 70.3 | 70.3 | −0.36 | −0.343 | −0.363 | −0.194 | −0.184 | −0.199 |
| 3rd codon position | 3736 | 3748 | 3722 | 87.8 | 92.8 | 90.7 | −0.057 | −0.107 | −0.121 | 0.7 | 0.642 | 0.673 |
| tRNA genes | 1445 | 1450 | 1415 | 81 | 81.2 | 82 | −0.001 | 0.019 | 0.007 | 0.182 | 0.161 | 0.157 |
| rRNA genes | 849 | 2195 | 866 | 84.3 | 85.1 | 80 | 0.095 | 0.071 | 0.062 | 0.338 | 0.331 | 0.260 |
| A + T-rich region | 364 | 345 | 91 | 92.1 | 89.6 | 75.9 | −0.092 | −0.043 | −0.130 | −0.379 | 0.333 | −0.182 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dan, Z.; Zhang, Y.; Chen, Z. Characterization of the Complete Mitochondrial Genome of Three Satyrid Butterfly Species (Satyrinae:Amathusiini) and Reconstructed Phylogeny of Satyrinae. Int. J. Mol. Sci. 2025, 26, 2609. https://doi.org/10.3390/ijms26062609
Dan Z, Zhang Y, Chen Z. Characterization of the Complete Mitochondrial Genome of Three Satyrid Butterfly Species (Satyrinae:Amathusiini) and Reconstructed Phylogeny of Satyrinae. International Journal of Molecular Sciences. 2025; 26(6):2609. https://doi.org/10.3390/ijms26062609
Chicago/Turabian StyleDan, Zhicuo, Ying Zhang, and Zhenning Chen. 2025. "Characterization of the Complete Mitochondrial Genome of Three Satyrid Butterfly Species (Satyrinae:Amathusiini) and Reconstructed Phylogeny of Satyrinae" International Journal of Molecular Sciences 26, no. 6: 2609. https://doi.org/10.3390/ijms26062609
APA StyleDan, Z., Zhang, Y., & Chen, Z. (2025). Characterization of the Complete Mitochondrial Genome of Three Satyrid Butterfly Species (Satyrinae:Amathusiini) and Reconstructed Phylogeny of Satyrinae. International Journal of Molecular Sciences, 26(6), 2609. https://doi.org/10.3390/ijms26062609


_Kim.png)



