Identification of Key Active Constituents in Eucommia ulmoides Oliv. Leaves Against Parkinson’s Disease and the Alleviative Effects via 4E-BP1 Up-Regulation
Abstract
1. Introductions
2. Results
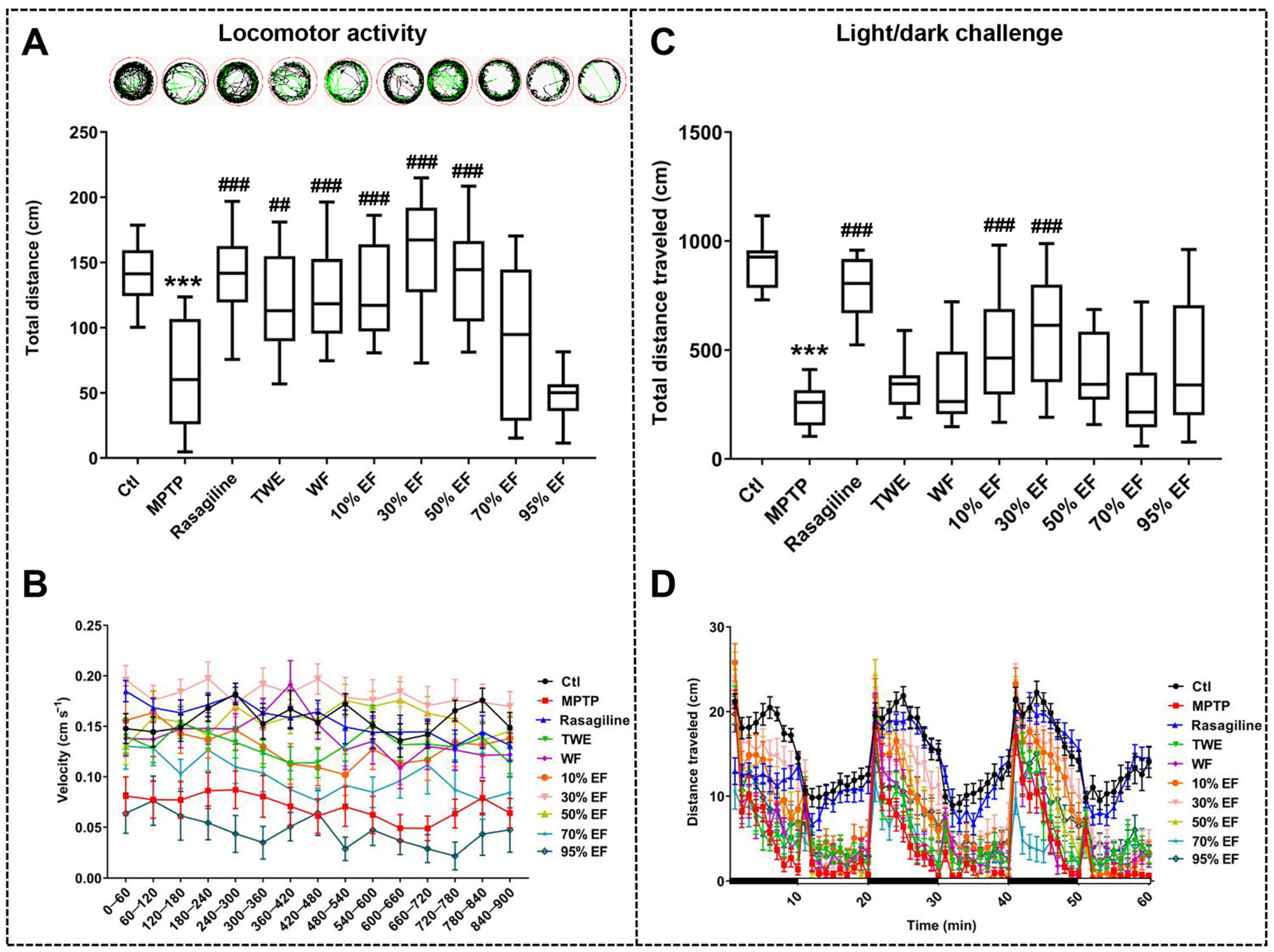
2.1. Neuroprotective Effect of Different Fractions of EUOL Extract on Zebrafish PD-like Behavior
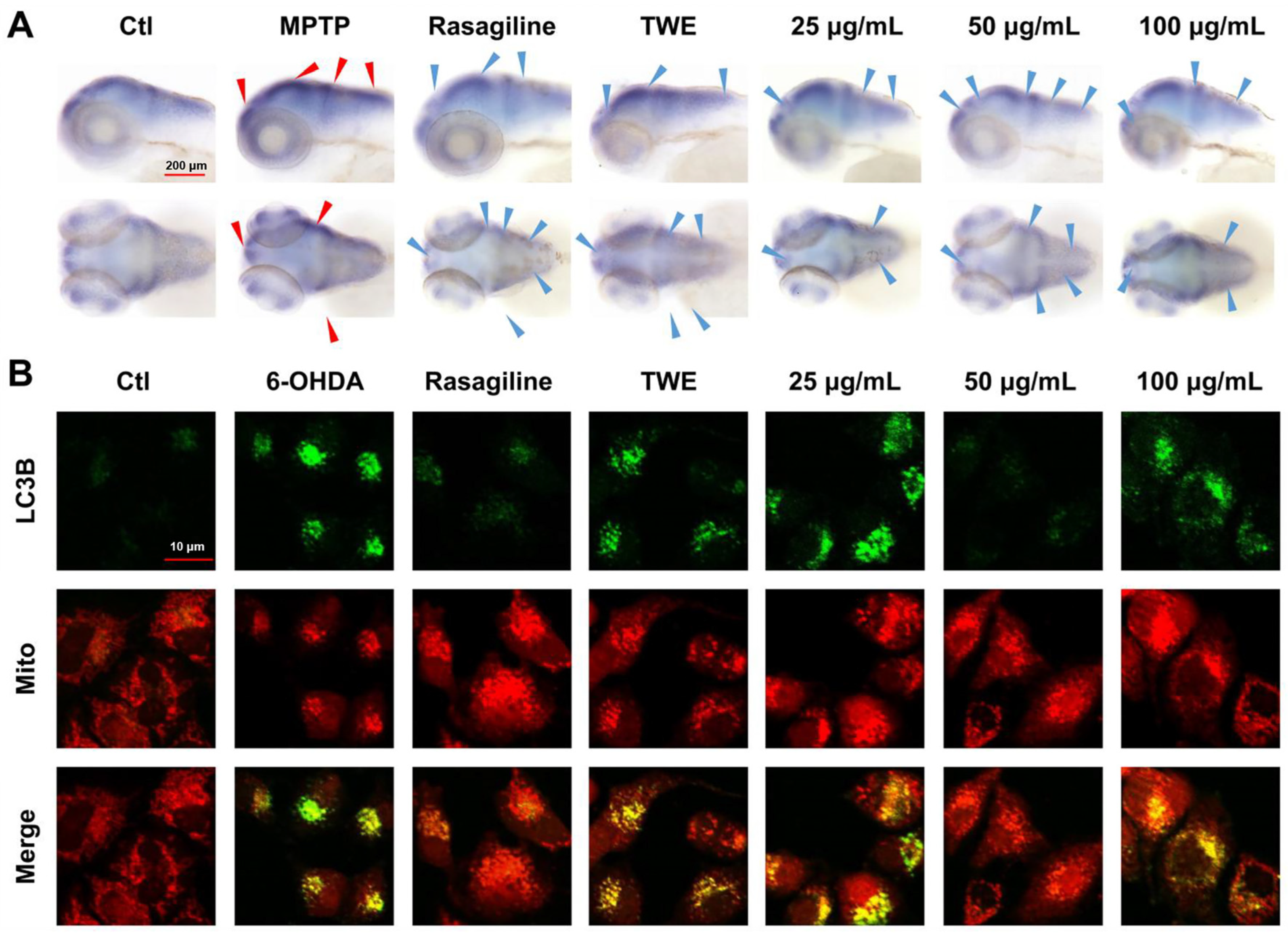
2.2. Effect of Different Fractions of EUOL Extract on PD Neuronal and Vascular Phenotype in Zebrafish PD Model
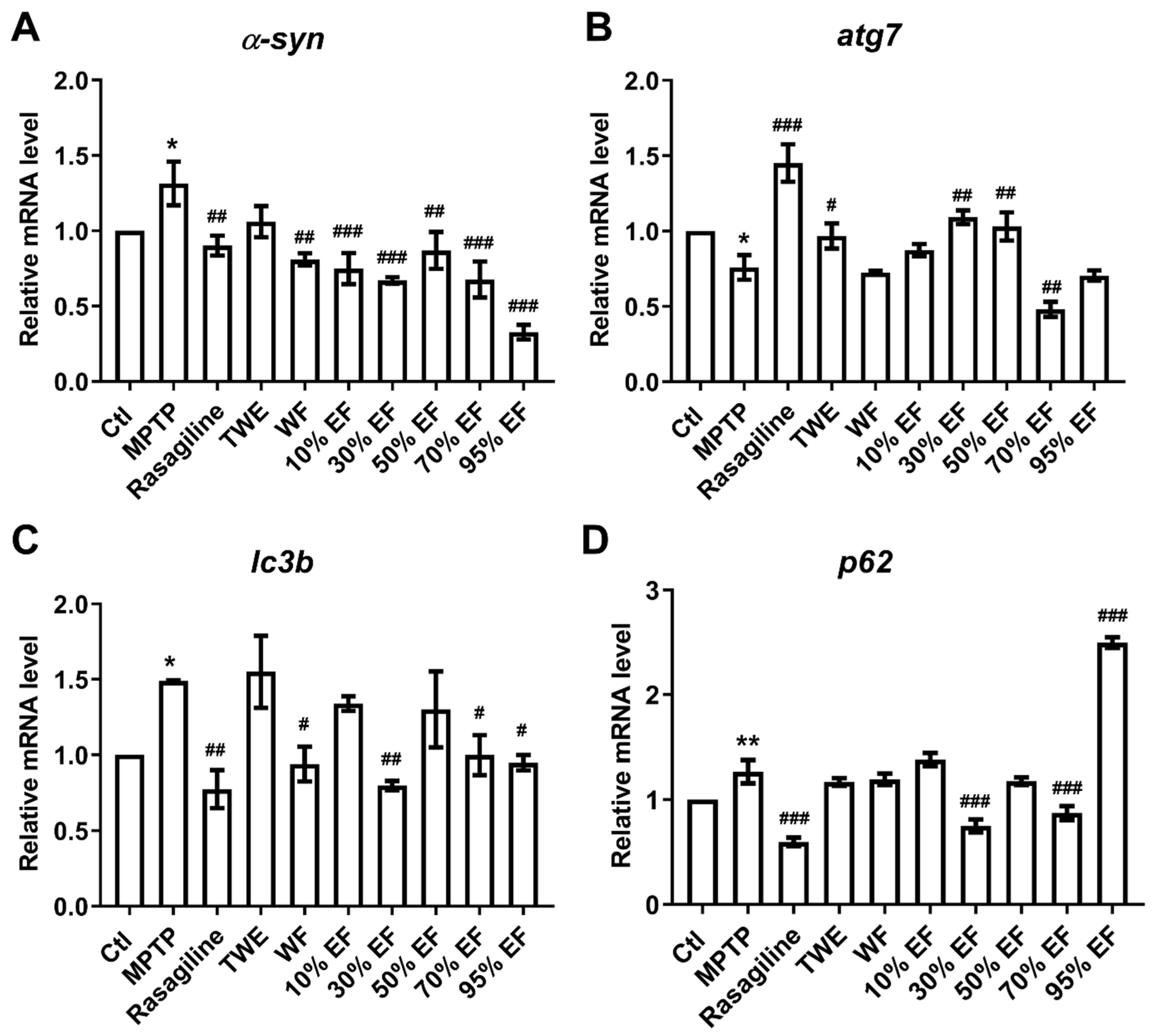
2.3. Regulatory Effect of Different Fractions of EUOL Extract on Core PD-Related Genes
2.4. Verification of the Main Active Constituent Against PD by Assaying the Key Regulators Involved in PD
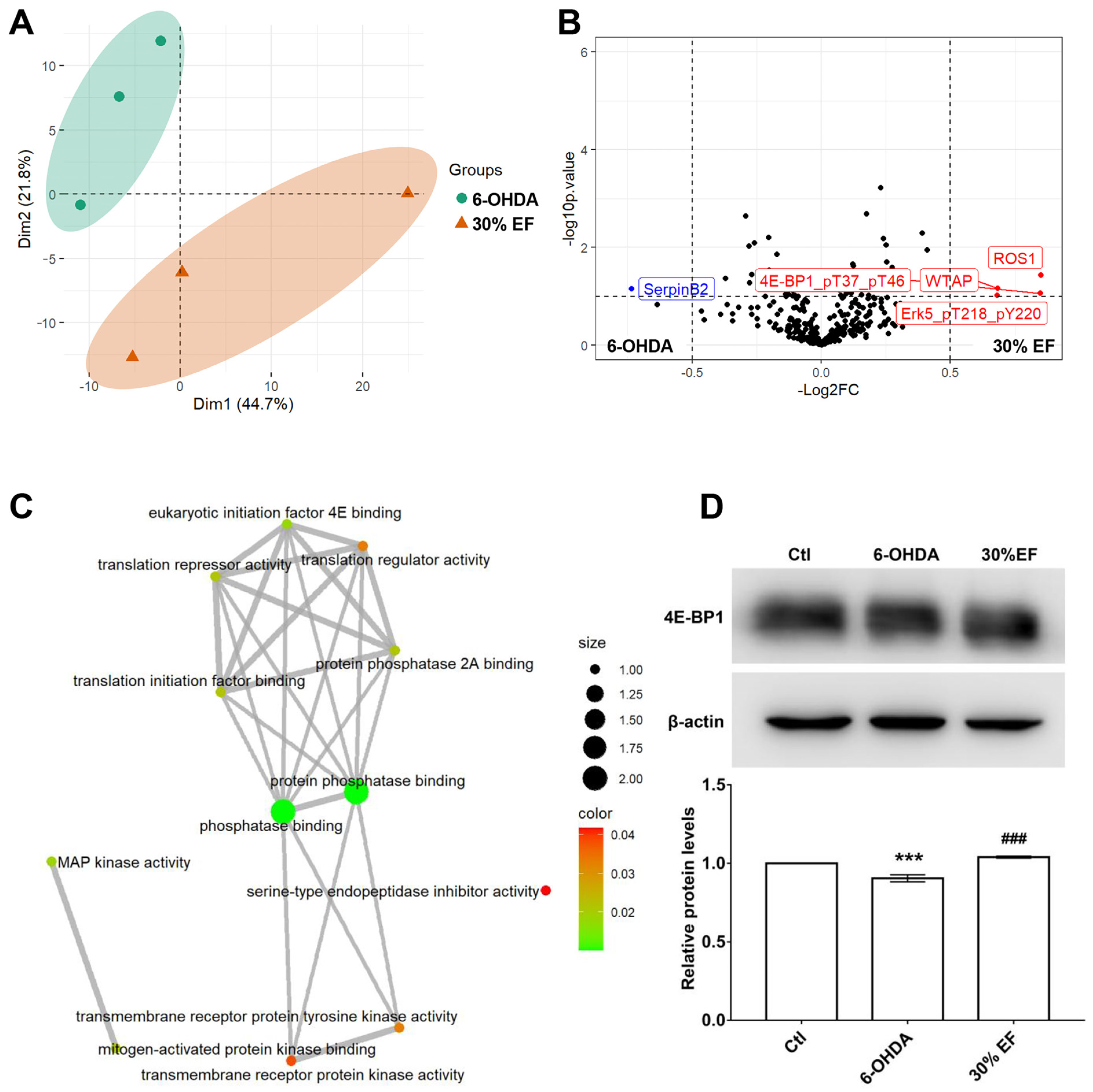
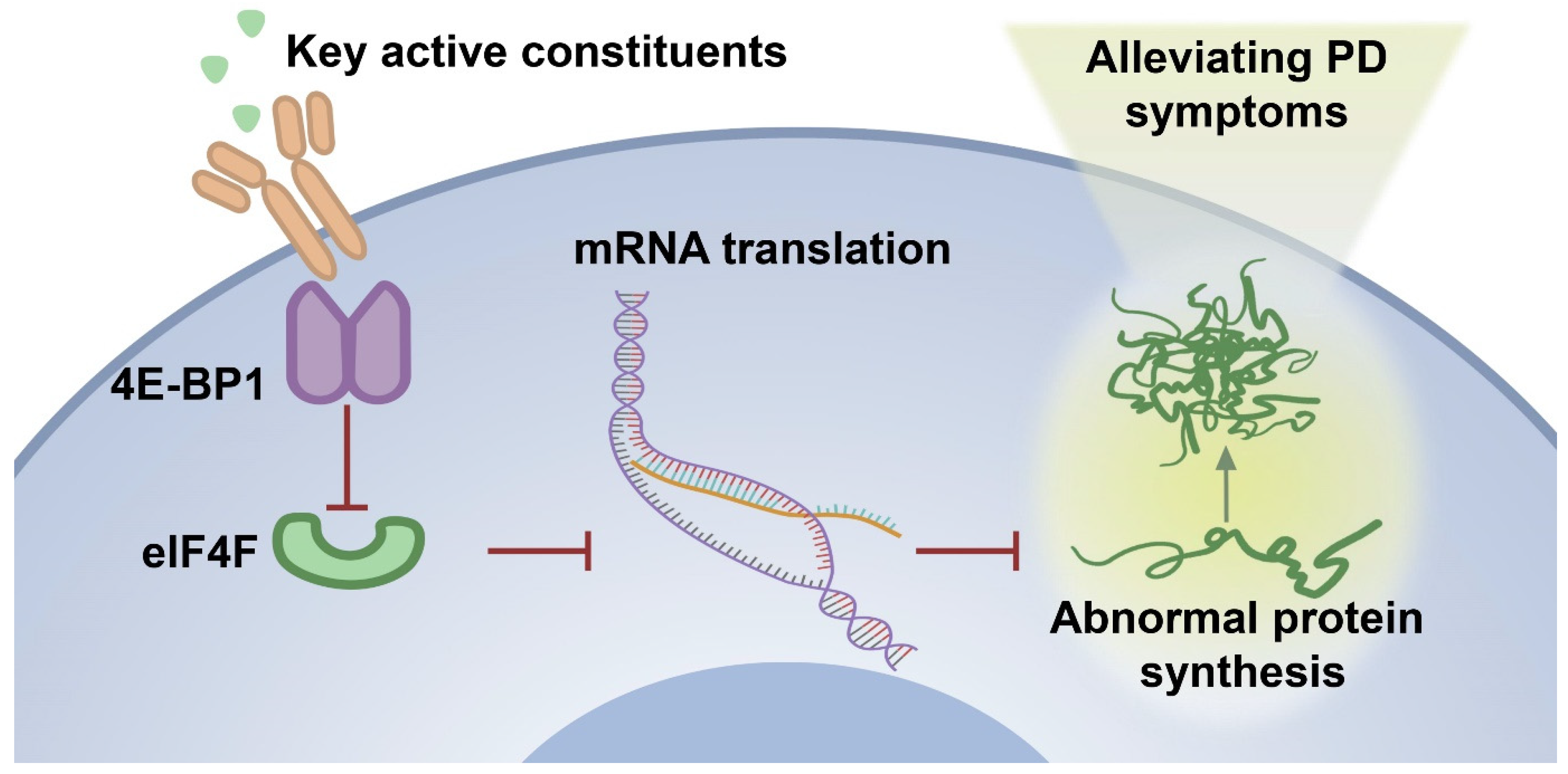
2.5. The Optimal Active Constituent 30% EF Exerts Anti-PD Action Through Up-Regulated 4E-BP1
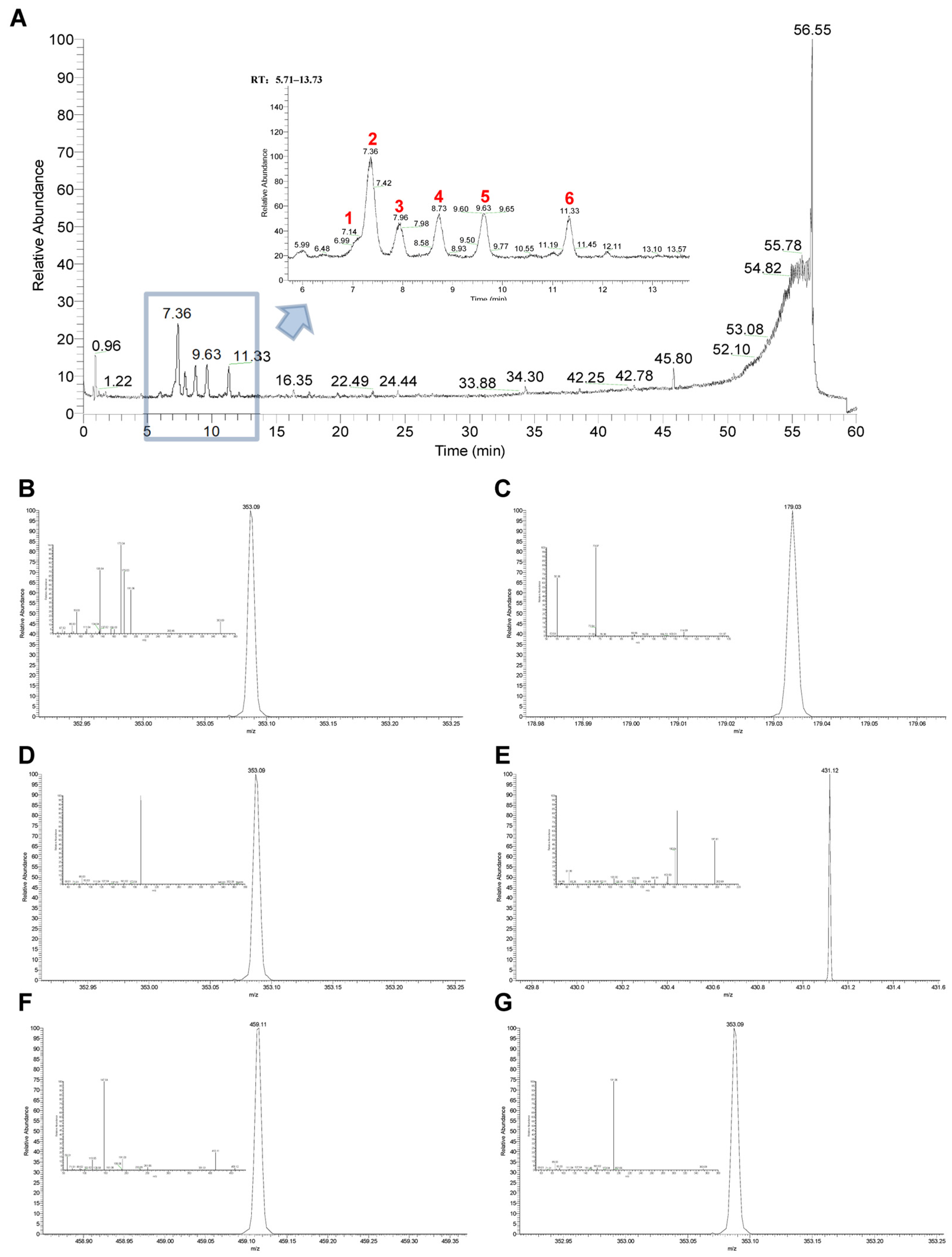
2.6. Identification of Main Chemical Components in the Optimal Active Constituent 30% EF Against PD
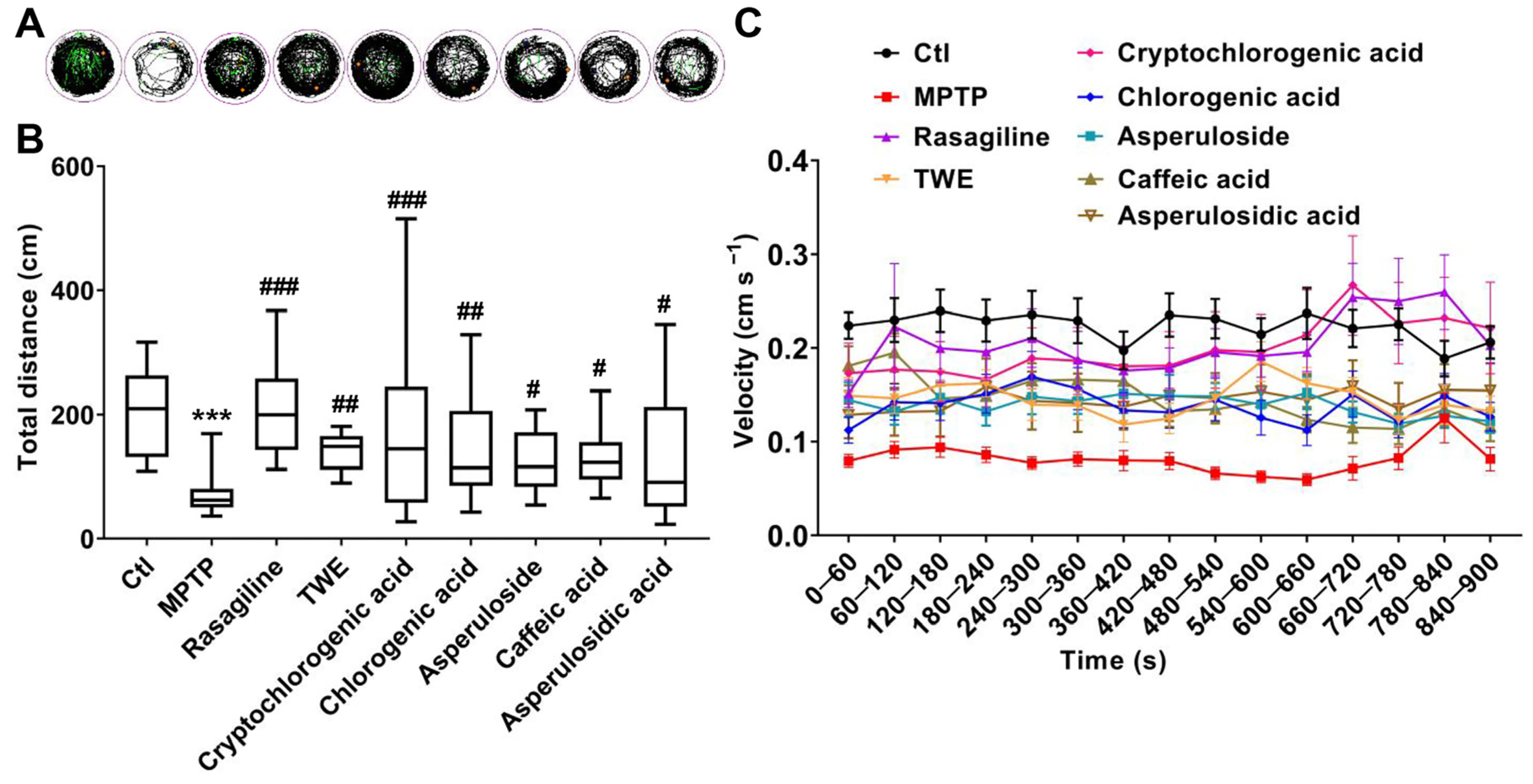
2.7. Molecular Docking Simulation, SPR, and Locomotion Assays Further Verify the Anti-PD Effect and 4E-BP1 Associated Mechanism of the Key Active Constituent 30% EF
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. EUOL Extract Preparation
4.3. Zebrafish Maintenance and Grouping
4.3.1. Zebrafish Maintenance
4.3.2. MPTP-Induced Zebrafish PD Model
4.3.3. Zebrafish Grouping
4.4. Cell Culture
4.5. Locomotion Analysis of Zebrafish
4.6. Assessment of the Morphology of Dopaminergic Neurons in Zebrafish
4.7. Evaluation of the Development of Neuronal Vasculature in Zebrafish
4.8. Real-Time Quantitative PCR (RT-qPCR) Analysis
4.9. Whole-Mount In Situ Hybridization in Zebrafish
4.10. Immunofluorescence
4.11. RPPA Analysis
4.12. Western Blotting Analysis
4.13. Identification and Characterization of Chemical Constituent in EUOL
4.14. Molecular Docking
4.15. SPR Assay
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Moustafa, A.A.; Chakravarthy, S.; Phillips, J.R.; Gupta, A.; Keri, S.; Polner, B.; Frank, M.J.; Jahanshahi, M. Motor symptoms in Parkinson’s disease: A unified framework. Neurosci. Biobehav. Rev. 2016, 68, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Blesa, J.; Foffani, G.; Dehay, B.; Bezard, E.; Obeso, J.A. Motor and non-motor circuit disturbances in early Parkinson disease: Which happens first? Nat. Rev. Neurosci. 2022, 23, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, E.; Paudel, Y.N.; Villa, C.; Shaikh, M.F.; Piperi, C. Lymphocyte-Activation Gene 3 (LAG3) Protein as a Possible Therapeutic Target for Parkinson’s Disease: Molecular Mechanisms Connecting Neuroinflammation to α-Synuclein Spreading Pathology. Biology 2020, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Tysnes, O.B.; Storstein, A. Epidemiology of Parkinson’s disease. J. Neural Transm. 2017, 124, 901–905. [Google Scholar] [CrossRef]
- Tan, E.K.; Chao, Y.X.; West, A.; Chan, L.L.; Poewe, W.; Jankovic, J. Parkinson disease and the immune system—Associations, mechanisms and therapeutics. Nat. Rev. Neurol. 2020, 16, 303–318. [Google Scholar] [CrossRef]
- Costa, C.A.D.; Manaa, W.E.; Duplan, E.; Checler, F. The Endoplasmic Reticulum Stress/Unfolded Protein Response and Their Contributions to Parkinson’s Disease Physiopathology. Cells 2020, 9, 2495. [Google Scholar] [CrossRef]
- Pellegrini, L.; Wetzel, A.; Granno, S.; Heaton, G.; Harvey, K. Back to the tubule: Microtubule dynamics in Parkinson’s disease. Cell. Mol. Life Sci. 2017, 74, 409–434. [Google Scholar] [CrossRef]
- Mancuso, C.; Scapagini, G.; Curro, D.; Giuffrida Stella, A.M.; De Marco, C.; Butterfield, D.A.; Calabrese, V. Mitochondrial dysfunction, free radical generation and cellular stress response in neurodegenerative disorders. Front. Biosci. 2007, 12, 1107–1123. [Google Scholar] [CrossRef]
- Garcia-Ptacek, S.; Kramberger, M.G. Parkinson Disease and Dementia. J. Geriatr. Psychiatry Neurol. 2016, 29, 261–270. [Google Scholar] [CrossRef]
- Su, W.; Liang, Z.; Mao, W.; Shao, M.; Hu, X.; Wu, Y.; Wei, W.; Liu, Z.; Zhang, K.; Tang, B.; et al. Safety and Effectiveness of Rasagiline in Chinese Patients with Parkinson’s Disease: A Prospective, Multicenter, Non-interventional Post-marketing Study. Drug Saf. 2023, 46, 637–646. [Google Scholar] [CrossRef]
- Richmond, A.M.; Lyons, K.E.; Pahwa, R. Safety review of current pharmacotherapies for levodopa-treated patients with Parkinson’s disease. Expert Opin. Drug Saf. 2023, 22, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Lecht, S.; Haroutiunian, S.; Hoffman, A.; Lazarovici, P. Rasagiline—A novel MAO B inhibitor in Parkinson’s disease therapy. Ther. Clin. Risk Manag. 2007, 3, 467–474. [Google Scholar] [PubMed]
- Malaty, I.A.; Fernandez, H.H. Role of rasagiline in treating Parkinson’s disease: Effect on disease progression. Ther. Clin. Risk Manag. 2009, 5, 413–419. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, Y.F.; Gao, F.; Gao, L.; Miao, J.D. Effects of rasagiline combined with levodopa and benserazide hydrochloride on motor function and homocysteine and IGF-1 levels in elderly patients with Parkinson’s disease. BMC Neurol. 2023, 23, 360. [Google Scholar] [CrossRef]
- Haddad, F.; Sawalha, M.; Khawaja, Y.; Najjar, A.; Karaman, R. Dopamine and Levodopa Prodrugs for the Treatment of Parkinson’s Disease. Molecules 2018, 23, 40. [Google Scholar] [CrossRef]
- Beckers, M.; Bloem, B.R.; Verbeek, M.M. Mechanisms of peripheral levodopa resistance in Parkinson’s disease. NPJ Park. Dis. 2022, 8, 56. [Google Scholar] [CrossRef]
- Eldeeb, M.A.; Thomas, R.A.; Ragheb, M.A.; Fallahi, A.; Fon, E.A. Mitochondrial Quality Control in Health and in Parkinson’s Disease. Physiol. Rev. 2022, 102, 1721–1755. [Google Scholar] [CrossRef]
- Themistokleous, C.; Bagnoli, E.; Parulekar, R.; Muqit, M.M.K. Role of Autophagy Pathway in Parkinson’s Disease and Related Genetic Neurological Disorders. J. Mol. Biol. 2023, 435, 168144. [Google Scholar] [CrossRef]
- Meng, X.; Zhou, J.; Zhao, C.N.; Gan, R.Y.; Li, H.B. Health Benefits and Molecular Mechanisms of Resveratrol: A Narrative Review. Foods 2020, 9, 340. [Google Scholar] [CrossRef]
- Chahine, L.M.; Stern, M.B. Rasagiline in Parkinson’s disease. Int. Rev. Neurobiol. 2011, 100, 151–168. [Google Scholar] [CrossRef]
- Picazio, S.; Ponzo, V.; Caltagirone, C.; Brusa, L.; Koch, G. Dysfunctional inhibitory control in Parkinson’s disease patients with levodopa-induced dyskinesias. J. Neurol. 2018, 265, 2088–2096. [Google Scholar] [CrossRef] [PubMed]
- Kaakkola, S. Clinical pharmacology, therapeutic use and potential of COMT inhibitors in Parkinson’s disease. Drugs 2000, 59, 1233–1250. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ye, H.; Jiang, P.; Liu, J.; Wang, B.; Zhang, S.; Sik, A.; Li, N.; Liu, K.; Jin, M. The alleviative effect of Calendula officinalis L. extract against Parkinson’s disease-like pathology in zebrafish via the involvement of autophagy activation. Front. Neurosci. 2023, 17, 1153889. [Google Scholar] [CrossRef]
- Li, X.; Gao, D.; Paudel, Y.N.; Li, X.; Zheng, M.; Liu, G.; Ma, Y.; Chu, L.; He, F.; Jin, M. Anti-Parkinson’s Disease Activity of Sanghuangprous vaninii Extracts in the MPTP-Induced Zebrafish Model. ACS Chem. Neurosci. 2022, 13, 330–339. [Google Scholar] [CrossRef]
- Li, M.; Zhang, J.; Jiang, L.; Wang, W.; Feng, X.; Liu, M.; Yang, D. Neuroprotective effects of morroniside from Cornus officinalis sieb. Et zucc against Parkinson’s disease via inhibiting oxidative stress and ferroptosis. BMC Complement. Med. Ther. 2023, 23, 218. [Google Scholar] [CrossRef]
- He, X.; Wang, J.; Li, M.; Hao, D.; Yang, Y.; Zhang, C.; He, R.; Tao, R. Eucommia ulmoides Oliv.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2014, 151, 78–92. [Google Scholar] [CrossRef]
- Xing, Y.F.; He, D.; Wang, Y.; Zeng, W.; Zhang, C.; Lu, Y.; Su, N.; Kong, Y.H.; Xing, X.H. Chemical constituents, biological functions and pharmacological effects for comprehensive utilization of Oliver. Food Sci. Hum. Wellness 2019, 8, 177–188. [Google Scholar] [CrossRef]
- Wu, M.F.; Zhuang, Q.L.; Lin, J.K.; Peng, Y.Y.; Luo, F.; Liu, Z.X.; Farooq, U.; Zhang, Q. Enrichment of the flavonoid fraction from leaves by a liquid antisolvent precipitation method and evaluation of antioxidant activities and. RSC Adv. 2023, 13, 17406–17419. [Google Scholar] [CrossRef]
- Hosoo, S.; Koyama, M.; Kato, M.; Hirata, T.; Yamaguchi, Y.; Yamasaki, H.; Wada, A.; Wada, K.; Nishibe, S.; Nakamura, K. The Restorative Effects of Oliver Leaf Extract on Vascular Function in Spontaneously Hypertensive Rats. Molecules 2015, 20, 21971–21981. [Google Scholar] [CrossRef]
- Kwan, C.Y.; Chen, C.X.; Deyama, T.; Nishibe, S. Endothelium-dependent vasorelaxant effects of the aqueous extracts of the Oliv. leaf and bark: Implications on their antihypertensive action. Vasc. Pharmacol. 2003, 40, 229–235. [Google Scholar] [CrossRef]
- Fu, G.M.; Tong, H.Y.; Zeng, H.L.; Zou, B.; Chai, J.X.; Zhang, L.N.; Xie, M.Y.; Chen, F.; Wan, Y. Antioxidant and xanthine oxidase inhibitory activity of Oliver leaf extracts. Pak. J. Pharm. Sci. 2018, 31, 1333–1339. [Google Scholar] [PubMed]
- Hao, S.; Xiao, Y.; Lin, Y.; Mo, Z.T.; Chen, Y.; Peng, X.F.; Xiang, C.H.; Li, Y.Q.; Li, W.N. Chlorogenic acid-enriched extract from leaves inhibits hepatic lipid accumulation through regulation of cholesterol metabolism in HepG2 cells. Pharm. Biol. 2016, 54, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Q.; Liang, M.; Li, W.Z.; Li, K.; Li, P.; Hu, Y.Z.; Yang, Z.L. Protective effects of Eucommia ulmoides Oliv. bark and leaf on amyloid beta-induced cytotoxicity. Environ. Toxicol. Pharmacol. 2009, 28, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.S.; Yu, Z.P.; Xia, J.Y.; Zhang, X.M.; Liu, K.C.; Sik, A.; Jin, M. Anti-Parkinson’s disease activity of phenolic acids from Oliver leaf extracts and their autophagy activation mechanism. Food Funct. 2020, 11, 1425–1440. [Google Scholar] [CrossRef]
- Razali, K.; Othman, N.; Nasir, M.H.M.; Doolaanea, A.; Kumar, J.; Ibrahim, W.N.; Ibrahim, N.M.; Mohamed, W.M.Y. The Promise of the Zebrafish Model for Parkinson’s Disease: Today’s Science and Tomorrow’s Treatment. Front. Genet. 2021, 12, 655550. [Google Scholar] [CrossRef]
- Doyle, J.M.; Croll, R.P. A Critical Review of Zebrafish Models of Parkinson’s Disease. Front. Pharmacol. 2022, 13, 835827. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef]
- Patton, E.E.; Zon, L.I.; Langenau, D.M. Zebrafish disease models in drug discovery: From preclinical modelling to clinical trials. Nat. Rev. Drug Discov. 2021, 20, 611–628. [Google Scholar] [CrossRef]
- Thirugnanam, T.; Santhakumar, K. Chemically induced models of Parkinson’s disease. Comp. Biochem. Phys. C 2022, 252, 109213. [Google Scholar] [CrossRef]
- Javitch, J.A.; D’Amato, R.J.; Strittmatter, S.M.; Snyder, S.H. Parkinsonism-inducing neurotoxin, N-methyl-4-phenyl-1,2,3,6 -tetrahydropyridine: Uptake of the metabolite N-methyl-4-phenylpyridine by dopamine neurons explains selective toxicity. Proc. Natl. Acad. Sci. USA 1985, 82, 2173–2177. [Google Scholar] [CrossRef]
- Nicklas, W.J.; Vyas, I.; Heikkila, R.E. Inhibition of NADH-linked oxidation in brain mitochondria by 1-methyl-4-phenyl-pyridine, a metabolite of the neurotoxin, 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. Life Sci. 1985, 36, 2503–2508. [Google Scholar] [CrossRef]
- Barnhill, L.M.; Murata, H.; Bronstein, J.M. Studying the Pathophysiology of Parkinson’s Disease Using Zebrafish. Biomedicines 2020, 8, 197. [Google Scholar] [CrossRef]
- Wen, S.; Aki, T.; Unuma, K.; Uemura, K. Chemically Induced Models of Parkinson’s Disease: History and Perspectives for the Involvement of Ferroptosis. Front. Cell. Neurosci. 2020, 14, 581191. [Google Scholar] [CrossRef]
- Dong, H.; Mao, L.; Bai, C.; Ye, K.; Wu, H.; Lei, Y.; Yu, S.; Liu, Y.; Tao, J.; Pan, W.; et al. Characterization of Developmental Neurobehavioral Toxicity in a Zebrafish MPTP-Induced Model: A Novel Mechanism Involving Anemia. ACS Chem. Neurosci. 2022, 13, 1877–1890. [Google Scholar] [CrossRef]
- Chen, X.; Guo, C.; Kong, J. Oxidative stress in neurodegenerative diseases. Neural Regen. Res. 2012, 7, 376–385. [Google Scholar] [CrossRef]
- Fonseca-Fonseca, L.A.; da Silva, V.D.A.; Wong-Guerra, M.; Ramirez-Sanchez, J.; Yaquis, A.S.P.; Ochoa-Rodriguez, E.; Verdecia-Reyes, Y.; de Araujo, F.M.; Santana, R.C.; Outeiro, T.F.; et al. JM-20 protects against 6-hydroxydopamine-induced neurotoxicity in models of Parkinson’s disease: Mitochondrial protection and antioxidant properties. Neurotoxicology 2021, 82, 89–98. [Google Scholar] [CrossRef]
- Xicoy, H.; Wieringa, B.; Martens, G.J. The SH-SY5Y cell line in Parkinson’s disease research: A systematic review. Mol. Neurodegener. 2017, 12, 10. [Google Scholar] [CrossRef]
- Wang, L.; Sheng, W.; Tan, Z.; Ren, Q.; Wang, R.; Stoika, R.; Liu, X.; Liu, K.; Shang, X.; Jin, M. Treatment of Parkinson’s disease in Zebrafish model with a berberine derivative capable of crossing blood brain barrier, targeting mitochondria, and convenient for bioimaging experiments. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2021, 249, 109151. [Google Scholar] [CrossRef]
- Akbani, R.; Becker, K.F.; Carragher, N.; Goldstein, T.; de Koning, L.; Korf, U.; Liotta, L.; Mills, G.B.; Nishizuka, S.S.; Pawlak, M.; et al. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: A workshop report: The RPPA (Reverse Phase Protein Array) society. Mol. Cell. Proteom. 2014, 13, 1625–1643. [Google Scholar] [CrossRef]
- Lu, Y.; Ling, S.; Hegde, A.M.; Byers, L.A.; Coombes, K.; Mills, G.B.; Akbani, R. Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer. Semin. Oncol. 2016, 43, 476–483. [Google Scholar] [CrossRef]
- Kuang, Z.; Huang, R.; Yang, Z.; Lv, Z.; Chen, X.; Xu, F.; Yi, Y.H.; Wu, J.; Huang, R.P. Quantitative screening of serum protein biomarkers by reverse phase protein arrays. Oncotarget 2018, 9, 32624–32641. [Google Scholar] [CrossRef]
- Jin, M.; Shi, R.D.; Gao, D.L.; Wang, B.K.; Li, N.; Li, X.; Sik, A.; Liu, K.C.; Zhang, X.J. ErbB2(pY)(-1248) as a predictive biomarker for Parkinson’s disease based on research with RPPA technology and in vivo verification. CNS Neurosci. Ther. 2023, 30, e14407. [Google Scholar] [CrossRef]
- Dastidar, S.G.; Pham, M.T.; Mitchell, M.B.; Yeom, S.G.; Jordan, S.; Chang, A.; Sopher, B.L.; La Spada, A.R. 4E-BP1 Protects Neurons from Misfolded Protein Stress and Parkinson’s Disease Toxicity by Inducing the Mitochondrial Unfolded Protein Response. J. Neurosci. 2020, 40, 8734–8745. [Google Scholar] [CrossRef]
- Tain, L.S.; Mortiboys, H.; Tao, R.N.; Ziviani, E.; Bandmann, O.; Whitworth, A.J. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci. 2009, 12, 1129–1135. [Google Scholar] [CrossRef]
- Vaz, R.L.; Outeiro, T.F.; Ferreira, J.J. Zebrafish as an Animal Model for Drug Discovery in Parkinson’s Disease and Other Movement Disorders: A Systematic Review. Front. Neurol. 2018, 9, 347. [Google Scholar] [CrossRef]
- Razali, K.; Nasir, M.H.M.; Othman, N.; Doolaanea, A.A.; Kumar, J.; Ibrahim, W.N.; Mohamed, W.M.Y. Characterization of neurobehavioral pattern in a zebrafish 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced model: A 96-hour behavioral study. PLoS ONE 2022, 17, e0274844. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, H.; Yang, R.; Tao, Y.; Wang, Z.; Zhang, S.; Sun, B.; Li, D.; Lu, B.; Liu, C. alpha-Synuclein amyloid fibril directly binds to LC3B and suppresses SQSTM1/p62-mediated selective autophagy. Cell Res. 2024, 35, 72–75. [Google Scholar] [CrossRef]
- Lynch-Day, M.A.; Mao, K.; Wang, K.; Zhao, M.; Klionsky, D.J. The role of autophagy in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009357. [Google Scholar] [CrossRef]
- Sveinbjornsdottir, S. The clinical symptoms of Parkinson’s disease. J. Neurochem. 2016, 139, 318–324. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Tavakol, S.; Ahmadi, Z.; Roomiani, S.; Mohammadinejad, R.; Samarghandian, S. Therapeutic effects of kaempferol affecting autophagy and endoplasmic reticulum stress. Phytother. Res. 2020, 34, 911–923. [Google Scholar] [CrossRef]
- Alexander, G.E. Biology of Parkinson’s disease: Pathogenesis and pathophysiology of a multisystem neurodegenerative disorder. Dialogues Clin. Neurosci. 2004, 6, 259–280. [Google Scholar] [CrossRef]
- Zhang, S.N.; Li, H.M.; Liu, Q.; Li, X.Z.; Yang, W.D.; Zhou, Y. Eucommiae Folium and Active Compounds Protect Against Mitochondrial Dysfunction-Calcium Overload in Epileptic Hippocampal Neurons Through the Hypertrophic Cardiomyopathy Pathway. Neurochem. Res. 2023, 48, 2674–2686. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, B.Y.; Zheng, Y.T.; Liu, X.C.; Rostyslav, P.; Finiuk, N.; Sik, A.; Stoika, R.; Liu, K.C.; Jin, M. Neuroprotective effect of chlorogenic acid on Parkinson’s disease like symptoms through boosting the autophagy in zebrafish. Eur. J. Pharmacol. 2023, 956, 175950. [Google Scholar] [CrossRef]
- Li, C.W.; Tang, B.Q.; Feng, Y.; Tang, F.; Hoi, M.P.M.; Su, Z.R.; Lee, S.M.Y. Pinostrobin Exerts Neuroprotective Actions in Neurotoxin-Induced Parkinson’s Disease Models through Nrf2 Induction. J. Agric. Food Chem. 2018, 66, 8307–8318. [Google Scholar] [CrossRef]
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef]
- Ren, Q.Y.; Jiang, X.; Paudel, Y.N.; Gao, X.; Gao, D.L.; Zhang, P.Y.; Sheng, W.L.; Shang, X.L.; Liu, K.C.; Zhang, X.J.; et al. Co-treatment with natural HMGB1 inhibitor Glycyrrhizin exerts neuroprotection and reverses Parkinson’s disease like pathology in Zebrafish. J. Ethnopharmacol. 2022, 292, 115234. [Google Scholar] [CrossRef]
- Yang, Z.G.; Lin, P.P.; Chen, B.; Zhang, X.Q.; Xiao, W.; Wu, S.L.; Huang, C.N.; Feng, D.; Zhang, W.Q.; Zhang, J.J. Autophagy alleviates hypoxia-induced blood-brain barrier injury via regulation of CLDN5 (claudin 5). Autophagy 2021, 17, 3048–3067. [Google Scholar] [CrossRef]
- Hubner, K.; Cabochette, P.; Dieguez-Hurtado, R.; Wiesner, C.; Wakayama, Y.; Grassme, K.S.; Hubert, M.; Guenther, S.; Belting, H.G.; Affolter, M.; et al. Wnt/beta-catenin signaling regulates VE-cadherin-mediated anastomosis of brain capillaries by counteracting S1pr1 signaling. Nat. Commun. 2018, 9, 4860. [Google Scholar] [CrossRef]
- Peker, N.; Gozuacik, D. Autophagy as a Cellular Stress Response Mechanism in the Nervous System. J. Mol. Biol. 2020, 432, 2560–2588. [Google Scholar] [CrossRef]
- Kessel, D. Apoptosis, Paraptosis and Autophagy: Death and Survival Pathways Associated with Photodynamic Therapy. Photochem. Photobiol. 2019, 95, 119–125. [Google Scholar] [CrossRef]
- Du, X.Y.; Xie, X.X.; Liu, R.T. The Role of alpha-Synuclein Oligomers in Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 8645. [Google Scholar] [CrossRef]
- Xilouri, M.; Brekk, O.R.; Stefanis, L. Autophagy and Alpha-Synuclein: Relevance to Parkinson’s Disease and Related Synucleopathies. Mov. Disord. 2016, 31, 178–192. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Niu, X.Y.; Huang, H.J.; Zhang, J.B.; Zhang, C.; Chen, W.G.; Sun, C.Y.; Ding, Y.Q.; Liao, M. Deletion of autophagy-related gene 7 in dopaminergic neurons prevents their loss induced by MPTP. Neuroscience 2016, 339, 22–31. [Google Scholar] [CrossRef]
- Wu, G.; Wang, X.; Feng, X.; Zhang, A.; Li, J.; Gu, K.; Huang, J.; Pang, S.; Dong, H.; Gao, H.; et al. Altered expression of autophagic genes in the peripheral leukocytes of patients with sporadic Parkinson’s disease. Brain Res. 2011, 1394, 105–111. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homolog of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728, Erratum in EMBO J. 2003, 22, 4577. [Google Scholar] [CrossRef]
- Jia, R.; Bonifacino, J.S. The ubiquitin isopeptidase USP10 deubiquitinates LC3B to increase LC3B levels and autophagic activity. J. Biol. Chem. 2021, 296, 100405. [Google Scholar] [CrossRef]
- Komatsu, M.; Ichimura, Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010, 584, 1374–1378. [Google Scholar] [CrossRef]
- Mathew, R.; Karp, C.M.; Beaudoin, B.; Vuong, N.; Chen, G.H.; Chen, H.Y.; Bray, K.; Reddy, A.; Bhanot, G.; Gelinas, C.; et al. Autophagy Suppresses Tumorigenesis through Elimination of p62. Cell 2009, 137, 1062–1075. [Google Scholar] [CrossRef]
- Imai, Y.; Gehrke, S.; Wang, H.Q.; Takahashi, R.; Hasegawa, K.; Oota, E.; Lu, B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008, 27, 2432–2443. [Google Scholar] [CrossRef]
- Wu, S.; Wagner, G. Computational inference of eIF4F complex function and structure in human cancers. Proc. Natl. Acad. Sci. USA 2024, 121, e2313589121. [Google Scholar] [CrossRef]
- Kaur, T.; Sidana, P.; Kaur, N.; Choubey, V.; Kaasik, A. Unraveling neuroprotection in Parkinson’s disease: Nrf2-Keap1 pathway’s vital role amidst pathogenic pathways. Inflammopharmacology 2024, 32, 2801–2820. [Google Scholar] [CrossRef]
- Chen, Q.X.; Zhou, L.; Long, T.; Qin, D.L.; Wang, Y.L.; Ye, Y.; Zhou, X.G.; Wu, J.M.; Wu, A.G. Galangin Exhibits Neuroprotective Effects in 6-OHDA-Induced Models of Parkinson’s Disease via the Nrf2/Keapl Pathway. Pharmaceuticals 2022, 15, 1014. [Google Scholar] [CrossRef]
- Rathore, A.S.; Singh, S.S.; Birla, H.; Zahra, W.; Keshri, P.K.; Dilnashin, H.; Singh, R.; Singh, S.; Singh, S.P. Curcumin Modulates p62-Keap1-Nrf2-Mediated Autophagy in Rotenone-Induced Parkinson?s Disease Mouse Models. ACS Chem. Neurosci. 2023, 14, 1412–1423. [Google Scholar] [CrossRef]
- Subramaniam, S.R.; Chesselet, M.F. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease. Prog. Neurobiol. 2013, 106, 17–32. [Google Scholar] [CrossRef]
- Malpartida, A.B.; Williamson, M.; Narendra, D.P.; Wade-Martins, R.; Ryan, B.J. Mitochondrial Dysfunction and Mitophagy in Parkinson’s Disease: From Mechanism to Therapy. Trends Biochem. Sci. 2021, 46, 329–343. [Google Scholar] [CrossRef]
- Li, X.Q.; Hu, M.L.; Zhou, X.G.; Yu, L.; Qin, D.L.; Wu, J.M.; Deng, L.; Huang, L.F.; Ren, F.; Liao, B.; et al. Hederagenin inhibits mitochondrial damage in Parkinson’s disease via mitophagy induction. Free. Radic. Biol. Med. 2024, 224, 740–756. [Google Scholar] [CrossRef]
- Bashirzade, A.A.O.; Cheresiz, S.V.; Belova, A.S.; Drobkov, A.V.; Korotaeva, A.D.; Azizi-Arani, S.; Azimirad, A.; Odle, E.; Gild, E.Y.V.; Ardashov, O.V.; et al. MPTP-Treated Zebrafish Recapitulate ’Late-Stage’ Parkinson’s-like Cognitive Decline. Toxics 2022, 10, 69. [Google Scholar] [CrossRef]
- Díaz-Casado, M.E.; Rusanova, I.; Aranda-Martínez, P.; Fernández-Ortiz, M.; Sayed, R.K.A.; Fernández-Gil, B.I.; Hidalgo-Gutiérrez, A.; Escames, G.; López, L.C.; Acuña-Castroviejo, D. Determination of Mitochondrial Respiration in 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine-Treated Zebrafish Reveals the Efficacy of Melatonin in Restoring Mitochondrial Normalcy. Zebrafish 2018, 15, 15–26. [Google Scholar] [CrossRef]
- Peterson, S.M.; Zhang, J.; Freeman, J.L. Developmental reelin expression and time point-specific alterations from lead exposure in zebrafish. Neurotoxicol. Teratol. 2013, 38, 53–60. [Google Scholar] [CrossRef] [PubMed]


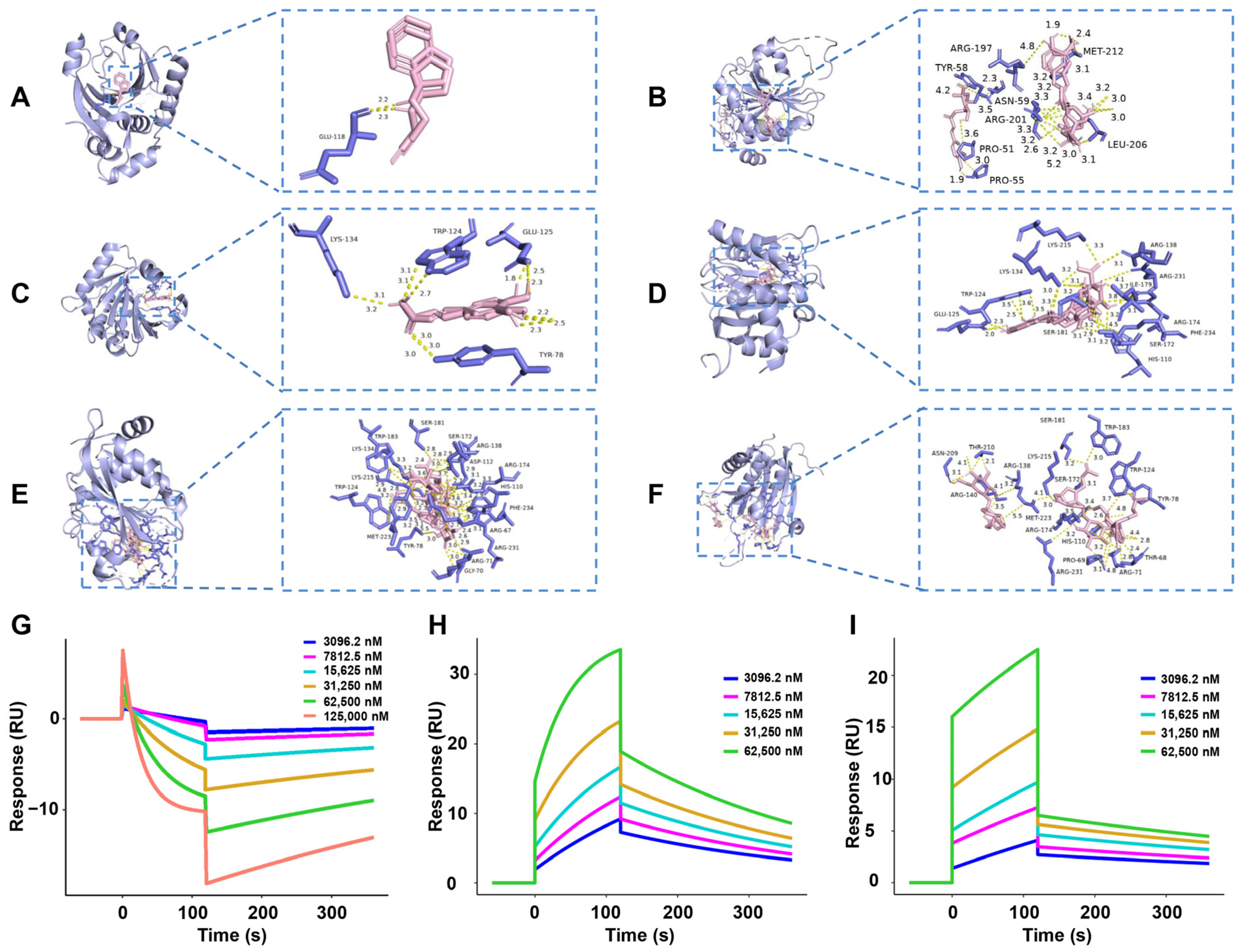
| Protein | Ligand | Affinity (kcal/mol) |
|---|---|---|
| 4E-BP1 (2jgb) | Rasagiline | −4.93 ± 0.12 |
| 4E-BP1 (2jgb) | Cryptochlorogenic acid | −9.26 ± 0.76 |
| 4E-BP1 (2jgb) | Caffeic acid | −7.98 ± 0.52 |
| 4E-BP1 (2jgb) | Chlorogenic acid | −7.79 ± 0.66 |
| 4E-BP1 (2jgb) | Asperulosidic acid | −7.71 ± 0.08 |
| 4E-BP1 (2jgb) | Asperuloside | −5.38 ± 0.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Shi, R.; Xia, L.; Zhang, X.; Zhang, P.; Liu, S.; Liu, K.; Sik, A.; Stoika, R.; Jin, M. Identification of Key Active Constituents in Eucommia ulmoides Oliv. Leaves Against Parkinson’s Disease and the Alleviative Effects via 4E-BP1 Up-Regulation. Int. J. Mol. Sci. 2025, 26, 2762. https://doi.org/10.3390/ijms26062762
Li Y, Shi R, Xia L, Zhang X, Zhang P, Liu S, Liu K, Sik A, Stoika R, Jin M. Identification of Key Active Constituents in Eucommia ulmoides Oliv. Leaves Against Parkinson’s Disease and the Alleviative Effects via 4E-BP1 Up-Regulation. International Journal of Molecular Sciences. 2025; 26(6):2762. https://doi.org/10.3390/ijms26062762
Chicago/Turabian StyleLi, Yuqing, Ruidie Shi, Lijie Xia, Xuanming Zhang, Pengyu Zhang, Siyuan Liu, Kechun Liu, Attila Sik, Rostyslav Stoika, and Meng Jin. 2025. "Identification of Key Active Constituents in Eucommia ulmoides Oliv. Leaves Against Parkinson’s Disease and the Alleviative Effects via 4E-BP1 Up-Regulation" International Journal of Molecular Sciences 26, no. 6: 2762. https://doi.org/10.3390/ijms26062762
APA StyleLi, Y., Shi, R., Xia, L., Zhang, X., Zhang, P., Liu, S., Liu, K., Sik, A., Stoika, R., & Jin, M. (2025). Identification of Key Active Constituents in Eucommia ulmoides Oliv. Leaves Against Parkinson’s Disease and the Alleviative Effects via 4E-BP1 Up-Regulation. International Journal of Molecular Sciences, 26(6), 2762. https://doi.org/10.3390/ijms26062762









