Characterization of the m6A Regulatory Gene Family in Phaseolus vulgaris L. and Functional Analysis of PvMTA in Response to BCMV Infection
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification and Characterization of m6A Regulatory Genes in Phaseolus vulgaris L.
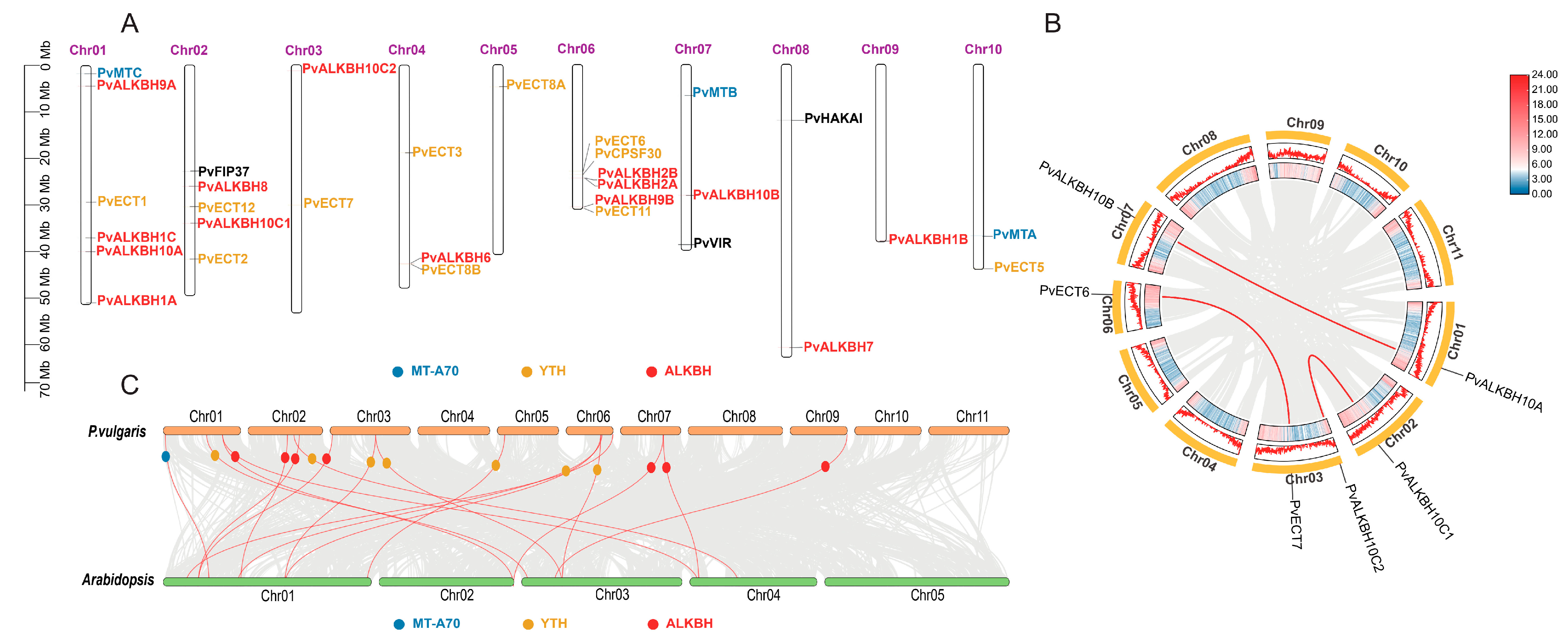
2.2. Chromosomal Location and Collinearity Analysis of m6A Regulatory Genes in Phaseolus vulgaris L.
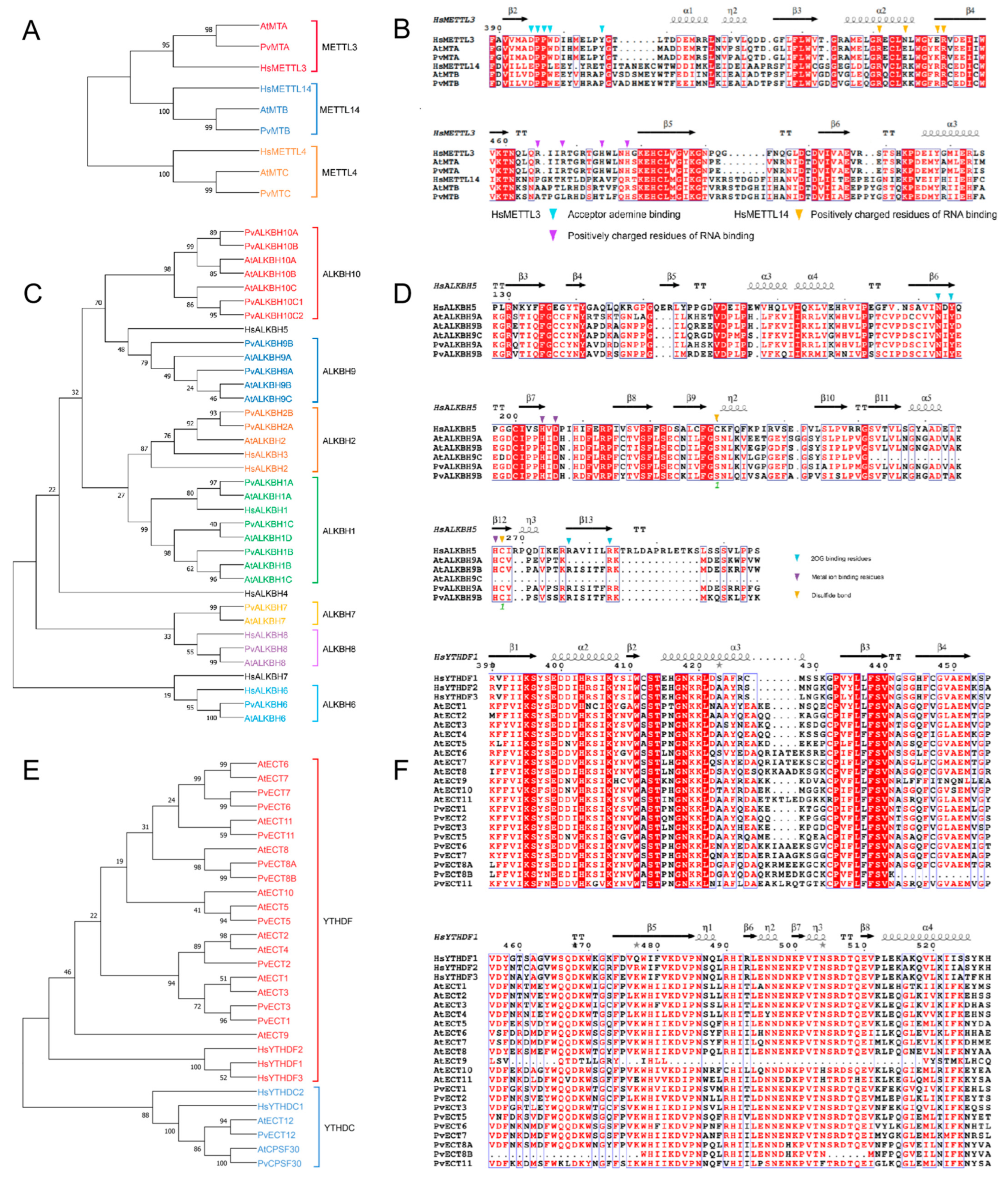
2.3. Evolutionary and Functional Key Residue Analyses of m6A Regulatory Gene Family in Phaseolus vulgaris L.
2.4. Structural Characterization of m6A Regulatory Genes in Phaseolus vulgaris L.
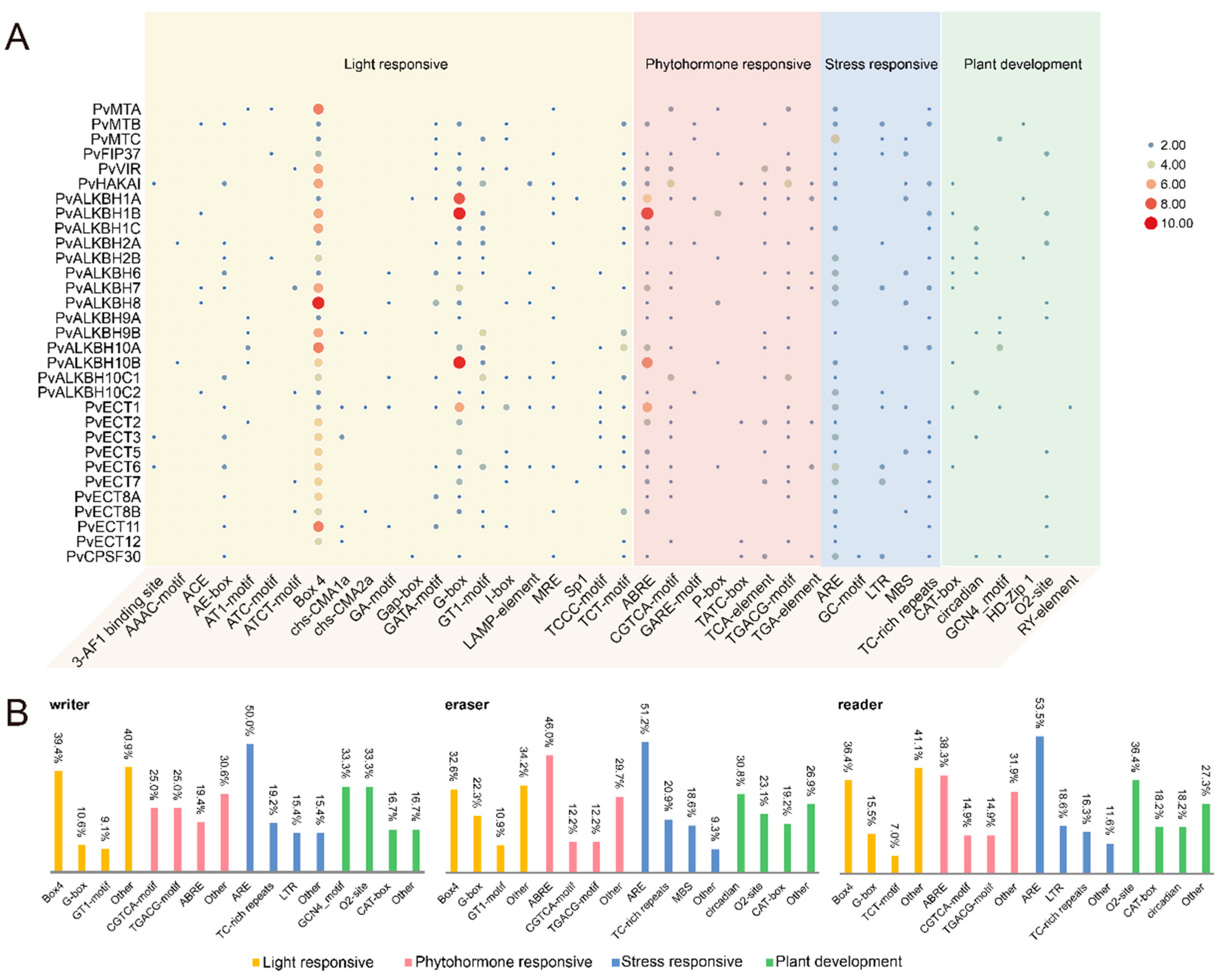
2.5. Detection of cis-Acting Elements in the Promoter Region of m6A Regulatory Genes in Phaseolus vulgaris L.
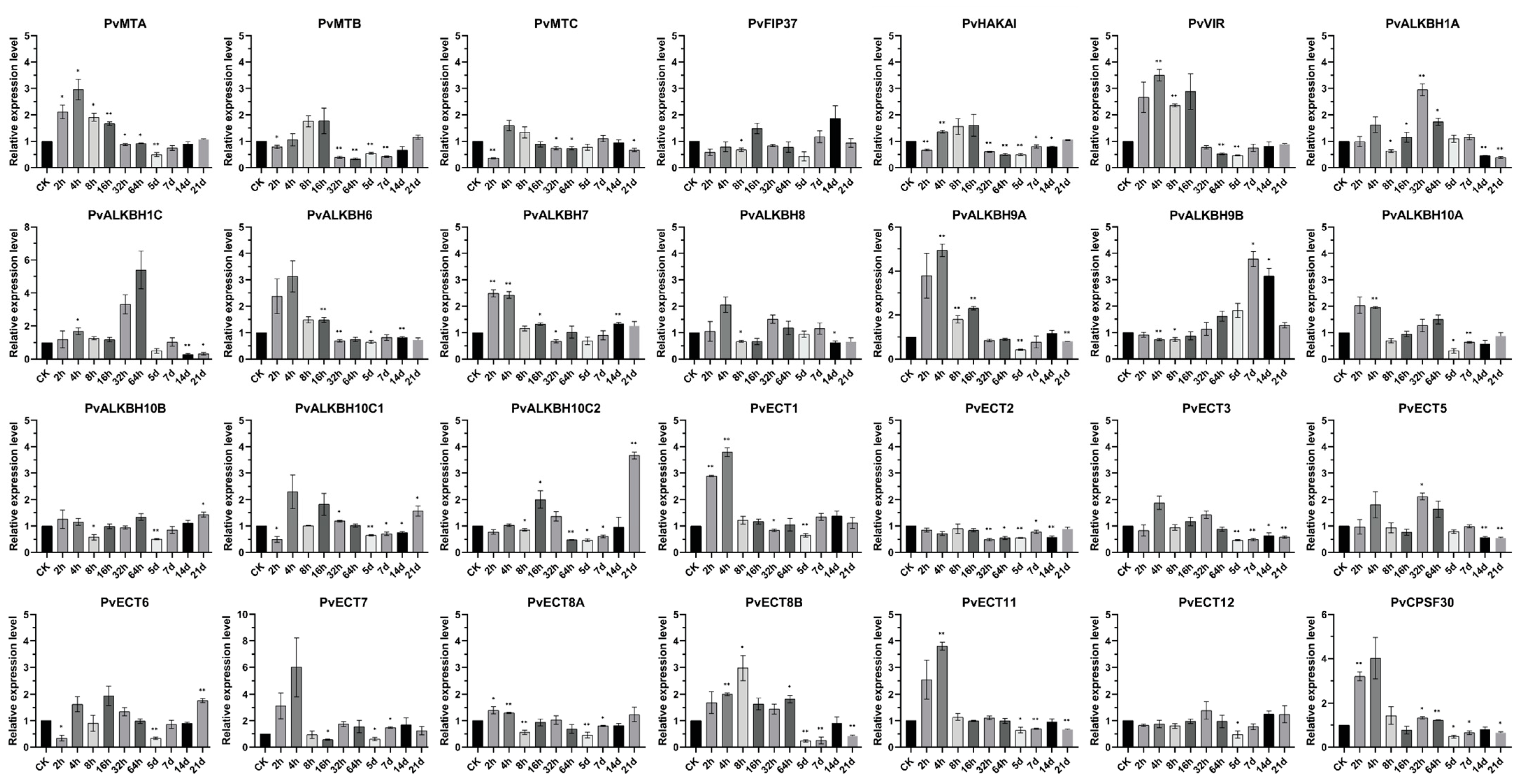
2.6. Alterations in the Expression of m6A Regulatory Genes in Common Bean Plants Infected with BCMV
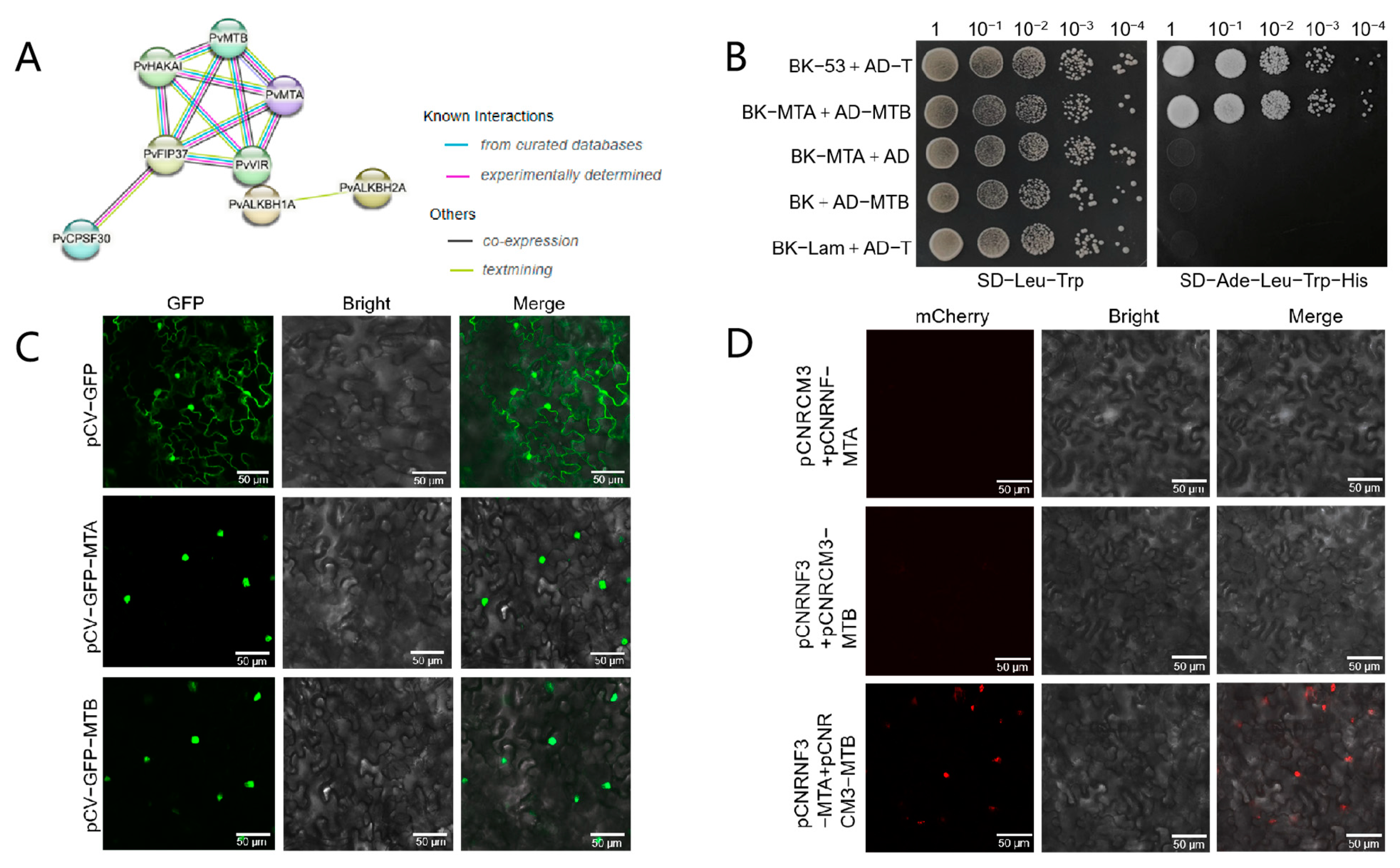
2.7. PvMTA Interacts with PvMTB in Phaseolus vulgaris L.
2.8. Characterizations of m6A Methyltransferases in Phaseolus vulgaris L.
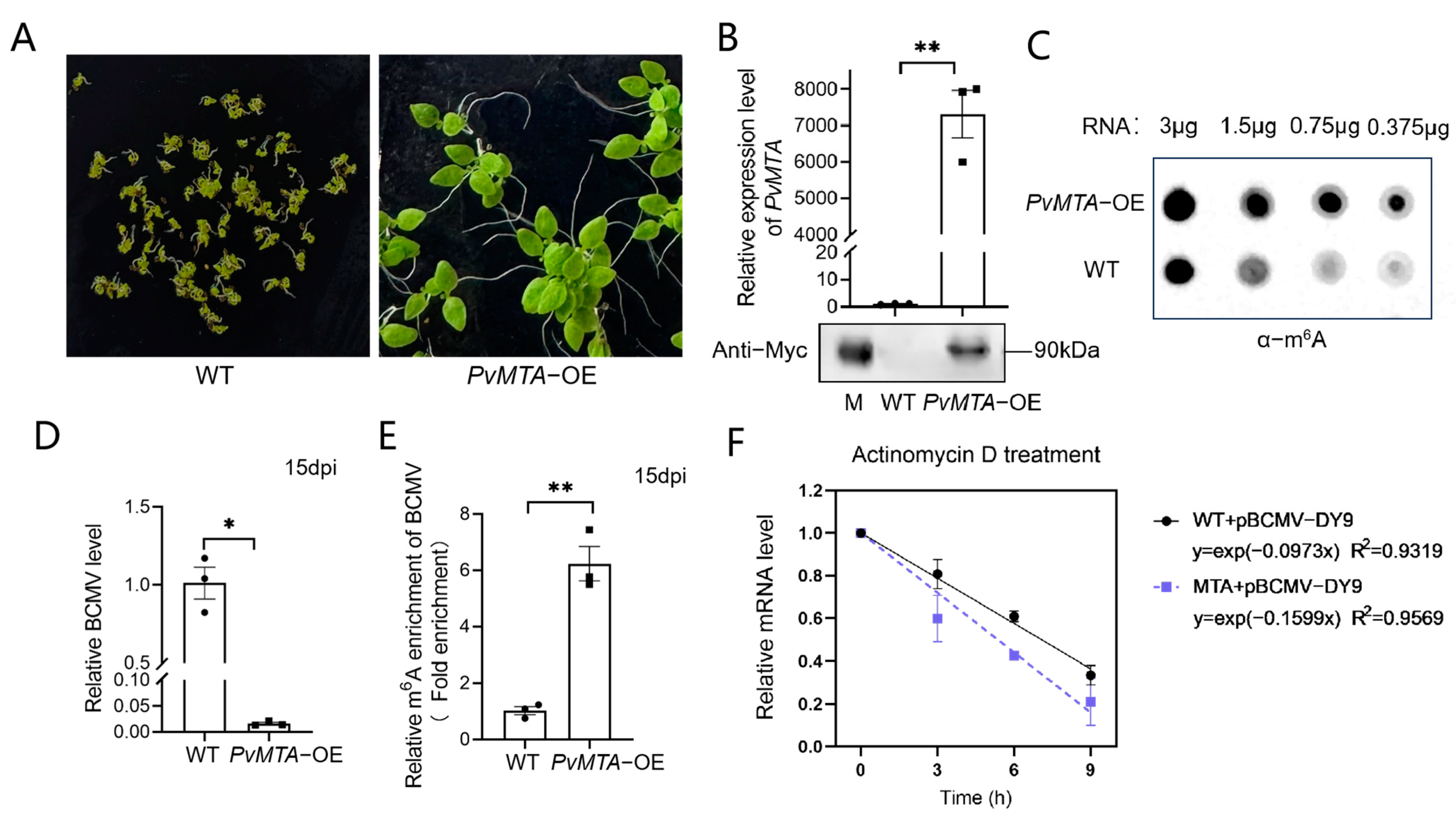
2.9. Overexpression of PvMTA Negatively Regulates BCMV Infection Through Mechanisms Mediated by m6A Modification
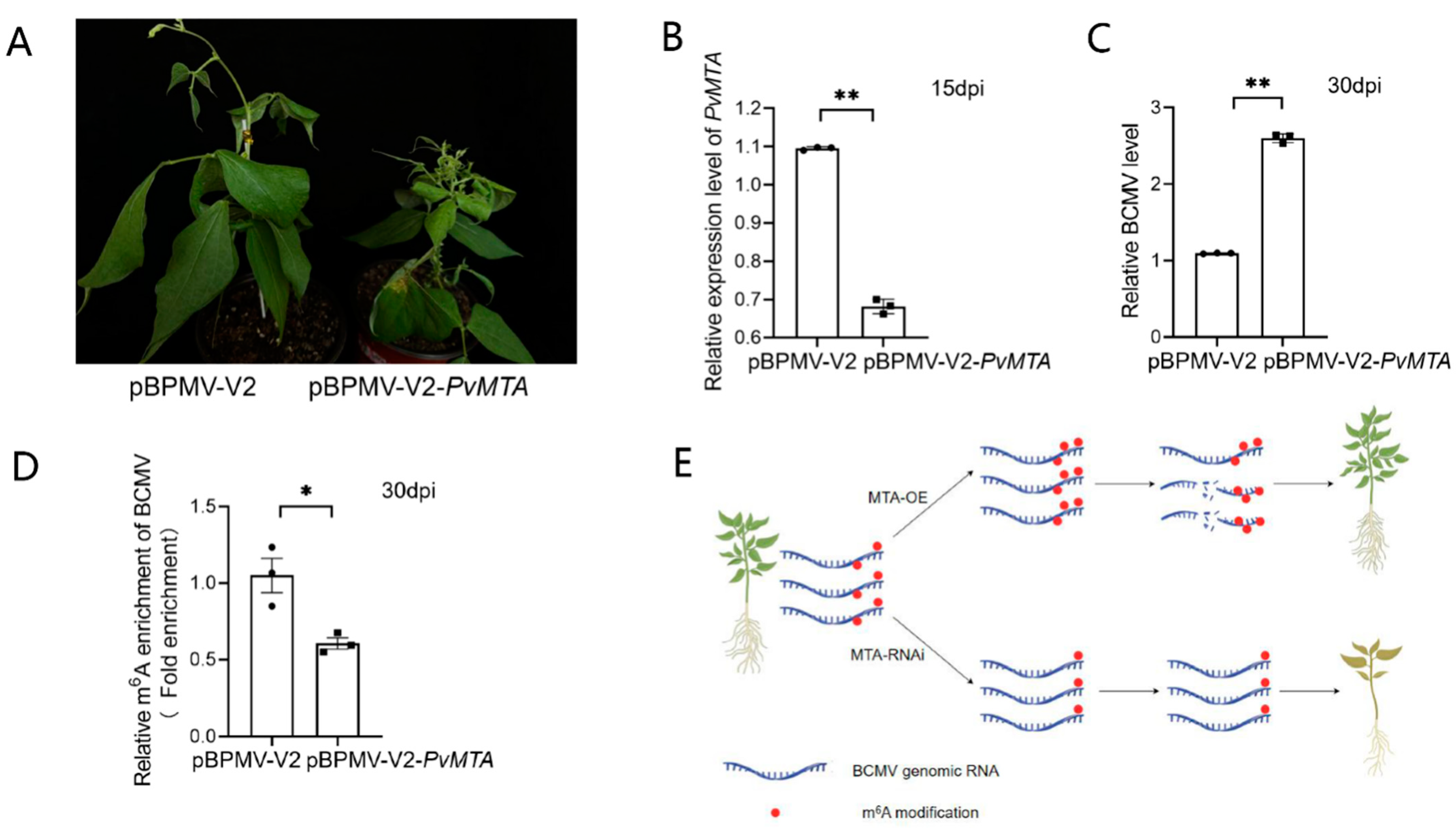
2.10. Silencing of PvMTA Promotes BCMV Infection Through Mechanisms Mediated by m6A Modification
3. Discussion
3.1. Features of the m6A Regulatory Gene Family
3.2. Roles of PvMTA in the Context of BCMV Infection in Common Bean
4. Materials and Methods
4.1. Identification and Characterization of m6A Regulatory Genes in Phaseolus vulgaris L.
4.2. Chromosome Location, Collinearity Relationships, Gene Duplication Events, and Evolutionary Selection
4.3. Phylogeny, Conserved Motifs, Domains, Gene Structure, cis-Elements, and the Protein–Protein Interactions Analyses
4.4. Plant Materials, Inoculation, and RT-qPCR Analysis
4.5. Y2H Analysis
4.6. BiFC and Subcellular Localization
4.7. Generation of Transgenic N. benthamiana Plants Overexpressing PvMTA
4.8. Western Blot Analysis
4.9. m6A Dot Blot Assay
4.10. m6A-IP-qPCR
4.11. RNA Stability Assay
4.12. BPMV-Based Silencing of PvMTA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cortés, A.J.; Monserrate, F.A.; Ramírez-Villegas, J.; Madriñán, S.; Blair, M.W. Drought tolerance in wild plant populations: The case of common beans (Phaseolus vulgaris L.). PLoS ONE 2013, 8, e62898. [Google Scholar] [CrossRef]
- Worrall, E.A.; Wamonje, F.O.; Mukeshimana, G.; Harvey, J.J.; Carr, J.P.; Mitter, N. Bean Common Mosaic Virus and Bean Common Mosaic Necrosis Virus: Relationships, Biology, and Prospects for Control. Adv. Virus Res. 2015, 93, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Flores-Estévez, N.; Acosta-Gallegos, J.A.; Silva-Rosales, L. Bean common mosaic virus and Bean common mosaic necrosis virus in Mexico. Plant Dis. 2003, 87, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Myers, J.R.; Karasev, A.V. Bean common mosaic virus Isolate Exhibits a Novel Pathogenicity Profile in Common Bean, Overcoming the bc-3 Resistance Allele Coding for the Mutated eIF4E Translation Initiation Factor. Phytopathology 2015, 105, 1487–1495. [Google Scholar] [CrossRef]
- Feng, X.; Poplawsky, A.R.; Nikolaeva, O.V.; Myers, J.R.; Karasev, A.V. Recombinants of bean common mosaic virus (BCMV) and genetic determinants of BCMV involved in overcoming resistance in common bean. Phytopathology 2014, 104, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sánchez-Navarro, J.A.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef]
- Zhang, T.; Shi, C.; Hu, H.; Zhang, Z.; Wang, Z.; Chen, Z.; Feng, H.; Liu, P.; Guo, J.; Lu, Q.; et al. N6-methyladenosine RNA modification promotes viral genomic RNA stability and infection. Nat. Commun. 2022, 13, 6576. [Google Scholar] [CrossRef]
- Han, C.; Zhang, F.; Qiao, X.; Zhao, Y.; Qiao, Q.; Huang, X.; Zhang, S. Multi-Omics Analysis Reveals the Dynamic Changes of RNA N6-Methyladenosine in Pear (Pyrus bretschneideri) Defense Responses to Erwinia amylovora Pathogen Infection. Front. Microbiol. 2021, 12, 803512. [Google Scholar] [CrossRef]
- Li, B.; Zhang, M.; Sun, W.; Yue, D.; Ma, Y.; Zhang, B.; Duan, L.; Wang, M.; Lindsey, K.; Nie, X.; et al. N6-methyladenosine RNA modification regulates cotton drought response in a Ca2+ and ABA-dependent manner. Plant Biotechnol. J. 2023, 21, 1270–1285. [Google Scholar] [CrossRef]
- Hu, J.; Cai, J.; Park, S.J.; Lee, K.; Li, Y.; Chen, Y.; Yun, J.Y.; Xu, T.; Kang, H. N6-Methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J. 2021, 106, 1759–1775. [Google Scholar] [CrossRef]
- Wang, S.; Wang, H.; Xu, Z.; Jiang, S.; Shi, Y.; Xie, H.; Wang, S.; Hua, J.; Wu, Y. m6A mRNA modification promotes chilling tolerance and modulates gene translation efficiency in Arabidopsis. Plant Physiol. 2023, 192, 1466–1482. [Google Scholar] [CrossRef] [PubMed]
- Tuck, M.T. The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int. J. Biochem. 1992, 24, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Bawankar, P.; Lence, T.; Paolantoni, C.; Haussmann, I.U.; Kazlauskiene, M.; Jacob, D.; Heidelberger, J.B.; Richter, F.M.; Nallasivan, M.P.; Morin, V.; et al. Hakai is required for stabilization of core components of the m6A mRNA methylation machinery. Nat. Commun. 2021, 12, 3778. [Google Scholar] [CrossRef]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e1026. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef]
- Růžička, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef]
- Parker, M.T.; Knop, K.; Zacharaki, V.; Sherwood, A.V.; Tomé, D.; Yu, X.; Martin, P.G.; Beynon, J.; Michaels, S.D.; Barton, G.J.; et al. Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA. elife 2021, 10, e65537. [Google Scholar] [CrossRef]
- Zhang, M.; Bodi, Z.; Mackinnon, K.; Zhong, S.; Archer, N.; Mongan, N.P.; Simpson, G.G.; Fray, R.G. Two zinc finger proteins with functions in m6A writing interact with HAKAI. Nat. Commun. 2022, 13, 1127. [Google Scholar] [CrossRef] [PubMed]
- Reichel, M.; Köster, T.; Staiger, D. Marking RNA: m6A writers, readers, and functions in Arabidopsis. J. Mol. Cell Biol. 2019, 11, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef]
- Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Li, X.; Zhang, Y.; Ouyang, Z.; Song, F. Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and Rice. Plant Mol. Biol. Rep. 2014, 32, 1169–1186. [Google Scholar] [CrossRef]
- Hou, Y.; Sun, J.; Wu, B.; Gao, Y.; Nie, H.; Nie, Z.; Quan, S.; Wang, Y.; Cao, X.; Li, S. CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis. Mol. Plant 2021, 14, 688–699. [Google Scholar] [CrossRef]
- Tian, S.; Wu, N.; Zhang, L.; Wang, X. RNA N6-methyladenosine modification suppresses replication of rice black streaked dwarf virus and is associated with virus persistence in its insect vector. Mol. Plant Pathol. 2021, 22, 1070–1081. [Google Scholar] [CrossRef]
- He, H.; Ge, L.; Li, Z.; Zhou, X.; Li, F. Pepino mosaic virus antagonizes plant m6A modification by promoting the autophagic degradation of the m6A writer HAKAI. aBIOTECH 2023, 4, 83–96. [Google Scholar] [CrossRef]
- He, H.; Ge, L.; Chen, Y.; Zhao, S.; Li, Z.; Zhou, X.; Li, F. m6A modification of plant virus enables host recognition by NMD factors in plants. Sci. China Life Sci. 2024, 67, 161–174. [Google Scholar] [CrossRef]
- Zhang, K.; Zhuang, X.; Dong, Z.; Xu, K.; Chen, X.; Liu, F.; He, Z. The dynamics of N6-methyladenine RNA modification in interactions between rice and plant viruses. Genome Biol. 2021, 22, 189. [Google Scholar] [CrossRef]
- Su, H.; Meng, L.; Qu, Z.; Zhang, W.; Liu, N.; Cao, P.; Jin, J. Genome-wide identification of the N6-methyladenosine regulatory genes reveals NtFIP37B increases drought resistance of tobacco (Nicotiana tabacum L.). BMC Plant Biol. 2024, 24, 134. [Google Scholar] [CrossRef]
- Zheng, H.; Gao, Y.; Dang, Y.; Wu, F.; Wang, X.; Zhang, F.; Sui, N. Characterization of the m6A gene family in sorghum and its function in growth, development and stress resistance. Ind. Crops Prod. 2023, 198, 116625. [Google Scholar] [CrossRef]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, Y.; Sun, T.; Zhang, C.; Liu, X.; Li, Y. Genome-Wide Classification and Evolutionary Analysis Reveal Diverged Patterns of Chalcone Isomerase in Plants. Biomolecules 2022, 12, 961. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiao, L.; Chi, J.; Li, R.; Han, Y.; Cui, F.; Peng, Z.; Wan, S.; Li, G. Genome-wide identification and expression of SAUR gene family in peanut (Arachis hypogaea L.) and functional identification of AhSAUR3 in drought tolerance. BMC Plant Biol. 2022, 22, 178. [Google Scholar] [CrossRef]
- Shen, L. Functional interdependence of N6-methyladenosine methyltransferase complex subunits in Arabidopsis. Plant Cell 2023, 35, 1901–1916. [Google Scholar] [CrossRef]
- Zheng, H.X.; Sun, X.; Zhang, X.S.; Sui, N. m6A Editing: New Tool to Improve Crop Quality? Trends Plant Sci. 2020, 25, 859–867. [Google Scholar] [CrossRef]
- Ren, Z.; Tang, B.; Xing, J.; Liu, C.; Cai, X.; Hendy, A.; Kamran, M.; Liu, H.; Zheng, L.; Huang, J.; et al. MTA1-mediated RNA m6 A modification regulates autophagy and is required for infection of the rice blast fungus. New Phytol. 2022, 235, 247–262. [Google Scholar] [CrossRef]
- Guo, T.; Liu, C.; Meng, F.; Hu, L.; Fu, X.; Yang, Z.; Wang, N.; Jiang, Q.; Zhang, X.; Ma, F. The m6 A reader MhYTP2 regulates MdMLO19 mRNA stability and antioxidant genes translation efficiency conferring powdery mildew resistance in apple. Plant Biotechnol. J. 2022, 20, 511–525. [Google Scholar] [CrossRef]
- Burow, M.D.; Chlan, C.A.; Sen, P.; Lisca, A.; Murai, N. High-frequency generation of transgenic tobacco plants after modified leaf disk cocultivation with Agrobacterium tumefaciens. Plant Mol. Bio. Rep. 1990, 8, 124–139. [Google Scholar] [CrossRef]
- Pflieger, S.; Blanchet, S.; Meziadi, C.; Richard, M.M.; Thareau, V.; Mary, F.; Mazoyer, C.; Geffroy, V. The “one-step” Bean pod mottle virus (BPMV)-derived vector is a functional genomics tool for efficient overexpression of heterologous protein, virus-induced gene silencing and genetic mapping of BPMV R-gene in common bean (Phaseolus vulgaris L.). BMC Plant Biol. 2014, 14, 232. [Google Scholar] [CrossRef]
- Zhang, C.; Bradshaw, J.D.; Whitham, S.A.; Hill, J.H. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 2010, 153, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Hou, N.; Li, C.; He, J.; Liu, Y.; Yu, S.; Malnoy, M.; Mobeen Tahir, M.; Xu, L.; Ma, F.; Guan, Q. MdMTA-mediated m6 A modification enhances drought tolerance by promoting mRNA stability and translation efficiency of genes involved in lignin deposition and oxidative stress. New Phytol. 2022, 234, 1294–1314. [Google Scholar] [CrossRef]
- Zhou, L.; Tang, R.; Li, X.; Tian, S.; Li, B.; Qin, G. N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner. Genome Biol. 2021, 22, 168. [Google Scholar] [CrossRef]
- Bodi, Z.; Zhong, S.; Mehra, S.; Song, J.; Graham, N.; Li, H.; May, S.; Fray, R.G. Adenosine Methylation in Arabidopsis mRNA is Associated with the 3′ End and Reduced Levels Cause Developmental Defects. Front. Plant Sci. 2012, 3, 48. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Y.C.; Liao, J.Y.; Yu, Y.; Zhou, Y.F.; Feng, Y.Z.; Yang, Y.W.; Lei, M.Q.; Bai, M.; Wu, H.; et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019, 15, e1008120. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Aik, W.; Scotti, J.S.; Choi, H.; Gong, L.; Demetriades, M.; Schofield, C.J.; McDonough, M.A. Structure of human RNA N⁶-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014, 42, 4741–4754. [Google Scholar] [CrossRef]
- Xu, C.; Liu, K.; Ahmed, H.; Loppnau, P.; Schapira, M.; Min, J. Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins. J. Biol. Chem. 2015, 290, 24902–24913. [Google Scholar] [CrossRef]
- Śledź, P.; Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. elife 2016, 5, e18434. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Salmon-Divon, M.; Amariglio, N.; Rechavi, G. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013, 8, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Ratel, D.; Ravanat, J.L.; Berger, F.; Wion, D. N6-methyladenine: The other methylated base of DNA. Bioessays 2006, 28, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Pflieger, S.; Blanchet, S.; Camborde, L.; Drugeon, G.; Rousseau, A.; Noizet, M.; Planchais, S.; Jupin, I. Efficient virus-induced gene silencing in Arabidopsis using a ‘one-step’ TYMV-derived vector. Plant J. 2008, 56, 678–690. [Google Scholar] [CrossRef]

| Gene Name | Locus ID | Group | Chr | Protein Property | ||
|---|---|---|---|---|---|---|
| Length (aa) | pI | MW (kDa) | ||||
| PvMTA | Phvul.010G102500.1 | MT-A70 | 10 | 761 | 6.01 | 84.32 |
| PvMTB | Phvul.007G073300.1 | 7 | 1086 | 6.86 | 120.67 | |
| PvMTC | Phvul.001G016200.1 | 1 | 427 | 8.04 | 48.64 | |
| PvALKBH1A | Phvul.001G262100.1 | ALKBH | 1 | 358 | 6.11 | 40.36 |
| PvALKBH1B | Phvul.009G262600.1 | 9 | 344 | 9.47 | 37.40 | |
| PvALKBH1C | Phvul.001G131400.1 | 1 | 443 | 8.89 | 49.26 | |
| PvALKBH2A | Phvul.006G137400.1 | 6 | 235 | 9.21 | 27.06 | |
| PvALKBH2B | Phvul.006G137611.1 | 6 | 235 | 9.21 | 27.02 | |
| PvALKBH6 | Phvul.004G131600.1 | 4 | 259 | 5.79 | 29.74 | |
| PvALKBH7 | Phvul.008G264300.1 | 8 | 224 | 6.36 | 25.44 | |
| PvALKBH8 | Phvul.002G123600.1 | 2 | 342 | 7.59 | 38.29 | |
| PvALKBH9A | Phvul.001G044000.2 | 1 | 476 | 5.92 | 53.60 | |
| PvALKBH9B | Phvul.006G214800.1 | 6 | 425 | 8.83 | 48.48 | |
| PvALKBH10A | Phvul.001G147800.1 | 1 | 516 | 5.89 | 56.99 | |
| PvALKBH10B | Phvul.007G168900.1 | 7 | 505 | 6.44 | 55.72 | |
| PvALKBH10C1 | Phvul.002G181800.1 | 2 | 671 | 6.36 | 73.15 | |
| PvALKBH10C2 | Phvul.003G014200.1 | 3 | 691 | 6.18 | 74.54 | |
| PvECT1 | Phvul.001G110200.1 | YTH | 1 | 658 | 7.96 | 72.22 |
| PvECT2 | Phvul.002G247000.1 | 2 | 677 | 6.02 | 74.03 | |
| PvECT3 | Phvul.004G080300.1 | 4 | 638 | 5.96 | 70.15 | |
| PvECT5 | Phvul.010G165400.1 | 10 | 634 | 5.25 | 69.57 | |
| PvECT6 | Phvul.006G121600.1 | 6 | 649 | 5.84 | 71.55 | |
| PvECT7 | Phvul.003G119300.1 | 3 | 698 | 7.59 | 77.24 | |
| PvECT8A | Phvul.005G045600.1 | 5 | 575 | 6.76 | 63.27 | |
| PvECT8B | Phvul.004G132700.1 | 4 | 231 | 9.34 | 26.62 | |
| PvECT11 | Phvul.006G218800.1 | 6 | 557 | 9.51 | 62.58 | |
| PvECT12 | Phvul.002G152600.1 | 2 | 379 | 5.38 | 42.68 | |
| PvCPSF30 | Phvul.006G130200.1 | 6 | 697 | 6.28 | 76.46 | |
| PvFIP37 | Phvul.002G107400.1 | 2 | 337 | 5.41 | 38.15 | |
| PvVIR | Phvul.007G267500.1 | 7 | 2188 | 5.35 | 240.79 | |
| PvHAKAI | Phvul.008G108800.1 | 8 | 436 | 6.16 | 47.61 | |
| Gene I | Location | Gene II | Location | Type of Duplication | Ka | Ks | Ka/Ks | T = Ks/2r(MYA) |
|---|---|---|---|---|---|---|---|---|
| PvALKBH10A | 1 | PvALKBH10B | 7 | Segmental/WGD | 0.126348392 | 0.852865956 | 0.148145662 | 28.42 |
| PvECT6 | 6 | PvECT7 | 3 | Segmental/WGD | 0.174929236 | 0.683184232 | 0.256049874 | 22.77 |
| PvALKBH10C1 | 2 | PvALKBH10C2 | 3 | Segmental/WGD | 0.189372849 | 0.669639206 | 0.28279833 | 22.32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Wang, X.; Liang, X.; Huang, X.; Nawaz, M.A.; Jing, C.; Fan, Y.; Niu, J.; Wu, J.; Feng, X. Characterization of the m6A Regulatory Gene Family in Phaseolus vulgaris L. and Functional Analysis of PvMTA in Response to BCMV Infection. Int. J. Mol. Sci. 2025, 26, 2748. https://doi.org/10.3390/ijms26062748
Wu W, Wang X, Liang X, Huang X, Nawaz MA, Jing C, Fan Y, Niu J, Wu J, Feng X. Characterization of the m6A Regulatory Gene Family in Phaseolus vulgaris L. and Functional Analysis of PvMTA in Response to BCMV Infection. International Journal of Molecular Sciences. 2025; 26(6):2748. https://doi.org/10.3390/ijms26062748
Chicago/Turabian StyleWu, Wenyan, Xinhua Wang, Xingrui Liang, Xinqi Huang, Muhammad Amjad Nawaz, Chenchen Jing, Yaru Fan, Jingya Niu, Jing Wu, and Xue Feng. 2025. "Characterization of the m6A Regulatory Gene Family in Phaseolus vulgaris L. and Functional Analysis of PvMTA in Response to BCMV Infection" International Journal of Molecular Sciences 26, no. 6: 2748. https://doi.org/10.3390/ijms26062748
APA StyleWu, W., Wang, X., Liang, X., Huang, X., Nawaz, M. A., Jing, C., Fan, Y., Niu, J., Wu, J., & Feng, X. (2025). Characterization of the m6A Regulatory Gene Family in Phaseolus vulgaris L. and Functional Analysis of PvMTA in Response to BCMV Infection. International Journal of Molecular Sciences, 26(6), 2748. https://doi.org/10.3390/ijms26062748







