Genome-Wide Analysis of the PERK Gene Family in Brassica napus L. and Their Potential Roles in Clubroot Disease
Abstract
1. Introduction
2. Results
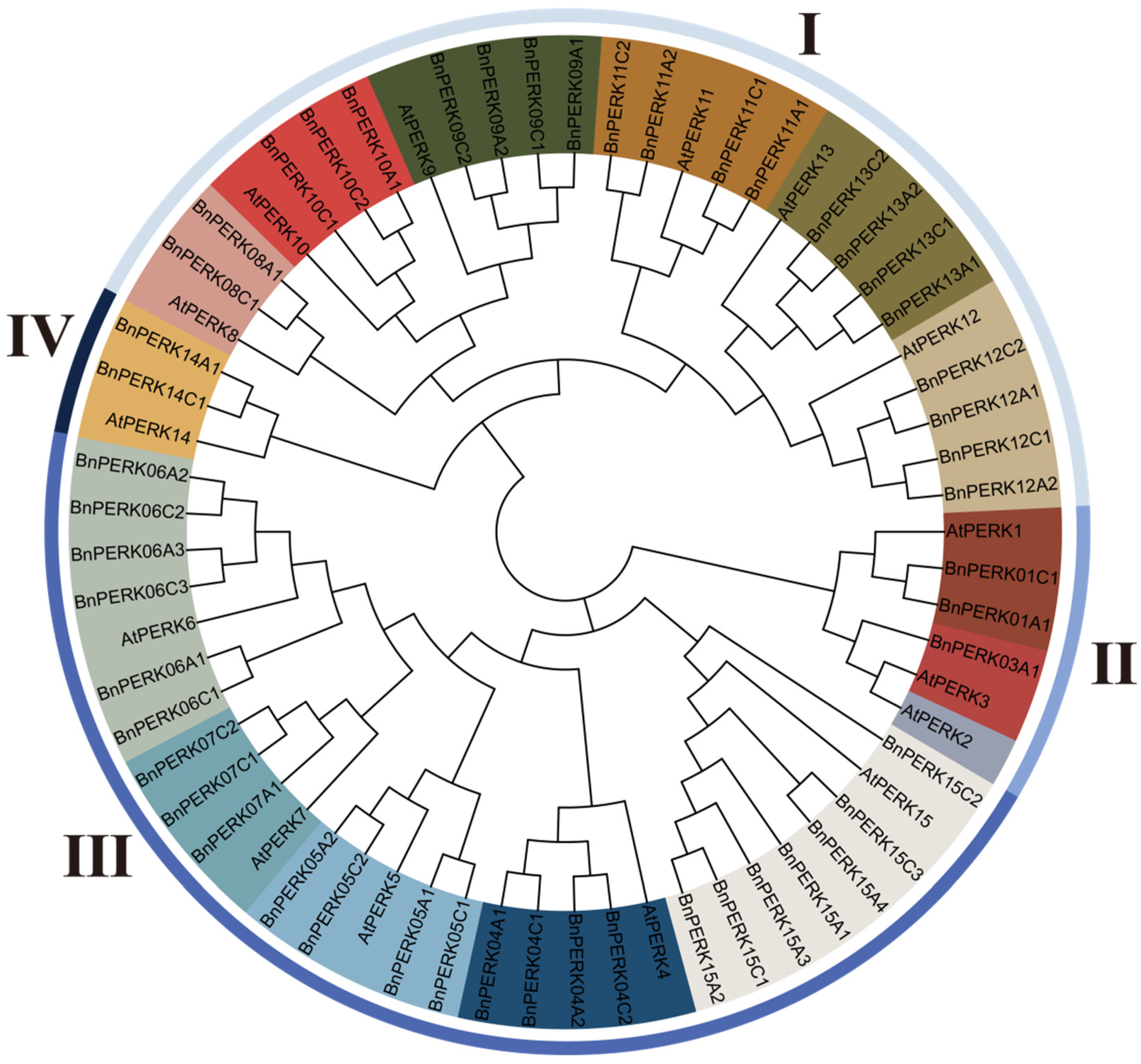
2.1. Identification and Phylogenetic Analysis of BnPERKs in Brassica napus
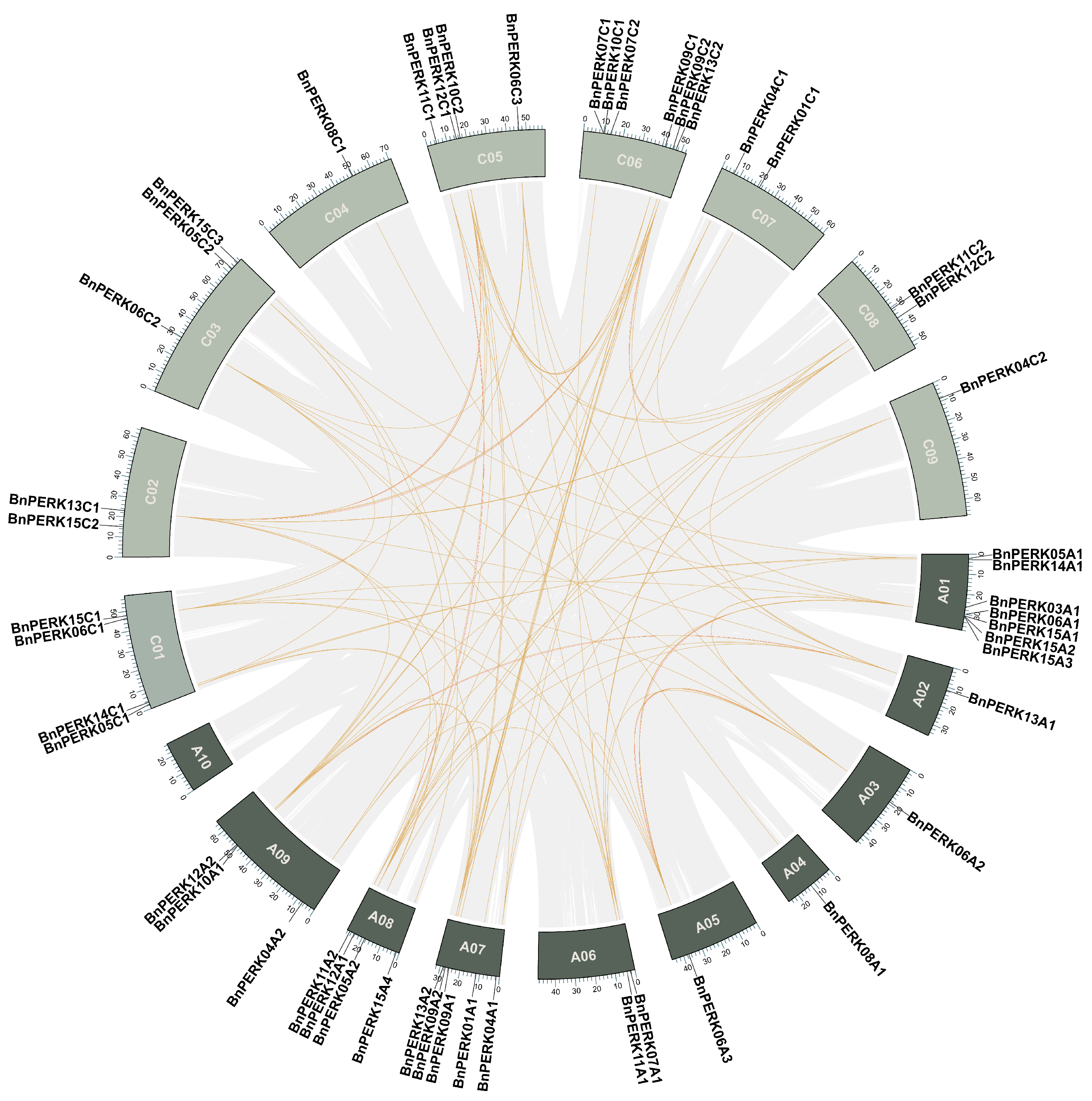
2.2. Chromosomal Distribution and Collinearity Analysis of BnPERK Genes
2.3. Gene Structure, Conserved Motifs, and Cis-Elements Analysis of BnPERK Genes
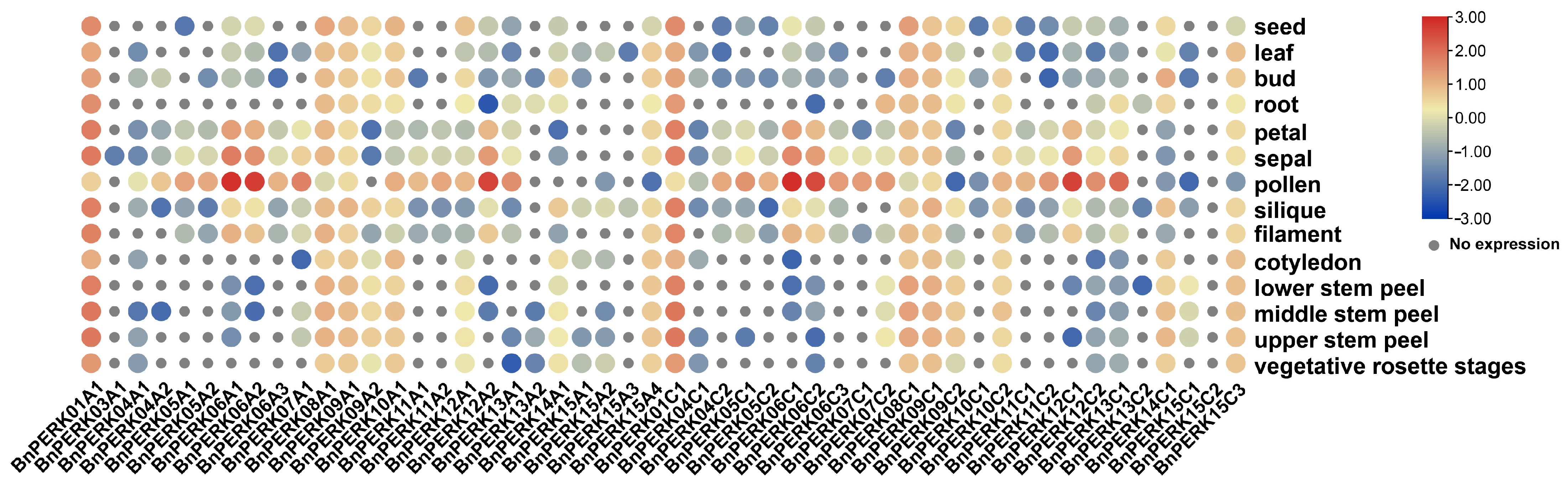
2.4. Expression Pattern of BnPERK Gene Family Across Various Tissues
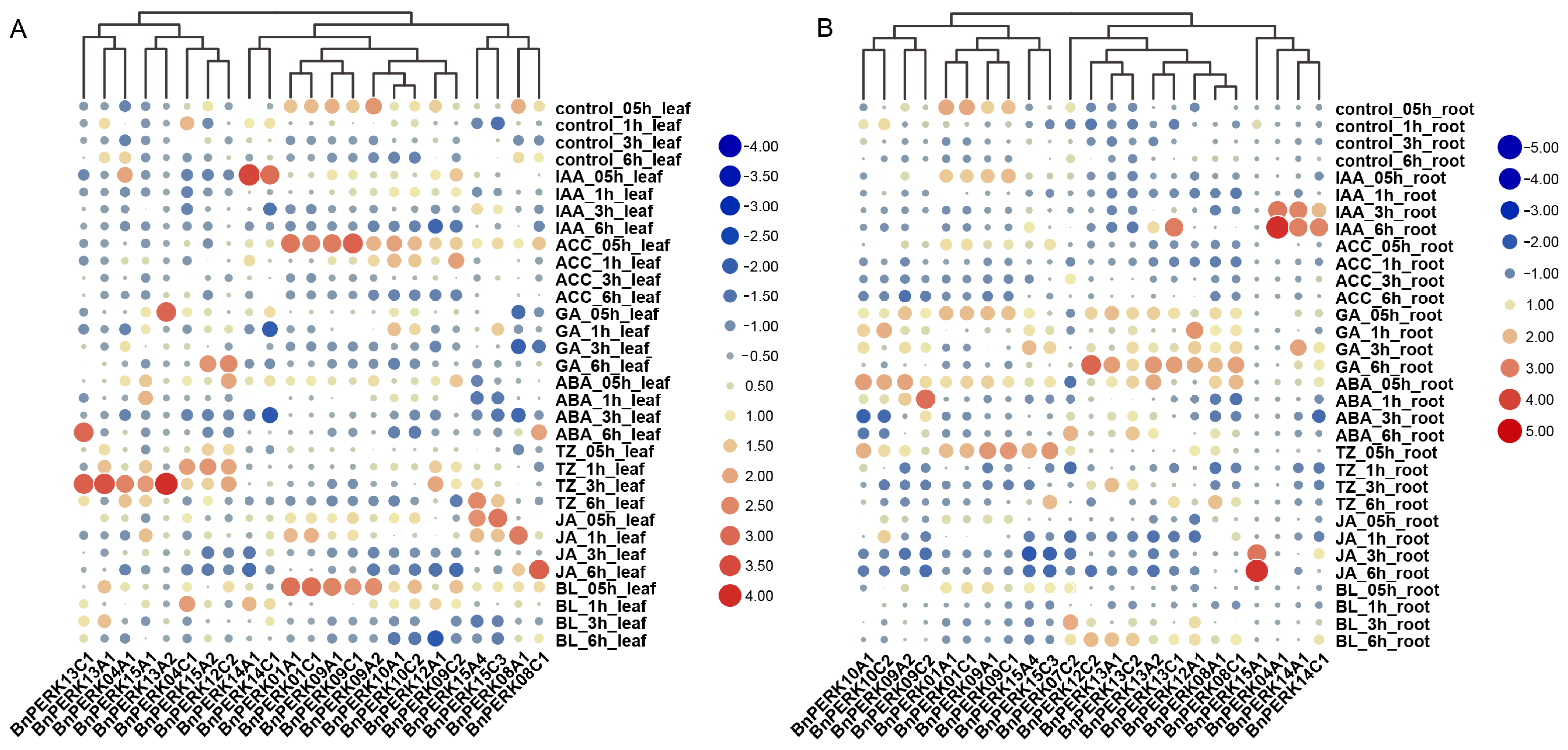
2.5. Expression Pattern of BnPERK Gene Family in Response to Hormone Treatments
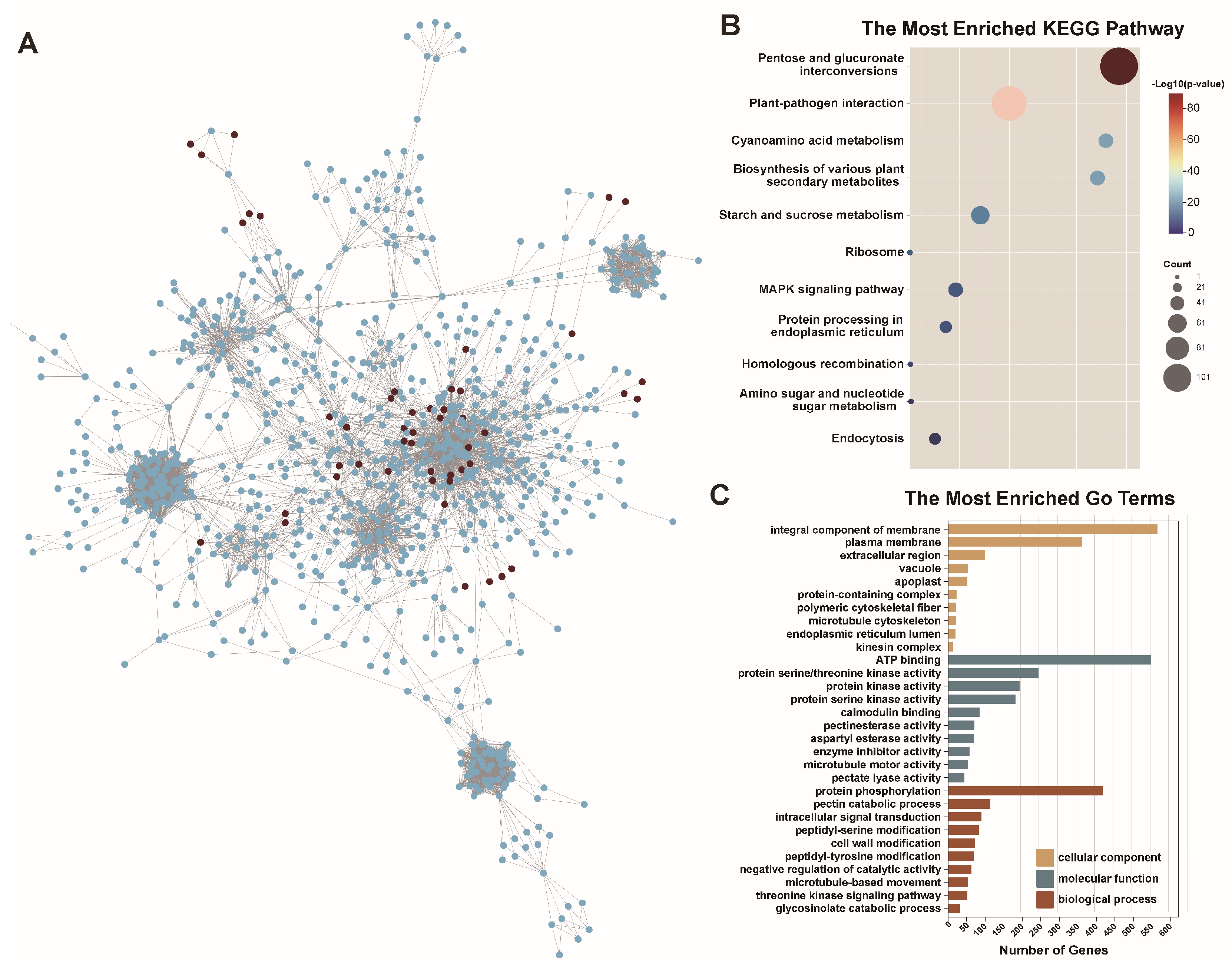
2.6. Protein–Protein Interaction Network Analysis of BnPERK Genes
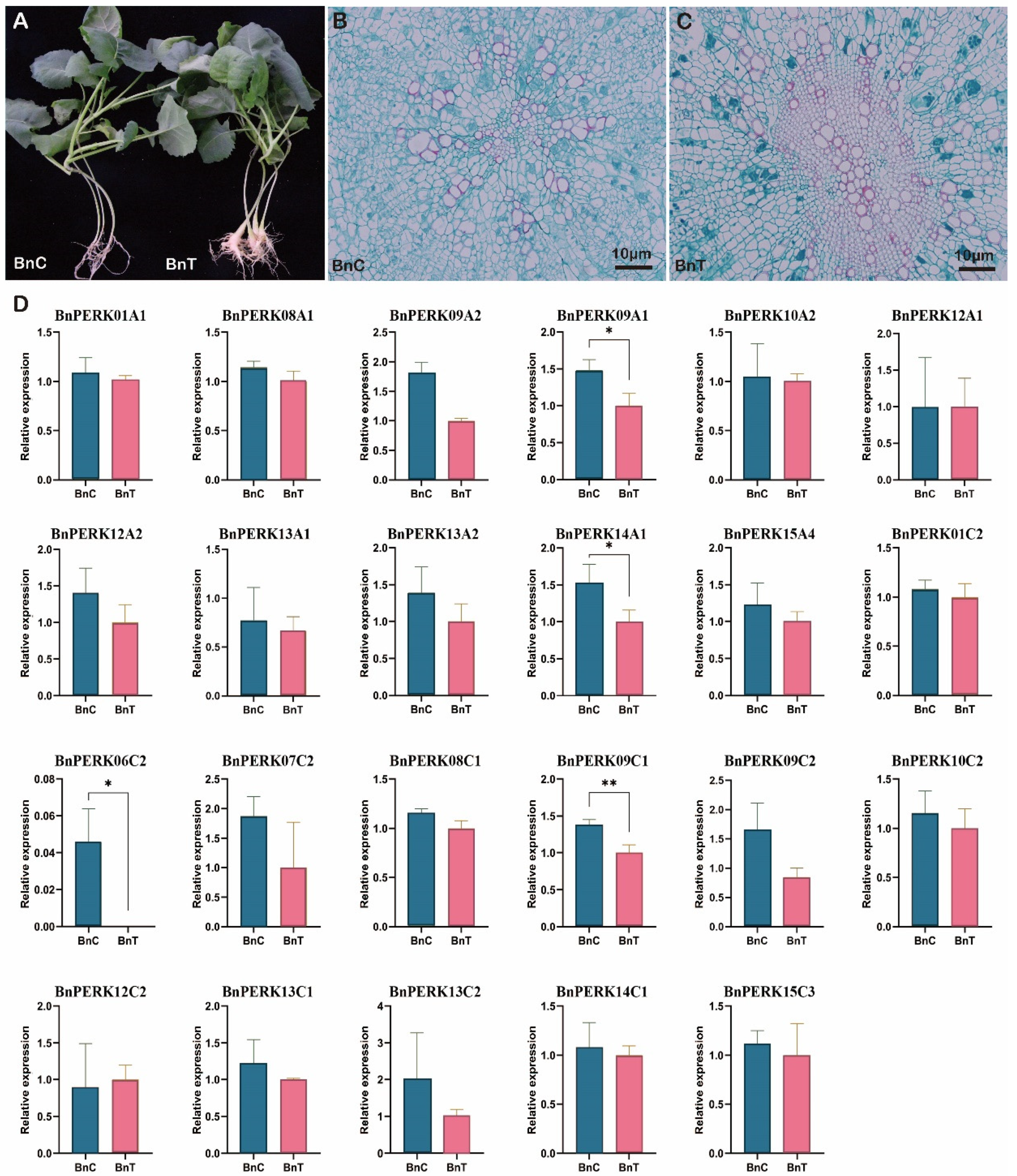
2.7. Root Gall Formation and BnPERK Gene Expression in B. napus upon Plasmodiophora brassicae Inoculation
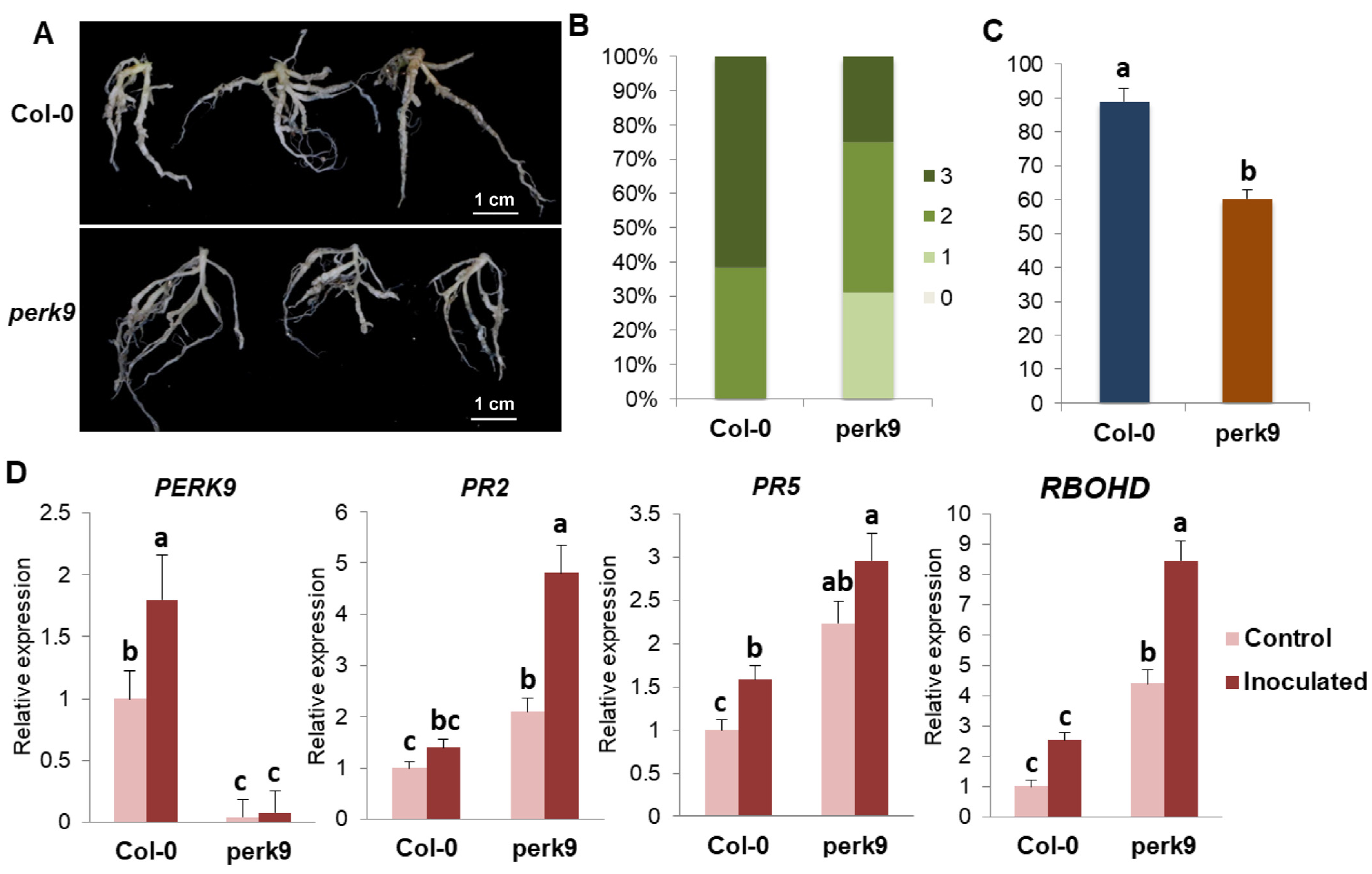
2.8. Function of AtPERK9 in Clubroot Disease
3. Discussion
3.1. Deciphering the Evolutionary Dynamics of the BnPERK Genes
3.2. Exploring the Structural Diversity and Functional Roles of BnPERKs
3.3. Revealing the Role of BnPERK in Clubroot Disease
4. Materials and Methods
4.1. Identification of BnPERK Genes in Brassica napus
4.2. Chromosomal Location, Gene Duplication, and Phylogenetic Analysis
4.3. Gene Structure, Conserved Motifs, and Cis-Elements Analysis
4.4. Expression Profiles of BnPERK Genes in Different Tissues and Under Hormone Treatments
4.5. Network-Based Prediction of Protein Interactions
4.6. Plasmodiophora Brassicae Inoculation, Clubroot Disease Investigation, and Histocytological Analysis
4.7. Transcriptome Analysis of BnPERK Genes in B. napus During Clubroot Disease
4.8. qRT-PCR Analysis of Pathogenesis-Related (PR) Genes in A. thaliana
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morris, E.R.; Walker, J.C. Receptor-like protein kinases: The keys to response. Curr. Opin. Plant Biol. 2003, 6, 339–342. [Google Scholar] [CrossRef]
- Cui, Y.W.; Lu, X.T.; Gou, X.P. Receptor-like protein kinases in plant reproduction: Current understanding and future perspectives. Plant Commun. 2022, 3, 100273. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, J.M. Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 2013, 55, 1271–1286. [Google Scholar] [CrossRef]
- Dievart, A.; Gottin, C.; Périn, C.; Ranwez, V.; Chantret, N. Origin and Diversity of Plant Receptor-Like Kinases. Annu. Rev. Plant Biol. 2020, 71, 131–156. [Google Scholar] [CrossRef]
- Invernizzi, M.; Hanemian, M.; Keller, J.; Libourel, C.; Roby, D. PERKing up our understanding of the proline-rich extensin-like receptor kinases, a forgotten plant receptor kinase family. New Phytol. 2022, 235, 875–884. [Google Scholar] [CrossRef]
- Bai, L.; Zhou, Y.; Song, C.P. Arabidopsis proline-rich extensin-like receptor kinase 4 modulates the early event toward abscisic acid response in root tip growth. Plant Signal. Behav. 2009, 4, 1075–1077. [Google Scholar] [CrossRef]
- Silva, N.F.; Goring, D.R. The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 2002, 50, 667–685. [Google Scholar] [CrossRef]
- Uemura, T.; Hachisu, M.; Desaki, Y.; Ito, A.; Hoshino, R.; Sano, Y.; Nozawa, A.; Mujiono, K.; Galis, I.; Yoshida, A.; et al. Soy and Arabidopsis receptor-like kinases respond to polysaccharide signals from Spodoptera species and mediate herbivore resistance. Commun. Biol. 2020, 3, 224. [Google Scholar] [CrossRef]
- Xue, C.W.; Li, W.F.; Shen, R.F.; Lan, P. PERK13 modulates phosphate deficiencyinduced root hair elongation in Arabidopsis. Plant Sci. 2021, 312, 111060. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhang, G.Z.; Zhou, Y.; Zhang, Z.P.; Wang, W.; Du, Y.Y.; Wu, Z.Y.; Song, C.P. Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in. Plant J. 2009, 60, 314–327. [Google Scholar] [CrossRef]
- Nakhamchik, A.; Zhao, Z.Y.; Provart, N.J.; Shiu, S.H.; Keatley, S.K.; Cameron, R.K.; Goring, D.R. A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol. 2004, 45, 1875–1881. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Routray, S.; Mohapatra, C.; Saha, D.; Ram, C.; Siddique, K.H.M.; Kumar, A.; et al. Genome-wide analysis and characterization of the proline-rich extensin-like receptor kinases (PERKs) gene family reveals their role in different developmental stages and stress conditions in wheat (Triticum aestivum L.). Plants 2022, 11, 496. [Google Scholar] [CrossRef]
- Qanmber, G.; Liu, J.; Yu, D.Q.; Liu, Z.; Lu, L.L.; Mo, H.J.; Ma, S.Y.; Wang, Z.; Yang, Z.R. Genome-wide identification and characterization of the PERK gene family in Gossypium hirsutum reveals gene duplication and functional divergence. Int. J. Mol. Sci. 2019, 20, 1750. [Google Scholar] [CrossRef]
- Feng, P.; Shi, J.Q.; Zhang, T.; Zhong, Y.Q.; Zhang, L.S.; Yu, G.L.; Zhang, T.Q.; Zhu, X.Y.; Xing, Y.D.; Yin, W.Z.; et al. Zebra leaf 15, a receptor-like protein kinase involved in moderate low temperature signaling pathway in rice. Rice 2019, 12, 83. [Google Scholar] [CrossRef]
- Haffani, Y.Z.; Silva-Gagliardi, N.F.; Sewter, S.K.; Aldea, M.G.; Zhao, Z.Y.; Nakhamchik, A.; Cameron, R.K.; Goring, D.R. Altered expression of PERK receptor kinases in leads to changes in Arabidopsis growth and floral organ formation. Plant Signal. Behav. 2006, 1, 251–260. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Katara, J.L.; Parameswaran, C.; Misra, N.; Kumar, M.; Chung, S.M.; Alamri, S.; Siddiqui, M.H. Genome-wide analysis of proline-rich extensin-like receptor kinases (PERKs) gene family reveals their roles in plant development and stress conditions in Oryza sativa L. Plant Sci. 2023, 334, 111749. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.Y.; Parkin, I.A.P.; Tang, H.B.; Wang, X.Y.; Chiquet, J.; Belcram, H.; Tong, C.B.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Song, X.M.; Wei, Y.P.; Xiao, D.; Gong, K.; Sun, P.C.; Ren, Y.M.; Yuan, J.Q.; Wu, T.; Yang, Q.H.; Li, X.Y.; et al. Genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 2021, 186, 388–406. [Google Scholar] [CrossRef]
- Chen, G.H.; Wang, J.; Wang, H.; Wang, C.G.; Tang, X.Y.; Li, J.; Zhang, L.; Song, J.H.; Hou, J.F.; Yuan, L.Y. Genome-wide analysis of proline-rich extension-like receptor protein kinase (PERK) in Brassica rapa and its association with the pollen development. BMC Genom. 2020, 21, 401. [Google Scholar] [CrossRef]
- Gu, B.J.; Tong, Y.K.; Wang, Y.Y.; Zhang, M.L.; Ma, G.J.; Wu, X.Q.; Zhang, J.F.; Xu, F.; Li, J.; Ren, F. Genome-wide evolution and expression analysis of the MYB-CC gene family in Brassica spp. PeerJ 2022, 10, e12882. [Google Scholar] [CrossRef]
- Roy, B.C.; Shukla, N.; Gachhui, R.; Mukherjee, A. Genome-wide analysis of glutamate receptor gene family in allopolyploid Brassica napus and its diploid progenitors. Genetica 2023, 151, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.L.; Zuo, R.; Bai, Z.T.; Yang, L.L.; Zhao, C.J.; Gao, F.; Cheng, X.H.; Huang, J.Y.; Liu, Y.Y.; Li, Y.; et al. Genome-wide characterization of serine/arginine-rich gene family and its genetic effects on agronomic traits of Brassica napus. Front. Plant Sci. 2022, 13, 829668. [Google Scholar] [CrossRef]
- Albalat, R.; Cañestro, C. Evolution by gene loss. Nat. Rev. Genet. 2016, 17, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.F.; Zhang, Y.; Lei, X.Y.; Zhang, Z.M. Natural selection of protein structural and functional properties: A single nucleotide polymorphism perspective. Genome Biol. 2008, 9, R69. [Google Scholar] [CrossRef]
- Barberis, E.; Marengo, E.; Manfredi, M. Protein subcellular localization prediction. Methods Mol. Biol. 2021, 2361, 197–212. [Google Scholar]
- Zhang, C.X.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 2017, 45, 291–299. [Google Scholar] [CrossRef]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Mangano, S.; Marzol, E.; Denita-Juarez, S.P.; Salter, J.D.S.; Velasquez, S.M.; Muschietti, J.P.; Estevez, J.M. Proline-rich extensin-like receptor kinases PERK5 and PERK12 are involved in pollen tube growth. FEBS Lett. 2021, 595, 2593–2607. [Google Scholar] [CrossRef]
- Parkin, I.A.P.; Gulden, S.M.; Sharpe, A.G.; Lukens, L.; Trick, M.; Osborn, T.C.; Lydiate, D.J. Segmental structure of the genome based on comparative analysis with Arabidopsis thaliana. Genetics 2005, 171, 765–781. [Google Scholar] [CrossRef]
- Lukens, L.N.; Quijada, P.A.; Udall, J.; Pires, J.C.; Schranz, M.E.; Osborn, T.C. Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linn. Soc. 2004, 82, 665–674. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Wu, H.C.; Jinn, T.L. Coordination of ABA and chaperone signaling in plant stress responses. Trends Plant Sci. 2019, 24, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Xu, H.; Gao, X.H.; Fu, X.D. New insights into gibberellin signaling in regulating plant growth-metabolic coordination. Curr. Opin. Plant Biol. 2021, 63, 102074. [Google Scholar] [CrossRef]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Wimalagunasekara, S.S.; Weeraman, J.W.J.K.; Tirimanne, S.; Fernando, P.C. Protein-protein interaction (PPI) network analysis reveals important hub proteins and sub-network modules for root development in rice (Oryza sativa). J. Genet. Eng. Biotechnol. 2023, 21, 69. [Google Scholar] [CrossRef]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Salter, J.D.S.; Marzol, E.; Muschietti, J.P.; Estevez, J.M. An update on cell surface proteins containing extensin-motifs. J. Exp. Bot. 2016, 67, 477–487. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Raja, V.; Majeed, U.; Kang, H.; Andrabi, K.I.; John, R. Abiotic stress: Interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Chen, W.; Li, Y.; Yan, R.B.; Ren, L.; Liu, F.; Zeng, L.Y.; Sun, S.N.; Yang, H.H.; Chen, K.R.; Xu, L.; et al. SnRK1.1-mediated resistance of Arabidopsis thaliana to clubroot disease is inhibited by the novel Plasmodiophora brassicae effector PBZF1. Mol. Plant Pathol. 2021, 22, 1057–1069. [Google Scholar] [CrossRef]
- Kemen, E.; Jones, J.D.G. Obligate biotroph parasitism: Can we link genomes to lifestyles? Trends Plant Sci. 2012, 17, 448–457. [Google Scholar] [CrossRef]
- Kong, L.; Li, X.; Zhan, Z.; Piao, Z. Sugar transporters in Plasmodiophora brassicae: Genome-wide identification and functional verification. Int. J. Mol. Sci. 2022, 23, 5264. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lal, N.K.; Lin, Z.J.D.; Ma, S.S.; Liu, J.; Castro, B.; Toruño, T.; Dinesh-Kumar, S.P.; Coaker, G. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 2020, 11, 1838. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Muller, J. Auxin homeostasis, signaling, and interaction with other growth hormones during the clubroot disease of Brassicaceae. Plant Signal. Behav. 2014, 9, e28593. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ren, L.; Chen, K.R.; Liu, F.; Fang, X.P. Putative role of IAA during the early response of Brassica napus L. to Plasmodiophora brassicae. Eur. J. Plant Pathol. 2016, 145, 601–613. [Google Scholar] [CrossRef]
- Wei, X.C.; Liao, R.J.; Zhang, X.W.; Zhao, Y.Y.; Xie, Z.Q.; Yang, S.J.; Su, H.A.; Wang, Z.Y.; Zhang, L.Y.; Tian, B.M.; et al. Integrative transcriptome, miRNAs, degradome, and phytohormone analysis of Brassica rapa L. in response to Plasmodiophora brassicae. Int. J. Mol. Sci. 2023, 24, 2652. [Google Scholar] [CrossRef]
- Yang, Z.Q.; Wang, S.B.; Wei, L.L.; Huang, Y.M.; Liu, D.X.; Jia, Y.P.; Luo, C.F.; Lin, Y.C.; Liang, C.Y.; Hu, Y.; et al. BnIR: A multi-omics database with various tools for research and breeding. Mol. Plant 2023, 16, 775–789. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS ONE 2010, 5, e9931. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Wang, X.Y.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2024, 19, 2206–2229. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, J.F.; Wu, J.Y.; Zhang, H.Y.; Liu, G.M.; Wang, X.M.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Cui, X.B.; Hu, M.; Yao, S.L.; Zhang, Y.Y.; Tang, M.Q.; Liu, L.J.; Cheng, X.H.; Tong, C.B.; Liu, S.Y. BnaOmics: A comprehensive platform combining pan-genome and multi-omics data from Brassica napus. Plant Commun. 2023, 4, 100609. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yang, H.; Ren, L.; Chen, W.; Liu, L.J.; Liu, F.; Zeng, L.Y.; Yan, R.B.; Chen, K.R.; Fang, X.P. Jasmonic acid-mediated aliphatic glucosinolate metabolism is involved in clubroot disease development in Brassica napus L. Front. Plant Sci. 2018, 9, 750. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Fu, T.; Zhou, C.; Liu, F.; Zeng, L.; Ren, L.; Tong, C.; Liu, L.; Xu, L. Genome-Wide Analysis of the PERK Gene Family in Brassica napus L. and Their Potential Roles in Clubroot Disease. Int. J. Mol. Sci. 2025, 26, 2685. https://doi.org/10.3390/ijms26062685
Zhang Z, Fu T, Zhou C, Liu F, Zeng L, Ren L, Tong C, Liu L, Xu L. Genome-Wide Analysis of the PERK Gene Family in Brassica napus L. and Their Potential Roles in Clubroot Disease. International Journal of Molecular Sciences. 2025; 26(6):2685. https://doi.org/10.3390/ijms26062685
Chicago/Turabian StyleZhang, Zeyu, Tongyu Fu, Cong Zhou, Fan Liu, Lingyi Zeng, Li Ren, Chaobo Tong, Lijiang Liu, and Li Xu. 2025. "Genome-Wide Analysis of the PERK Gene Family in Brassica napus L. and Their Potential Roles in Clubroot Disease" International Journal of Molecular Sciences 26, no. 6: 2685. https://doi.org/10.3390/ijms26062685
APA StyleZhang, Z., Fu, T., Zhou, C., Liu, F., Zeng, L., Ren, L., Tong, C., Liu, L., & Xu, L. (2025). Genome-Wide Analysis of the PERK Gene Family in Brassica napus L. and Their Potential Roles in Clubroot Disease. International Journal of Molecular Sciences, 26(6), 2685. https://doi.org/10.3390/ijms26062685





