A New Perspective on the Role of Alterations in Mitochondrial Proteins Involved in ATP Synthesis and Mobilization in Cardiomyopathies
Abstract
:1. Introduction
2. Cardiomyopathies: Generalities
2.1. Hypertrophic Cardiomyopathy (HCM)
2.2. Dilated Cardiomyopathy (DCM)
2.3. Arrhythmogenic Right-Ventricular Cardiomyopathy (ARVC)
2.4. Restrictive Cardiomyopathy (RCM)
2.5. Cardiomyopathies Associated with Mitochondrial Dysfunction
2.6. Other Cardiomyopathies
3. Mutations in Mitochondrial Respiratory Chain and Their Relationship with Cardiovascular Diseases
3.1. Complex I
3.2. Complex II
3.3. Complex III
3.4. Complex IV
3.5. Complex V
4. ATP/ADP Antiporter or Carrier (ACC or ANT)
5. Mitochondrial Phosphate Transporter (PiC)
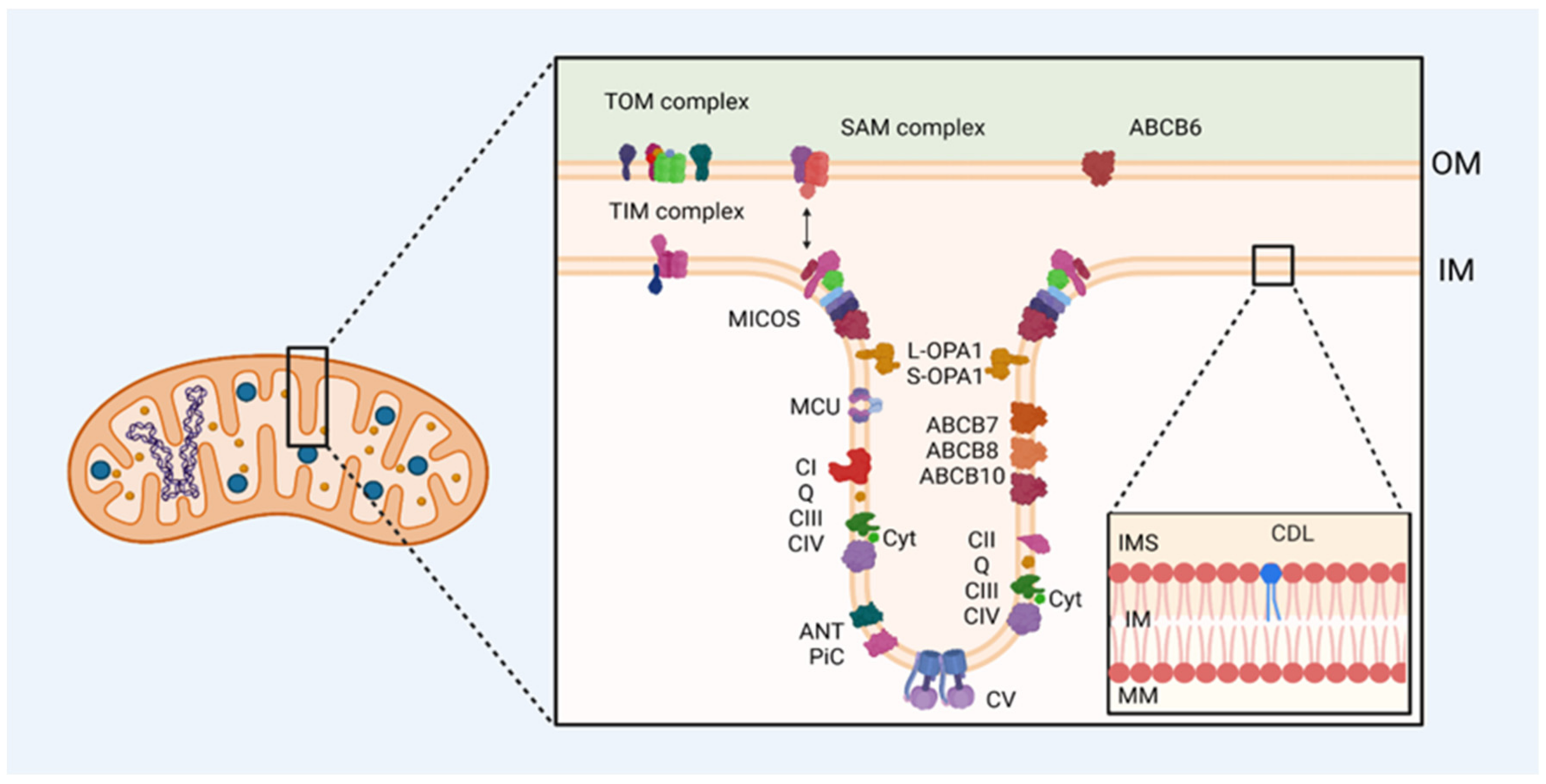
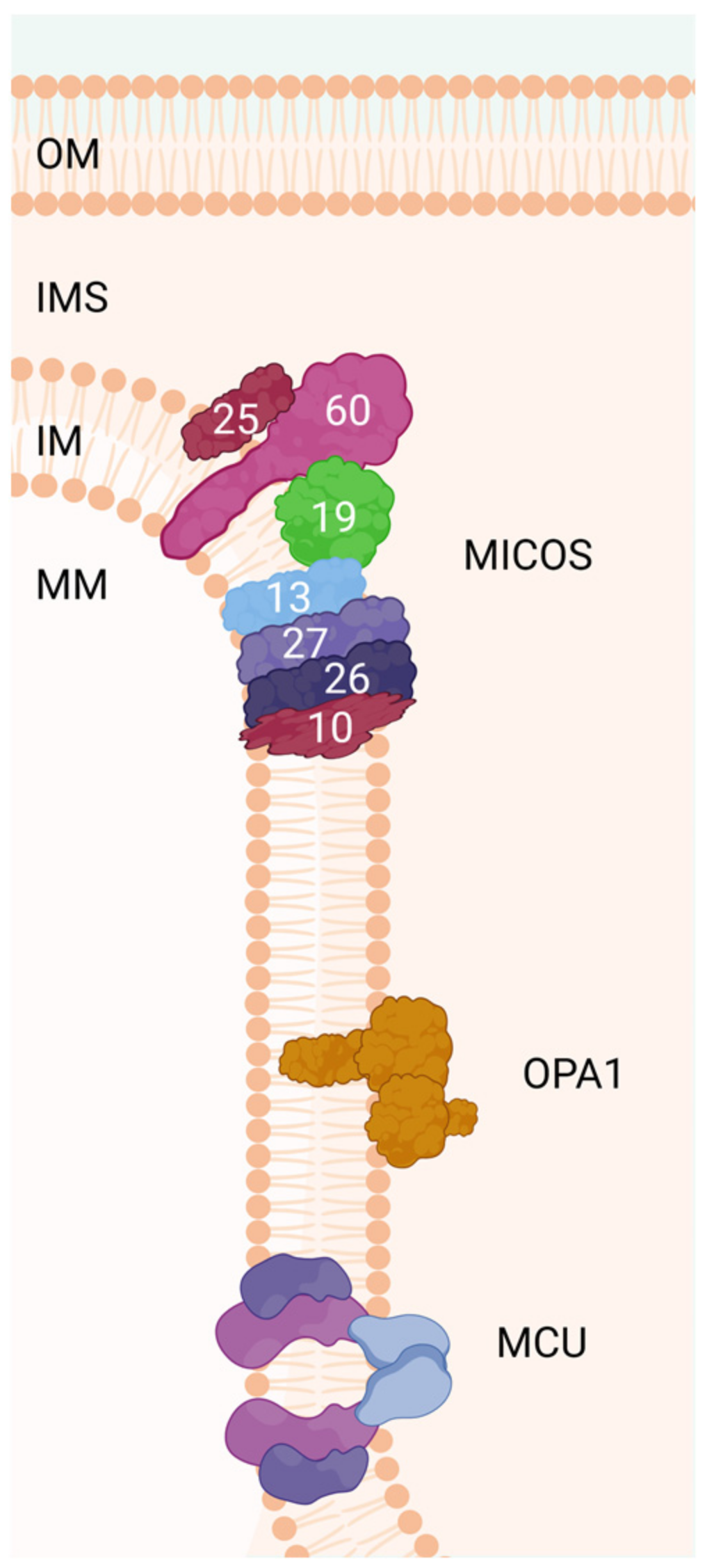
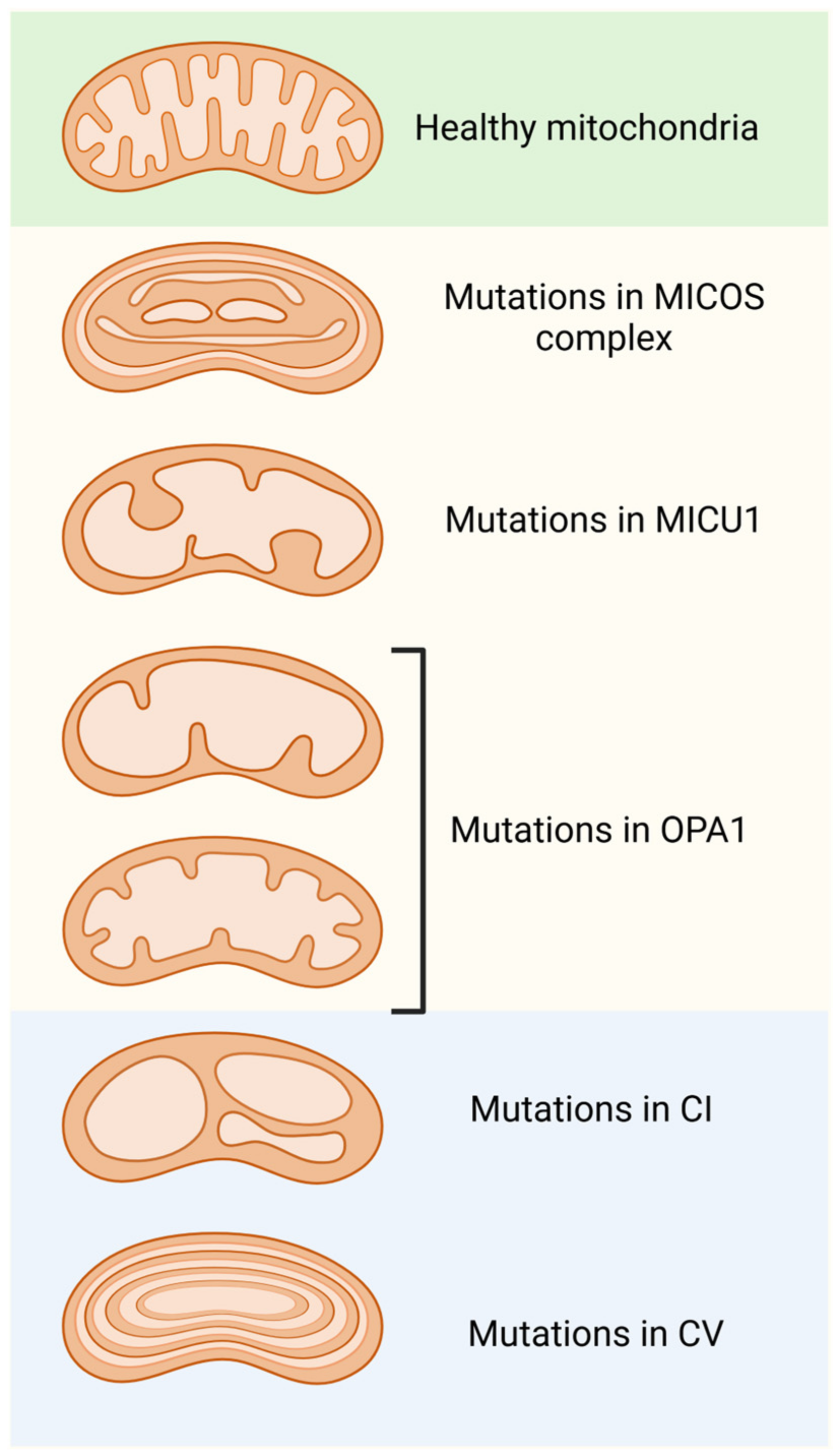
6. Mitochondrial Contact Site and Cristae-Organizing System (MICOS Complex)
6.1. Aberrant Mitochondrial Cristae and Related Diseases
6.2. Other Disorders Related to Abnormal Mitochondrial Cristae
- (A)
- CHCHD10 deficiency (CHCHD10, monoallelic variants, autosomal dominant inheritance, MIM #615903). CHCHD10-related diseases include mtDNA instability disorder, the clinical spectrum of frontotemporal dementia–amyotrophic lateral sclerosis (FTD-ALS), late-onset spinal motor neuropathy (SMAJ), and Charcot–Marie–Tooth disease type 2 (CMT2) [184,215,216].
- (B)
- CHCHD2 deficiency (CHCHD2, monoallelic variants, autosomal dominant inheritance, MIM #616244). CHCHD2 was the first mitochondrial gene reported to cause PD [217]; however, mutations have also been described in Alzheimer’s disease and frontotemporal dementia. As with CHCHD10 defects, a gain-of-function mechanism with misfolded protein toxicity has been suggested as the pathogenetic mechanism of neurodegeneration [216].
- (C)
- SLC25A46 deficiency (SLC25A46, biallelic variants, autosomal-recessive inheritance, MIM #610826). SLC25A46 deficiency causes Leigh syndrome, optic atrophy spectrum disorder (variably associated with PD), severe sensorimotor axonal neuropathy, cerebellar ataxia, and lethal pontocerebellar hypoplasia [218,219].
- (D)
- ATAD3A deficiency (ATAD3, MIM #612316). ATAD3A participates in mtDNA maintenance through the cholesterol metabolic pathway [220]. ATAD3A interacts with the MICOS complex, and its formation is reduced in ATAD3A-knockout mice [221]. This suggests that mtDNA maintenance may be regulated by interacting with the MICOS complex and ATAD3A. The oligomerization of ATAD3 is necessary for nucleoid mobility [222,223]. ATAD3A deficiency has been associated with a syndrome of neurodevelopmental delay with truncal hypotonia, spasticity, and inherited peripheral neuropathy in an AD or AR form (Harel–Yoon syndrome, MIM #617183) and with a syndrome of lethal AR pontocerebellar hypoplasia, hypotonia, and respiratory failure (MIM #618810). Interestingly, the ATAD3A-associated phenotype has shown enhanced type I IFN signaling, although the pathogenetic mechanisms remain to be determined [224].
- (E)
- Transmembrane protein 70 (TMEM70) deficiency (TMEM70, autosomal-recessive, MIM #614052). This is the most frequently reported cause of nuclear-encoded ATP synthase deficiency, resulting in neonatal encephalic cardiomyopathy with lactic acidosis and hyperammonemia [225]. TMEM70 functions to transport the c subunit of ATP synthase from the TIM complex to OXA1L, a protein required for the insertion of integral membrane proteins into the IM [226].
- (F)
- CLPB deficiency (CLPB, SKD3, biallelic variants, 3-methylglutaconic aciduria, type VIIA, autosomal dominant, 3-methylglutaconic aciduria, type VIIB, autosomal-recessive, neutropenia, severe congenital, 9, autosomal dominant, MIM #616254). It causes autosomal-recessive or -dominant 3MGA, variable neurological disease [227], and neutropenia. CLPB is an ATP-driven disaggregase protein that plays a key role in maintaining mitochondrial cristae stability. The presence of 3MGA associated with isolated severe congenital neutropenia, particularly when a distinct mutation is located in the ATP-binding site (position 105), suggests that this disease may represent a link between defects in membrane biosynthesis and cristae remodeling disorders.
7. Cardiolipin
8. ABC Transporters
8.1. Iron Metabolism Dysregulation in Cardiovascular Diseases
8.2. Mitochondrial ABC Transporters
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Biro, G.P. Oxygen and ATP: The Energy Economy of the Cell. In Blood Substitutes and Oxygen Biotherapeutics; Liu, H., Kaye, A.D., Jahr, J.S., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 21–32. [Google Scholar]
- Jeyaseelan, R.; Poizat, C.; Wu, H.Y.; Kedes, L. Molecular mechanisms of doxorubicin-induced cardiomyopathy. Selective suppression of Reiske iron-sulfur protein, ADP/ATP translocase, and phosphofructokinase genes is associated with ATP depletion in rat cardiomyocytes. J. Biol. Chem. 1997, 272, 5828–5832. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, K.; Siddiqi, N.; Singh, S.; Neil, C.J.; Dawson, D.K.; Frenneaux, M.P. The breathing heart—Mitochondrial respiratory chain dysfunction in cardiac disease. Int. J. Cardiol. 2014, 171, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Protasoni, M.; Zeviani, M. Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions. Int. J. Mol. Sci. 2021, 22, 586. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Chan, D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016, 212, 379–387. [Google Scholar] [CrossRef]
- Suen, D.F.; Norris, K.L.; Youle, R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008, 22, 1577–1590. [Google Scholar] [CrossRef]
- Marin-Garcia, J.; Goldenthal, M.J. Understanding the impact of mitochondrial defects in cardiovascular disease: A review. J. Card. Fail. 2002, 8, 347–361. [Google Scholar] [CrossRef]
- Ramaccini, D.; Montoya-Uribe, V.; Aan, F.J.; Modesti, L.; Potes, Y.; Wieckowski, M.R.; Krga, I.; Glibetic, M.; Pinton, P.; Giorgi, C.; et al. Mitochondrial Function and Dysfunction in Dilated Cardiomyopathy. Front. Cell Dev. Biol. 2020, 8, 624216. [Google Scholar] [CrossRef]
- Schaefer, A.M.; McFarland, R.; Blakely, E.L.; He, L.; Whittaker, R.G.; Taylor, R.W.; Chinnery, P.F.; Turnbull, D.M. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 2008, 63, 35–39. [Google Scholar] [CrossRef]
- Wallace, D.C.; Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a021220. [Google Scholar] [CrossRef]
- Limongelli, G.; Tome-Esteban, M.; Dejthevaporn, C.; Rahman, S.; Hanna, M.G.; Elliott, P.M. Prevalence and natural history of heart disease in adults with primary mitochondrial respiratory chain disease. Eur. J. Heart Fail. 2010, 12, 114–121. [Google Scholar] [CrossRef]
- Schon, E.A.; DiMauro, S.; Hirano, M. Human mitochondrial DNA: Roles of inherited and somatic mutations. Nat. Rev. Genet. 2012, 13, 878–890. [Google Scholar] [CrossRef] [PubMed]
- Russell, O.M.; Gorman, G.S.; Lightowlers, R.N.; Turnbull, D.M. Mitochondrial Diseases: Hope for the Future. Cell 2020, 181, 168–188. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Rizzuto, R.; Hajnoczky, G.; Su, T.P. MAM: More than just a housekeeper. Trends Cell Biol. 2009, 19, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Raturi, A.; Simmen, T. Where the endoplasmic reticulum and the mitochondrion tie the knot: The mitochondria-associated membrane (MAM). Biochim. Biophys. Acta 2013, 1833, 213–224. [Google Scholar] [CrossRef]
- Vance, J.E. Inter-organelle membrane contact sites: Implications for lipid metabolism. Biol. Direct 2020, 15, 24. [Google Scholar] [CrossRef]
- Area-Gomez, E.; Del Carmen Lara Castillo, M.; Tambini, M.D.; Guardia-Laguarta, C.; de Groof, A.J.; Madra, M.; Ikenouchi, J.; Umeda, M.; Bird, T.D.; Sturley, S.L.; et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 2012, 31, 4106–4123. [Google Scholar] [CrossRef]
- Gomez-Suaga, P.; Bravo-San Pedro, J.M.; Gonzalez-Polo, R.A.; Fuentes, J.M.; Niso-Santano, M. ER-mitochondria signaling in Parkinson’s disease. Cell Death Dis. 2018, 9, 337. [Google Scholar] [CrossRef]
- Lau, D.H.W.; Hartopp, N.; Welsh, N.J.; Mueller, S.; Glennon, E.B.; Morotz, G.M.; Annibali, A.; Gomez-Suaga, P.; Stoica, R.; Paillusson, S.; et al. Disruption of ER-mitochondria signalling in fronto-temporal dementia and related amyotrophic lateral sclerosis. Cell Death Dis. 2018, 9, 327. [Google Scholar] [CrossRef]
- Watanabe, S.; Ilieva, H.; Tamada, H.; Nomura, H.; Komine, O.; Endo, F.; Jin, S.; Mancias, P.; Kiyama, H.; Yamanaka, K. Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol. Med. 2016, 8, 1421–1437. [Google Scholar] [CrossRef]
- Chappell, J.B. Systems used for the transport of substrates into mitochondria. Br. Med. Bull. 1968, 24, 150–157. [Google Scholar] [CrossRef]
- Kumar, V.; Santhosh Kumar, T.R.; Kartha, C.C. Mitochondrial membrane transporters and metabolic switch in heart failure. Heart Fail. Rev. 2019, 24, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Ciarambino, T.; Menna, G.; Sansone, G.; Giordano, M. Cardiomyopathies: An Overview. Int. J. Mol. Sci. 2021, 22, 7722. [Google Scholar] [CrossRef] [PubMed]
- Bang, M.L.; Bogomolovas, J.; Chen, J. Understanding the molecular basis of cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 2022, 322, H181–H233. [Google Scholar] [CrossRef]
- Franz, W.M.; Muller, O.J.; Katus, H.A. Cardiomyopathies: From genetics to the prospect of treatment. Lancet 2001, 358, 1627–1637. [Google Scholar] [CrossRef]
- Vio, R.; Angelini, A.; Basso, C.; Cipriani, A.; Zorzi, A.; Melacini, P.; Thiene, G.; Rampazzo, A.; Corrado, D.; Calore, C. Hypertrophic Cardiomyopathy and Primary Restrictive Cardiomyopathy: Similarities, Differences and Phenocopies. J. Clin. Med. 2021, 10, 1954. [Google Scholar] [CrossRef]
- Precone, V.; Krasi, G.; Guerri, G.; Madureri, A.; Piazzani, M.; Michelini, S.; Barati, S.; Maniscalchi, T.; Bressan, S.; Bertelli, M. Cardiomyopathies. Acta Biomed. 2019, 90, 32–43. [Google Scholar] [CrossRef]
- Nollet, E.E.; Duursma, I.; Rozenbaum, A.; Eggelbusch, M.; Wust, R.C.I.; Schoonvelde, S.A.C.; Michels, M.; Jansen, M.; van der Wel, N.N.; Bedi, K.C.; et al. Mitochondrial dysfunction in human hypertrophic cardiomyopathy is linked to cardiomyocyte architecture disruption and corrected by improving NADH-driven mitochondrial respiration. Eur. Heart J. 2023, 44, 1170–1185. [Google Scholar] [CrossRef]
- Ranjbarvaziri, S.; Kooiker, K.B.; Ellenberger, M.; Fajardo, G.; Zhao, M.; Vander Roest, A.S.; Woldeyes, R.A.; Koyano, T.T.; Fong, R.; Ma, N.; et al. Altered Cardiac Energetics and Mitochondrial Dysfunction in Hypertrophic Cardiomyopathy. Circulation 2021, 144, 1714–1731. [Google Scholar] [CrossRef]
- Chan, K.; Harper, A.R.; Ashrafian, H.; Yavari, A. Cardiomyopathies. Medicine 2018, 46, 606–617. [Google Scholar] [CrossRef]
- Aguilera, B.; Suárez-Mier, M.P.; Guzzo-Merello, G.; García-Pavía, P. Cardiomyopathies. In Clinico-Pathological Atlas of Cardiovascular Diseases; Lucena, J.S., García-Pavía, P., Suarez-Mier, M.P., Alonso-Pulpon, L.A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 201–231. [Google Scholar]
- Braunwald, E. Cardiomyopathies: An Overview. Circ. Res. 2017, 121, 711–721. [Google Scholar] [CrossRef]
- Brieler, J.; Breeden, M.A.; Tucker, J. Cardiomyopathy: An Overview. Am. Fam. Physician 2017, 96, 640–646. [Google Scholar]
- Hershberger, R.E.; Cowan, J.; Jordan, E.; Kinnamon, D.D. The Complex and Diverse Genetic Architecture of Dilated Cardiomyopathy. Circ. Res. 2021, 128, 1514–1532. [Google Scholar] [CrossRef] [PubMed]
- Puckelwartz, M.J.; Pesce, L.L.; Dellefave-Castillo, L.M.; Wheeler, M.T.; Pottinger, T.D.; Robinson, A.C.; Kearns, S.D.; Gacita, A.M.; Schoppen, Z.J.; Pan, W.; et al. Genomic Context Differs Between Human Dilated Cardiomyopathy and Hypertrophic Cardiomyopathy. J. Am. Heart Assoc. 2021, 10, e019944. [Google Scholar] [CrossRef] [PubMed]
- Alblaihed, L.; Kositz, C.; Brady, W.J.; Al-Salamah, T.; Mattu, A. Diagnosis and management of arrhythmogenic right ventricular cardiomyopathy. Am. J. Emerg. Med. 2023, 65, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Gerull, B.; Brodehl, A. Insights Into Genetics and Pathophysiology of Arrhythmogenic Cardiomyopathy. Curr. Heart Fail. Rep. 2021, 18, 378–390. [Google Scholar] [CrossRef]
- Bosman, L.P.; Te Riele, A. Arrhythmogenic right ventricular cardiomyopathy: A focused update on diagnosis and risk stratification. Heart 2022, 108, 90–97. [Google Scholar] [CrossRef]
- Rapezzi, C.; Aimo, A.; Barison, A.; Emdin, M.; Porcari, A.; Linhart, A.; Keren, A.; Merlo, M.; Sinagra, G. Restrictive cardiomyopathy: Definition and diagnosis. Eur. Heart J. 2022, 43, 4679–4693. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Shkurat, T.P.; Melnichenko, A.A.; Grechko, A.V.; Orekhov, A.N. The role of mitochondrial dysfunction in cardiovascular disease: A brief review. Ann. Med. 2018, 50, 121–127. [Google Scholar] [CrossRef]
- Manolis, A.S.; Manolis, A.A.; Manolis, T.A.; Apostolaki, N.E.; Apostolopoulos, E.J.; Melita, H.; Katsiki, N. Mitochondrial dysfunction in cardiovascular disease: Current status of translational research/clinical and therapeutic implications. Med. Res. Rev. 2021, 41, 275–313. [Google Scholar] [CrossRef]
- Morciano, G.; Boncompagni, C.; Ramaccini, D.; Pedriali, G.; Bouhamida, E.; Tremoli, E.; Giorgi, C.; Pinton, P. Comprehensive Analysis of Mitochondrial Dynamics Alterations in Heart Diseases. Int. J. Mol. Sci. 2023, 24, 3414. [Google Scholar] [CrossRef]
- Piano, M.R.; Phillips, S.A. Alcoholic cardiomyopathy: Pathophysiologic insights. Cardiovasc. Toxicol. 2014, 14, 291–308. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, Y.; Ren, J. Epigenetic modification in alcohol use disorder and alcoholic cardiomyopathy: From pathophysiology to therapeutic opportunities. Metabolism 2021, 125, 154909. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.L.; Lang, C.H. Etiology of alcoholic cardiomyopathy: Mitochondria, oxidative stress and apoptosis. Int. J. Biochem. Cell Biol. 2017, 89, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Andersson, C.; Schou, M.; Gustafsson, F.; Torp-Pedersen, C. Alcohol Intake in Patients With Cardiomyopathy and Heart Failure: Consensus and Controversy. Circ. Heart Fail. 2022, 15, e009459. [Google Scholar] [CrossRef] [PubMed]
- Duncan, J.G. Mitochondrial dysfunction in diabetic cardiomyopathy. Biochim. Biophys. Acta 2011, 1813, 1351–1359. [Google Scholar] [CrossRef]
- Grubic Rotkvic, P.; Planinic, Z.; Liberati Prso, A.M.; Sikic, J.; Galic, E.; Rotkvic, L. The Mystery of Diabetic Cardiomyopathy: From Early Concepts and Underlying Mechanisms to Novel Therapeutic Possibilities. Int. J. Mol. Sci. 2021, 22, 5973. [Google Scholar] [CrossRef]
- Salvatore, T.; Pafundi, P.C.; Galiero, R.; Albanese, G.; Di Martino, A.; Caturano, A.; Vetrano, E.; Rinaldi, L.; Sasso, F.C. The Diabetic Cardiomyopathy: The Contributing Pathophysiological Mechanisms. Front. Med. 2021, 8, 695792. [Google Scholar] [CrossRef]
- El-Hattab, A.W.; Scaglia, F. Mitochondrial Cardiomyopathies. Front. Cardiovasc. Med. 2016, 3, 25. [Google Scholar] [CrossRef]
- Brunel-Guitton, C.; Levtova, A.; Sasarman, F. Mitochondrial Diseases and Cardiomyopathies. Can. J. Cardiol. 2015, 31, 1360–1376. [Google Scholar] [CrossRef]
- Rosca, M.G.; Hoppel, C.L. Mitochondrial dysfunction in heart failure. Heart Fail. Rev. 2013, 18, 607–622. [Google Scholar] [CrossRef]
- Detta, N.; Frisso, G.; Salvatore, F. The multi-faceted aspects of the complex cardiac Nav1.5 protein in membrane function and pathophysiology. Biochim. Biophys. Acta 2015, 1854, 1502–1509. [Google Scholar] [CrossRef] [PubMed]
- Gray, B.; Semsarian, C.; Sy, R.W. Brugada syndrome: a heterogeneous disease with a common ECG phenotype? J. Cardiovasc. Electrophysiol. 2014, 25, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Leme, A.M.; Salemi, V.M.; Parga, J.R.; Ianni, B.M.; Mady, C.; Weiss, R.G.; Kalil-Filho, R. Evaluation of the metabolism of high energy phosphates in patients with Chagas’ disease. Arq. Bras. Cardiol. 2010, 95, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.P.S.; Andrieux, P.; Brochet, P.; Almeida, R.R.; Kitano, E.; Honda, A.K.; Iwai, L.K.; Andrade-Silva, D.; Goudenege, D.; Alcantara Silva, K.D.; et al. Co-Exposure of Cardiomyocytes to IFN-gamma and TNF-alpha Induces Mitochondrial Dysfunction and Nitro-Oxidative Stress: Implications for the Pathogenesis of Chronic Chagas Disease Cardiomyopathy. Front. Immunol. 2021, 12, 755862. [Google Scholar] [CrossRef]
- Nunes, J.P.S.; Roda, V.M.P.; Andrieux, P.; Kalil, J.; Chevillard, C.; Cunha-Neto, E. Inflammation and mitochondria in the pathogenesis of chronic Chagas disease cardiomyopathy. Exp. Biol. Med. 2023, 248, 2062–2071. [Google Scholar] [CrossRef]
- Teixeira, P.C.; Ducret, A.; Langen, H.; Nogoceke, E.; Santos, R.H.B.; Silva Nunes, J.P.; Benvenuti, L.; Levy, D.; Bydlowski, S.P.; Bocchi, E.A.; et al. Impairment of Multiple Mitochondrial Energy Metabolism Pathways in the Heart of Chagas Disease Cardiomyopathy Patients. Front. Immunol. 2021, 12, 755782. [Google Scholar] [CrossRef]
- Teixeira, P.C.; Santos, R.H.; Fiorelli, A.I.; Bilate, A.M.; Benvenuti, L.A.; Stolf, N.A.; Kalil, J.; Cunha-Neto, E. Selective decrease of components of the creatine kinase system and ATP synthase complex in chronic Chagas disease cardiomyopathy. PLoS Negl. Trop. Dis. 2011, 5, e1205. [Google Scholar] [CrossRef]
- Deshpande, O.A.; Mohiuddin, S.S. Biochemistry, Oxidative Phosphorylation; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Caruana, N.J.; Stroud, D.A. The road to the structure of the mitochondrial respiratory chain supercomplex. Biochem. Soc. Trans. 2020, 48, 621–629. [Google Scholar] [CrossRef]
- Lobo-Jarne, T.; Ugalde, C. Respiratory chain supercomplexes: Structures, function and biogenesis. Semin. Cell Dev. Biol. 2018, 76, 179–190. [Google Scholar] [CrossRef]
- Salazar, C.; Elorza, A.A.; Cofre, G.; Ruiz-Hincapie, P.; Shirihai, O.; Ruiz, L.M. The OXPHOS supercomplex assembly factor HIG2A responds to changes in energetic metabolism and cell cycle. J. Cell. Physiol. 2019, 234, 17405–17419. [Google Scholar] [CrossRef]
- Acin-Perez, R.; Enriquez, J.A. The function of the respiratory supercomplexes: The plasticity model. Biochim. Biophys. Acta 2014, 1837, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, D.; Blaza, J.N.; Larsson, N.G.; Hirst, J. The Enigma of the Respiratory Chain Supercomplex. Cell Metab. 2017, 25, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Stuart, R.A. Chapter 11 Supercomplex organization of the yeast respiratory chain complexes and the ADP/ATP carrier proteins. Methods Enzymol. 2009, 456, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Duanmu, X.; Zeng, L.; Liu, B.; Song, Z. Mitochondrial DNA: Distribution, Mutations, and Elimination. Cells 2019, 8, 379. [Google Scholar] [CrossRef]
- Dimauro, S.; Davidzon, G. Mitochondrial DNA and disease. Ann. Med. 2005, 37, 222–232. [Google Scholar] [CrossRef]
- Wilkins, H.M.; Carl, S.M.; Swerdlow, R.H. Cytoplasmic hybrid (cybrid) cell lines as a practical model for mitochondriopathies. Redox Biol. 2014, 2, 619–631. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Genova, M.L.; Baracca, A.; Biondi, A.; Casalena, G.; Faccioli, M.; Falasca, A.I.; Formiggini, G.; Sgarbi, G.; Solaini, G.; Lenaz, G. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim. Biophys. Acta 2008, 1777, 740–746. [Google Scholar] [CrossRef]
- Bartosz, G. Reactive oxygen species: Destroyers or messengers? Biochem. Pharmacol. 2009, 77, 1303–1315. [Google Scholar] [CrossRef]
- Halliwell, B. Reactive oxygen species in living systems: Source, biochemistry, and role in human disease. Am. J. Med. 1991, 91, 14S–22S. [Google Scholar] [CrossRef]
- Nathan, C.; Ding, A. SnapShot: Reactive Oxygen Intermediates (ROI). Cell 2010, 140, 952–952.e2. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Ahuir, A.; Manzanares-Estreder, S.; Proft, M. Pro- and Antioxidant Functions of the Peroxisome-Mitochondria Connection and Its Impact on Aging and Disease. Oxidative Med. Cell. Longev. 2017, 2017, 9860841. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Larosa, V.; Remacle, C. Insights into the respiratory chain and oxidative stress. Biosci. Rep. 2018, 38, BSR20171492. [Google Scholar] [CrossRef]
- Tarafdar, A.; Pula, G. The Role of NADPH Oxidases and Oxidative Stress in Neurodegenerative Disorders. Int. J. Mol. Sci. 2018, 19, 3824. [Google Scholar] [CrossRef]
- Shokolenko, I.; Venediktova, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548. [Google Scholar] [CrossRef]
- van Oven, M.; Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009, 30, E386–E394. [Google Scholar] [CrossRef]
- Giordano, C.; D’Amati, G. Cardiomyopathies due to defective energy metabolism: Morphological and functional features. Pathologica 2005, 97, 361–368. [Google Scholar]
- Benson, D.W.; Silberbach, G.M.; Kavanaugh-McHugh, A.; Cottrill, C.; Zhang, Y.; Riggs, S.; Smalls, O.; Johnson, M.C.; Watson, M.S.; Seidman, J.G.; et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Investig. 1999, 104, 1567–1573. [Google Scholar] [CrossRef]
- Nakamura, M.; Yabe, I.; Sudo, A.; Hosoki, K.; Yaguchi, H.; Saitoh, S.; Sasaki, H. MERRF/MELAS overlap syndrome: A double pathogenic mutation in mitochondrial tRNA genes. J. Med. Genet. 2010, 47, 659–664. [Google Scholar] [CrossRef]
- Catarino, C.B.; Ahting, U.; Gusic, M.; Iuso, A.; Repp, B.; Peters, K.; Biskup, S.; von Livonius, B.; Prokisch, H.; Klopstock, T. Characterization of a Leber’s hereditary optic neuropathy (LHON) family harboring two primary LHON mutations m.11778G>A and m.14484T>C of the mitochondrial DNA. Mitochondrion 2017, 36, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Zarrouk, S.; Finsterer, J.; Mehri, S.; Ourda, F.; Ben Arab, S.; Boussada, R. Dilated Cardiomyopathy due to the Novel MT-CYB Missense Mutation m.14757T>C. J. Med. Cases 2021, 12, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Maio, N.; Ghezzi, D.; Verrigni, D.; Rizza, T.; Bertini, E.; Martinelli, D.; Zeviani, M.; Singh, A.; Carrozzo, R.; Rouault, T.A. Disease-Causing SDHAF1 Mutations Impair Transfer of Fe-S Clusters to SDHB. Cell Metab. 2016, 23, 292–302. [Google Scholar] [CrossRef]
- Brand, M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016, 100, 14–31. [Google Scholar] [CrossRef]
- Antonicka, H.; Leary, S.C.; Guercin, G.H.; Agar, J.N.; Horvath, R.; Kennaway, N.G.; Harding, C.O.; Jaksch, M.; Shoubridge, E.A. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 2003, 12, 2693–2702. [Google Scholar] [CrossRef]
- Halperin, D.; Drabkin, M.; Wormser, O.; Yogev, Y.; Dolgin, V.; Shorer, Z.; Gradstein, L.; Shelef, I.; Flusser, H.; Birk, O.S. Phenotypic variability and mutation hotspot in COX15-related Leigh syndrome. Am. J. Med. Genet. A 2020, 182, 1506–1512. [Google Scholar] [CrossRef]
- Weraarpachai, W.; Antonicka, H.; Sasarman, F.; Seeger, J.; Schrank, B.; Kolesar, J.E.; Lochmuller, H.; Chevrette, M.; Kaufman, B.A.; Horvath, R.; et al. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 2009, 41, 833–837. [Google Scholar] [CrossRef]
- Wei, Y.L.; Yu, C.A.; Yang, P.; Li, A.L.; Wen, J.Y.; Zhao, S.M.; Liu, H.X.; Ke, Y.N.; Campbell, W.; Zhang, Y.G.; et al. Novel mitochondrial DNA mutations associated with Chinese familial hypertrophic cardiomyopathy. Clin. Exp. Pharmacol. Physiol. 2009, 36, 933–939. [Google Scholar] [CrossRef]
- Fragaki, K.; Chaussenot, A.; Serre, V.; Acquaviva, C.; Bannwarth, S.; Rouzier, C.; Chabrol, B.; Paquis-Flucklinger, V. A novel variant m.8561C>T in the overlapping region of MT-ATP6 and MT-ATP8 in a child with early-onset severe neurological signs. Mol. Genet. Metab. Rep. 2019, 21, 100543. [Google Scholar] [CrossRef]
- Tsao, C.Y.; Mendell, J.R.; Bartholomew, D. High mitochondrial DNA T8993G mutation (<90%) without typical features of Leigh’s and NARP syndromes. J. Child. Neurol. 2001, 16, 533–535. [Google Scholar] [CrossRef]
- Na, J.H.; Lee, Y.M. Genotype-phenotype analysis of MT-ATP6-associated Leigh syndrome. Acta Neurol. Scand. 2022, 145, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Escamilla, I.; Benedicto, C.; Perez-Carrillo, L.; Delgado-Arija, M.; Gonzalez-Torrent, I.; Vilchez, R.; Martinez-Dolz, L.; Portoles, M.; Tarazon, E.; Rosello-Lleti, E. Alterations in Mitochondrial Oxidative Phosphorylation System: Relationship of Complex V and Cardiac Dysfunction in Human Heart Failure. Antioxidants 2024, 13, 285. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, Z.; Zhu, M.; Shen, Y.; Leon, L.J.; Chi, L.; Spinozzi, S.; Tan, C.; Gu, Y.; Nguyen, A.; et al. Cardiolipin Remodeling Defects Impair Mitochondrial Architecture and Function in a Murine Model of Barth Syndrome Cardiomyopathy. Circ. Heart Fail. 2021, 14, e008289. [Google Scholar] [CrossRef] [PubMed]
- Claypool, S.M.; McCaffery, J.M.; Koehler, C.M. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J. Cell Biol. 2006, 174, 379–390. [Google Scholar] [CrossRef]
- Vieira Neto, E.; Wang, M.; Szuminsky, A.J.; Ferraro, L.; Koppes, E.; Wang, Y.; Van’t Land, C.; Mohsen, A.W.; Zanatta, G.; El-Gharbawy, A.H.; et al. Mitochondrial bioenergetics and cardiolipin remodeling abnormalities in mitochondrial trifunctional protein deficiency. JCI Insight 2024, 9, e176887. [Google Scholar] [CrossRef]
- Popoiu, T.A.; Dudek, J.; Maack, C.; Bertero, E. Cardiac Involvement in Mitochondrial Disorders. Curr. Heart Fail. Rep. 2023, 20, 76–87. [Google Scholar] [CrossRef]
- Davey, K.M.; Parboosingh, J.S.; McLeod, D.R.; Chan, A.; Casey, R.; Ferreira, P.; Snyder, F.F.; Bridge, P.J.; Bernier, F.P. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J. Med. Genet. 2006, 43, 385–393. [Google Scholar] [CrossRef]
- Hoshino, A.; Wang, W.J.; Wada, S.; McDermott-Roe, C.; Evans, C.S.; Gosis, B.; Morley, M.P.; Rathi, K.S.; Li, J.; Li, K.; et al. The ADP/ATP translocase drives mitophagy independent of nucleotide exchange. Nature 2019, 575, 375–379. [Google Scholar] [CrossRef]
- Walther, T.; Tschope, C.; Sterner-Kock, A.; Westermann, D.; Heringer-Walther, S.; Riad, A.; Nikolic, A.; Wang, Y.; Ebermann, L.; Siems, W.E.; et al. Accelerated mitochondrial adenosine diphosphate/adenosine triphosphate transport improves hypertension-induced heart disease. Circulation 2007, 115, 333–344. [Google Scholar] [CrossRef]
- Mayr, J.A.; Zimmermann, F.A.; Horvath, R.; Schneider, H.C.; Schoser, B.; Holinski-Feder, E.; Czermin, B.; Freisinger, P.; Sperl, W. Deficiency of the mitochondrial phosphate carrier presenting as myopathy and cardiomyopathy in a family with three affected children. Neuromuscul. Disord. 2011, 21, 803–808. [Google Scholar] [CrossRef]
- Moraes, C.T. A magic bullet to specifically eliminate mutated mitochondrial genomes from patients’ cells. EMBO Mol. Med. 2014, 6, 434–435. [Google Scholar] [CrossRef] [PubMed]
- Viscomi, C.; Bottani, E.; Zeviani, M. Emerging concepts in the therapy of mitochondrial disease. Biochim. Biophys. Acta 2015, 1847, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Bacman, S.R.; Williams, S.L.; Duan, D.; Moraes, C.T. Manipulation of mtDNA heteroplasmy in all striated muscles of newborn mice by AAV9-mediated delivery of a mitochondria-targeted restriction endonuclease. Gene Ther. 2012, 19, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Silva-Pinheiro, P.; Minczuk, M. The potential of mitochondrial genome engineering. Nat. Rev. Genet. 2022, 23, 199–214. [Google Scholar] [CrossRef]
- Papworth, M.; Kolasinska, P.; Minczuk, M. Designer zinc-finger proteins and their applications. Gene 2006, 366, 27–38. [Google Scholar] [CrossRef]
- Choo, Y.; Klug, A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc. Natl. Acad. Sci. USA 1994, 91, 11168–11172. [Google Scholar] [CrossRef]
- Bacman, S.R.; Williams, S.L.; Pinto, M.; Peralta, S.; Moraes, C.T. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med. 2013, 19, 1111–1113. [Google Scholar] [CrossRef]
- Hashimoto, M.; Bacman, S.R.; Peralta, S.; Falk, M.J.; Chomyn, A.; Chan, D.C.; Williams, S.L.; Moraes, C.T. MitoTALEN: A General Approach to Reduce Mutant mtDNA Loads and Restore Oxidative Phosphorylation Function in Mitochondrial Diseases. Mol. Ther. 2015, 23, 1592–1599. [Google Scholar] [CrossRef]
- Yin, Z.; Burger, N.; Kula-Alwar, D.; Aksentijevic, D.; Bridges, H.R.; Prag, H.A.; Grba, D.N.; Viscomi, C.; James, A.M.; Mottahedin, A.; et al. Structural basis for a complex I mutation that blocks pathological ROS production. Nat. Commun. 2021, 12, 707. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Pell, V.R.; Gaude, E.; Aksentijevic, D.; Sundier, S.Y.; Robb, E.L.; Logan, A.; Nadtochiy, S.M.; Ord, E.N.J.; Smith, A.C.; et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014, 515, 431–435. [Google Scholar] [CrossRef]
- Robb, E.L.; Hall, A.R.; Prime, T.A.; Eaton, S.; Szibor, M.; Viscomi, C.; James, A.M.; Murphy, M.P. Control of mitochondrial superoxide production by reverse electron transport at complex I. J. Biol. Chem. 2018, 293, 9869–9879. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; Bayir, H.; Belousov, V.; Chang, C.J.; Davies, K.J.A.; Davies, M.J.; Dick, T.P.; Finkel, T.; Forman, H.J.; Janssen-Heininger, Y.; et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 2022, 4, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Benit, P.; Beugnot, R.; Chretien, D.; Giurgea, I.; De Lonlay-Debeney, P.; Issartel, J.P.; Corral-Debrinski, M.; Kerscher, S.; Rustin, P.; Rotig, A.; et al. Mutant NDUFV2 subunit of mitochondrial complex I causes early onset hypertrophic cardiomyopathy and encephalopathy. Hum. Mutat. 2003, 21, 582–586. [Google Scholar] [CrossRef]
- Bezawork-Geleta, A.; Rohlena, J.; Dong, L.; Pacak, K.; Neuzil, J. Mitochondrial Complex II: At the Crossroads. Trends Biochem. Sci. 2017, 42, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Rutter, J.; Winge, D.R.; Schiffman, J.D. Succinate dehydrogenase—Assembly, regulation and role in human disease. Mitochondrion 2010, 10, 393–401. [Google Scholar] [CrossRef]
- Chandel, N.S. Mitochondrial complex III: An essential component of universal oxygen sensing machinery? Respir. Physiol. Neurobiol. 2010, 174, 175–181. [Google Scholar] [CrossRef]
- Fernandez-Vizarra, E.; Zeviani, M. Mitochondrial complex III Rieske Fe-S protein processing and assembly. Cell Cycle 2018, 17, 681–687. [Google Scholar] [CrossRef]
- Kramer, D.M.; Roberts, A.G.; Muller, F.; Cape, J.; Bowman, M.K. Q-cycle bypass reactions at the Qo site of the cytochrome bc1 (and related) complexes. Methods Enzymol. 2004, 382, 21–45. [Google Scholar] [CrossRef]
- Yu, C.A.; Cen, X.; Ma, H.W.; Yin, Y.; Yu, L.; Esser, L.; Xia, D. Domain conformational switch of the iron-sulfur protein in cytochrome bc1 complex is induced by the electron transfer from cytochrome bL to bH. Biochim. Biophys. Acta 2008, 1777, 1038–1043. [Google Scholar] [CrossRef]
- Gutowski, M.; Kowalczyk, S. A study of free radical chemistry: Their role and pathophysiological significance. Acta Biochim. Pol. 2013, 60, 1–16. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef] [PubMed]
- Takac, I.; Schroder, K.; Brandes, R.P. The Nox family of NADPH oxidases: Friend or foe of the vascular system? Curr. Hypertens. Rep. 2012, 14, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Kadenbach, B. Regulation of cytochrome c oxidase contributes to health and optimal life. World J. Biol. Chem. 2020, 11, 52–61. [Google Scholar] [CrossRef]
- Li, X.Y.; Yang, Y.L. Mitochondrial disorders associated with mitochondrial respiratory chain complex V deficiency. Zhongguo Dang Dai Er Ke Za Zhi 2013, 15, 596–600. [Google Scholar]
- Jonckheere, A.I.; Smeitink, J.A.; Rodenburg, R.J. Mitochondrial ATP synthase: Architecture, function and pathology. J. Inherit. Metab. Dis. 2012, 35, 211–225. [Google Scholar] [CrossRef]
- Junge, W.; Nelson, N. ATP synthase. Annu. Rev. Biochem. 2015, 84, 631–657. [Google Scholar] [CrossRef]
- Nirody, J.A.; Budin, I.; Rangamani, P. ATP synthase: Evolution, energetics, and membrane interactions. J. Gen. Physiol. 2020, 152, e201912475. [Google Scholar] [CrossRef]
- Aksimentiev, A.; Balabin, I.A.; Fillingame, R.H.; Schulten, K. Insights into the molecular mechanism of rotation in the Fo sector of ATP synthase. Biophys. J. 2004, 86, 1332–1344. [Google Scholar] [CrossRef]
- Neupane, P.; Bhuju, S.; Thapa, N.; Bhattarai, H.K. ATP Synthase: Structure, Function and Inhibition. Biomol. Concepts 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Senior, A.E. Two ATPases. J. Biol. Chem. 2012, 287, 30049–30062. [Google Scholar] [CrossRef]
- Blum, T.B.; Hahn, A.; Meier, T.; Davies, K.M.; Kuhlbrandt, W. Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc. Natl. Acad. Sci. USA 2019, 116, 4250–4255. [Google Scholar] [CrossRef] [PubMed]
- Colina-Tenorio, L.; Dautant, A.; Miranda-Astudillo, H.; Giraud, M.F.; Gonzalez-Halphen, D. The Peripheral Stalk of Rotary ATPases. Front. Physiol. 2018, 9, 1243. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Y.P.J. Enzymatic regeneration and conservation of ATP: Challenges and opportunities. Crit. Rev. Biotechnol. 2021, 41, 16–33. [Google Scholar] [CrossRef]
- Luft, F.C. ATP-ADP translocase and effects on ischemic heart. J. Mol. Med. 2016, 94, 609–611. [Google Scholar] [CrossRef]
- Dupont, P.Y.; Stepien, G. Computational analysis of the transcriptional regulation of the adenine nucleotide translocator isoform 4 gene and its role in spermatozoid glycolytic metabolism. Gene 2011, 487, 38–45. [Google Scholar] [CrossRef]
- Ogunbona, O.B.; Baile, M.G.; Claypool, S.M. Cardiomyopathy-associated mutation in the ADP/ATP carrier reveals translation-dependent regulation of cytochrome c oxidase activity. Mol. Biol. Cell 2018, 29, 1449–1464. [Google Scholar] [CrossRef]
- Pellegrino-Coppola, D. Regulation of the mitochondrial permeability transition pore and its effects on aging. Microb. Cell 2020, 7, 222–233. [Google Scholar] [CrossRef]
- Sharer, J.D. The adenine nucleotide translocase type 1 (ANT1): A new factor in mitochondrial disease. IUBMB Life 2005, 57, 607–614. [Google Scholar] [CrossRef]
- Rosca, M.G.; Okere, I.A.; Sharma, N.; Stanley, W.C.; Recchia, F.A.; Hoppel, C.L. Altered expression of the adenine nucleotide translocase isoforms and decreased ATP synthase activity in skeletal muscle mitochondria in heart failure. J. Mol. Cell. Cardiol. 2009, 46, 927–935. [Google Scholar] [CrossRef]
- Narula, N.; Zaragoza, M.V.; Sengupta, P.P.; Li, P.; Haider, N.; Verjans, J.; Waymire, K.; Vannan, M.; Wallace, D.C. Adenine nucleotide translocase 1 deficiency results in dilated cardiomyopathy with defects in myocardial mechanics, histopathological alterations, and activation of apoptosis. JACC Cardiovasc. Imaging 2011, 4, 1–10. [Google Scholar] [CrossRef]
- Palmieri, L.; Alberio, S.; Pisano, I.; Lodi, T.; Meznaric-Petrusa, M.; Zidar, J.; Santoro, A.; Scarcia, P.; Fontanesi, F.; Lamantea, E.; et al. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum. Mol. Genet. 2005, 14, 3079–3088. [Google Scholar] [CrossRef] [PubMed]
- Dorner, A.; Lynetskiy, O.; Euler, G.; Landmesser, U.; Schluter, K.D.; Heger, J. Mitochondria Isolated from Hearts Subjected to Ischemia/Reperfusion Benefit from Adenine Nucleotide Translocase 1 Overexpression. Membranes 2021, 11, 836. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhao, S.; Liu, J.; Liu, X.; Sha, X.; Huang, C.; Hu, L.; Sun, S.; Gao, Y.; Chen, H.; et al. Mitochondrial GSNOR Alleviates Cardiac Dysfunction via ANT1 Denitrosylation. Circ. Res. 2023, 133, 220–236. [Google Scholar] [CrossRef] [PubMed]
- Seifert, E.L.; Ligeti, E.; Mayr, J.A.; Sondheimer, N.; Hajnoczky, G. The mitochondrial phosphate carrier: Role in oxidative metabolism, calcium handling and mitochondrial disease. Biochem. Biophys. Res. Commun. 2015, 464, 369–375. [Google Scholar] [CrossRef]
- Bauer, T.M.; Murphy, E. Role of Mitochondrial Calcium and the Permeability Transition Pore in Regulating Cell Death. Circ. Res. 2020, 126, 280–293. [Google Scholar] [CrossRef]
- Seifert, E.L.; Gal, A.; Acoba, M.G.; Li, Q.; Anderson-Pullinger, L.; Golenar, T.; Moffat, C.; Sondheimer, N.; Claypool, S.M.; Hajnoczky, G. Natural and Induced Mitochondrial Phosphate Carrier Loss: DIFFERENTIAL DEPENDENCE OF MITOCHONDRIAL METABOLISM AND DYNAMICS AND CELL SURVIVAL ON THE EXTENT OF DEPLETION. J. Biol. Chem. 2016, 291, 26126–26137. [Google Scholar] [CrossRef]
- Bhoj, E.J.; Li, M.; Ahrens-Nicklas, R.; Pyle, L.C.; Wang, J.; Zhang, V.W.; Clarke, C.; Wong, L.J.; Sondheimer, N.; Ficicioglu, C.; et al. Pathologic Variants of the Mitochondrial Phosphate Carrier SLC25A3: Two New Patients and Expansion of the Cardiomyopathy/Skeletal Myopathy Phenotype With and Without Lactic Acidosis. JIMD Rep. 2015, 19, 59–66. [Google Scholar] [CrossRef]
- Kwong, J.Q.; Davis, J.; Baines, C.P.; Sargent, M.A.; Karch, J.; Wang, X.; Huang, T.; Molkentin, J.D. Genetic deletion of the mitochondrial phosphate carrier desensitizes the mitochondrial permeability transition pore and causes cardiomyopathy. Cell Death Differ. 2014, 21, 1209–1217. [Google Scholar] [CrossRef]
- Peoples, J.N.; Ghazal, N.; Duong, D.M.; Hardin, K.R.; Manning, J.R.; Seyfried, N.T.; Faundez, V.; Kwong, J.Q. Loss of the mitochondrial phosphate carrier SLC25A3 induces remodeling of the cardiac mitochondrial protein acylome. Am. J. Physiol. Cell Physiol. 2021, 321, C519–C534. [Google Scholar] [CrossRef]
- Li, S.; Zhang, J.; Fu, W.; Cao, J.; Li, Z.; Tian, X.; Yang, M.; Zhao, J.; Wang, C.; Liu, Y.; et al. Mitochondrial transplantation rescues Ca2+ homeostasis imbalance and myocardial hypertrophy in SLC25A3-related hypertrophic cardiomyopathy. Cell Rep. 2024, 43, 115065. [Google Scholar] [CrossRef]
- Fogo, G.M.; Raghunayakula, S.; Emaus, K.J.; Torres Torres, F.J.; Wider, J.M.; Sanderson, T.H. Mitochondrial membrane potential and oxidative stress interact to regulate Oma1-dependent processing of Opa1 and mitochondrial dynamics. FASEB J. 2024, 38, e70066. [Google Scholar] [CrossRef] [PubMed]
- Janer, A.; Prudent, J.; Paupe, V.; Fahiminiya, S.; Majewski, J.; Sgarioto, N.; Des Rosiers, C.; Forest, A.; Lin, Z.Y.; Gingras, A.C.; et al. SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol. Med. 2016, 8, 1019–1038. [Google Scholar] [CrossRef] [PubMed]
- Barbot, M.; Jans, D.C.; Schulz, C.; Denkert, N.; Kroppen, B.; Hoppert, M.; Jakobs, S.; Meinecke, M. Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 2015, 21, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Guarani, V.; Jardel, C.; Chretien, D.; Lombes, A.; Benit, P.; Labasse, C.; Lacene, E.; Bourillon, A.; Imbard, A.; Benoist, J.F.; et al. QIL1 mutation causes MICOS disassembly and early onset fatal mitochondrial encephalopathy with liver disease. Elife 2016, 5, e17163. [Google Scholar] [CrossRef]
- Weber, T.A.; Koob, S.; Heide, H.; Wittig, I.; Head, B.; van der Bliek, A.; Brandt, U.; Mittelbronn, M.; Reichert, A.S. APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS ONE 2013, 8, e63683. [Google Scholar] [CrossRef]
- Anand, R.; Reichert, A.S.; Kondadi, A.K. Emerging Roles of the MICOS Complex in Cristae Dynamics and Biogenesis. Biology 2021, 10, 600. [Google Scholar] [CrossRef]
- Kondadi, A.K.; Reichert, A.S. Mitochondrial Dynamics at Different Levels: From Cristae Dynamics to Interorganellar Cross Talk. Annu. Rev. Biophys. 2024, 53, 147–168. [Google Scholar] [CrossRef]
- Caron, C.; Bertolin, G. Cristae shaping and dynamics in mitochondrial function. J. Cell Sci. 2024, 137, jcs260986. [Google Scholar] [CrossRef]
- Cogliati, S.; Enriquez, J.A.; Scorrano, L. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 2016, 41, 261–273. [Google Scholar] [CrossRef]
- Dlaskova, A.; Spacek, T.; Engstova, H.; Spackova, J.; Schrofel, A.; Holendova, B.; Smolkova, K.; Plecita-Hlavata, L.; Jezek, P. Mitochondrial cristae narrowing upon higher 2-oxoglutarate load. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 659–678. [Google Scholar] [CrossRef]
- Feng, Y.; Madungwe, N.B.; Bopassa, J.C. Mitochondrial inner membrane protein, Mic60/mitofilin in mammalian organ protection. J. Cell. Physiol. 2019, 234, 3383–3393. [Google Scholar] [CrossRef] [PubMed]
- Matsuhashi, T.; Sato, T.; Kanno, S.I.; Suzuki, T.; Matsuo, A.; Oba, Y.; Kikusato, M.; Ogasawara, E.; Kudo, T.; Suzuki, K.; et al. Mitochonic Acid 5 (MA-5) Facilitates ATP Synthase Oligomerization and Cell Survival in Various Mitochondrial Diseases. EBioMedicine 2017, 20, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Sakowska, P.; Jans, D.C.; Mohanraj, K.; Riedel, D.; Jakobs, S.; Chacinska, A. The Oxidation Status of Mic19 Regulates MICOS Assembly. Mol. Cell. Biol. 2015, 35, 4222–4237. [Google Scholar] [CrossRef] [PubMed]
- Darshi, M.; Mendiola, V.L.; Mackey, M.R.; Murphy, A.N.; Koller, A.; Perkins, G.A.; Ellisman, M.H.; Taylor, S.S. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 2011, 286, 2918–2932. [Google Scholar] [CrossRef]
- Ott, C.; Ross, K.; Straub, S.; Thiede, B.; Gotz, M.; Goosmann, C.; Krischke, M.; Mueller, M.J.; Krohne, G.; Rudel, T.; et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol. 2012, 32, 1173–1188. [Google Scholar] [CrossRef]
- Utsumi, T.; Matsuzaki, K.; Kiwado, A.; Tanikawa, A.; Kikkawa, Y.; Hosokawa, T.; Otsuka, A.; Iuchi, Y.; Kobuchi, H.; Moriya, K. Identification and characterization of protein N-myristoylation occurring on four human mitochondrial proteins, SAMM50, TOMM40, MIC19, and MIC25. PLoS ONE 2018, 13, e0206355. [Google Scholar] [CrossRef]
- Godiker, J.; Gruneberg, M.; DuChesne, I.; Reunert, J.; Rust, S.; Westermann, C.; Wada, Y.; Classen, G.; Langhans, C.D.; Schlingmann, K.P.; et al. QIL1-dependent assembly of MICOS complex-lethal mutation in C19ORF70 resulting in liver disease and severe neurological retardation. J. Hum. Genet. 2018, 63, 707–716. [Google Scholar] [CrossRef]
- Kishita, Y.; Shimura, M.; Kohda, M.; Akita, M.; Imai-Okazaki, A.; Yatsuka, Y.; Nakajima, Y.; Ito, T.; Ohtake, A.; Murayama, K.; et al. A novel homozygous variant in MICOS13/QIL1 causes hepato-encephalopathy with mitochondrial DNA depletion syndrome. Mol. Genet. Genom. Med. 2020, 8, e1427. [Google Scholar] [CrossRef]
- Ikon, N.; Ryan, R.O. Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1156–1163. [Google Scholar] [CrossRef]
- Tang, X.; Huang, Z.; Wang, F.; Chen, J.; Qin, D.; Peng, D.; Yu, B. Macrophage-specific deletion of MIC26 (APOO) mitigates advanced atherosclerosis by increasing efferocytosis. Atherosclerosis 2023, 386, 117374. [Google Scholar] [CrossRef]
- Anand, R.; Kondadi, A.K.; Meisterknecht, J.; Golombek, M.; Nortmann, O.; Riedel, J.; Peifer-Weiss, L.; Brocke-Ahmadinejad, N.; Schlutermann, D.; Stork, B.; et al. MIC26 and MIC27 cooperate to regulate cardiolipin levels and the landscape of OXPHOS complexes. Life Sci. Alliance 2020, 3, e202000711. [Google Scholar] [CrossRef] [PubMed]
- von der Malsburg, K.; Muller, J.M.; Bohnert, M.; Oeljeklaus, S.; Kwiatkowska, P.; Becker, T.; Loniewska-Lwowska, A.; Wiese, S.; Rao, S.; Milenkovic, D.; et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 2011, 21, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Kondadi, A.K.; Anand, R.; Reichert, A.S. Cristae Membrane Dynamics—A Paradigm Change. Trends Cell Biol. 2020, 30, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Stephan, T.; Bruser, C.; Deckers, M.; Steyer, A.M.; Balzarotti, F.; Barbot, M.; Behr, T.S.; Heim, G.; Hubner, W.; Ilgen, P.; et al. MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 2020, 39, e104105. [Google Scholar] [CrossRef]
- Klecker, T.; Westermann, B. Pathways shaping the mitochondrial inner membrane. Open Biol. 2021, 11, 210238. [Google Scholar] [CrossRef]
- Wiedemann, N.; Kozjak, V.; Chacinska, A.; Schonfisch, B.; Rospert, S.; Ryan, M.T.; Pfanner, N.; Meisinger, C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 2003, 424, 565–571. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, K.; Dong, J.; Yan, C.; Hu, C.; Ji, H.; Chen, L.; Chen, S.; Zhao, H.; Song, Z. Sam50-Mic19-Mic60 axis determines mitochondrial cristae architecture by mediating mitochondrial outer and inner membrane contact. Cell Death Differ. 2020, 27, 146–160. [Google Scholar] [CrossRef]
- Xie, J.; Marusich, M.F.; Souda, P.; Whitelegge, J.; Capaldi, R.A. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007, 581, 3545–3549. [Google Scholar] [CrossRef]
- Ding, C.; Wu, Z.; Huang, L.; Wang, Y.; Xue, J.; Chen, S.; Deng, Z.; Wang, L.; Song, Z.; Chen, S. Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci. Rep. 2015, 5, 16064. [Google Scholar] [CrossRef]
- John, G.B.; Shang, Y.; Li, L.; Renken, C.; Mannella, C.A.; Selker, J.M.; Rangell, L.; Bennett, M.J.; Zha, J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 2005, 16, 1543–1554. [Google Scholar] [CrossRef]
- Genin, E.C.; Plutino, M.; Bannwarth, S.; Villa, E.; Cisneros-Barroso, E.; Roy, M.; Ortega-Vila, B.; Fragaki, K.; Lespinasse, F.; Pinero-Martos, E.; et al. CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol. Med. 2016, 8, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Koob, S.; Barrera, M.; Anand, R.; Reichert, A.S. The non-glycosylated isoform of MIC26 is a constituent of the mammalian MICOS complex and promotes formation of crista junctions. Biochim. Biophys. Acta 2015, 1853, 1551–1563. [Google Scholar] [CrossRef]
- Kozjak-Pavlovic, V.; Prell, F.; Thiede, B.; Gotz, M.; Wosiek, D.; Ott, C.; Rudel, T. C1orf163/RESA1 is a novel mitochondrial intermembrane space protein connected to respiratory chain assembly. J. Mol. Biol. 2014, 426, 908–920. [Google Scholar] [CrossRef] [PubMed]
- Ott, C.; Dorsch, E.; Fraunholz, M.; Straub, S.; Kozjak-Pavlovic, V. Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLoS ONE 2015, 10, e0120213. [Google Scholar] [CrossRef]
- Li, H.; Ruan, Y.; Zhang, K.; Jian, F.; Hu, C.; Miao, L.; Gong, L.; Sun, L.; Zhang, X.; Chen, S.; et al. Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ. 2016, 23, 380–392. [Google Scholar] [CrossRef]
- Yang, R.F.; Sun, L.H.; Zhang, R.; Zhang, Y.; Luo, Y.X.; Zheng, W.; Zhang, Z.Q.; Chen, H.Z.; Liu, D.P. Suppression of Mic60 compromises mitochondrial transcription and oxidative phosphorylation. Sci. Rep. 2015, 5, 7990. [Google Scholar] [CrossRef]
- Paumard, P.; Vaillier, J.; Coulary, B.; Schaeffer, J.; Soubannier, V.; Mueller, D.M.; Brethes, D.; di Rago, J.P.; Velours, J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002, 21, 221–230. [Google Scholar] [CrossRef]
- Quintana-Cabrera, R.; Mehrotra, A.; Rigoni, G.; Soriano, M.E. Who and how in the regulation of mitochondrial cristae shape and function. Biochem. Biophys. Res. Commun. 2018, 500, 94–101. [Google Scholar] [CrossRef]
- Gu, J.; Wu, M.; Guo, R.; Yan, K.; Lei, J.; Gao, N.; Yang, M. The architecture of the mammalian respirasome. Nature 2016, 537, 639–643. [Google Scholar] [CrossRef]
- Guo, R.; Gu, J.; Wu, M.; Yang, M. Amazing structure of respirasome: Unveiling the secrets of cell respiration. Protein Cell 2016, 7, 854–865. [Google Scholar] [CrossRef]
- Khacho, M.; Tarabay, M.; Patten, D.; Khacho, P.; MacLaurin, J.G.; Guadagno, J.; Bergeron, R.; Cregan, S.P.; Harper, M.E.; Park, D.S.; et al. Acidosis overrides oxygen deprivation to maintain mitochondrial function and cell survival. Nat. Commun. 2014, 5, 3550. [Google Scholar] [CrossRef] [PubMed]
- Colina-Tenorio, L.; Horten, P.; Pfanner, N.; Rampelt, H. Shaping the mitochondrial inner membrane in health and disease. J. Intern. Med. 2020, 287, 645–664. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Deng, K.; Wu, M. Mitochondrial cristae in health and disease. Int. J. Biol. Macromol. 2023, 235, 123755. [Google Scholar] [CrossRef]
- Panicker, N.; Ge, P.; Dawson, V.L.; Dawson, T.M. The cell biology of Parkinson’s disease. J. Cell Biol. 2021, 220, e202012095. [Google Scholar] [CrossRef]
- Pickrell, A.M.; Youle, R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef]
- Akabane, S.; Uno, M.; Tani, N.; Shimazaki, S.; Ebara, N.; Kato, H.; Kosako, H.; Oka, T. PKA Regulates PINK1 Stability and Parkin Recruitment to Damaged Mitochondria through Phosphorylation of MIC60. Mol. Cell 2016, 62, 371–384. [Google Scholar] [CrossRef]
- Tsai, P.I.; Lin, C.H.; Hsieh, C.H.; Papakyrikos, A.M.; Kim, M.J.; Napolioni, V.; Schoor, C.; Couthouis, J.; Wu, R.M.; Wszolek, Z.K.; et al. PINK1 Phosphorylates MIC60/Mitofilin to Control Structural Plasticity of Mitochondrial Crista Junctions. Mol. Cell 2018, 69, 744–756.e6. [Google Scholar] [CrossRef]
- Lake, N.J.; Compton, A.G.; Rahman, S.; Thorburn, D.R. Leigh syndrome: One disorder, more than 75 monogenic causes. Ann. Neurol. 2016, 79, 190–203. [Google Scholar] [CrossRef]
- Schubert Baldo, M.; Vilarinho, L. Molecular basis of Leigh syndrome: A current look. Orphanet J. Rare Dis. 2020, 15, 31. [Google Scholar] [CrossRef]
- Barca, E.; Ganetzky, R.D.; Potluri, P.; Juanola-Falgarona, M.; Gai, X.; Li, D.; Jalas, C.; Hirsch, Y.; Emmanuele, V.; Tadesse, S.; et al. USMG5 Ashkenazi Jewish founder mutation impairs mitochondrial complex V dimerization and ATP synthesis. Hum. Mol. Genet. 2018, 27, 3305–3312. [Google Scholar] [CrossRef]
- Ohsakaya, S.; Fujikawa, M.; Hisabori, T.; Yoshida, M. Knockdown of DAPIT (diabetes-associated protein in insulin-sensitive tissue) results in loss of ATP synthase in mitochondria. J. Biol. Chem. 2011, 286, 20292–20296. [Google Scholar] [CrossRef] [PubMed]
- Del Dotto, V.; Carelli, V. Dominant Optic Atrophy (DOA): Modeling the Kaleidoscopic Roles of OPA1 in Mitochondrial Homeostasis. Front. Neurol. 2021, 12, 681326. [Google Scholar] [CrossRef] [PubMed]
- Frezza, C.; Cipolat, S.; Martins de Brito, O.; Micaroni, M.; Beznoussenko, G.V.; Rudka, T.; Bartoli, D.; Polishuck, R.S.; Danial, N.N.; De Strooper, B.; et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 2006, 126, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, R.; Lartigue, L.; Perkins, G.; Scott, R.T.; Dixit, A.; Kushnareva, Y.; Kuwana, T.; Ellisman, M.H.; Newmeyer, D.D. Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization. Mol. Cell 2008, 31, 557–569. [Google Scholar] [CrossRef]
- Miller, M.A.; Zachary, J.F. Mechanisms and Morphology of Cellular Injury, Adaptation, and Death. In Pathologic Basis of Veterinary Disease; Elsevier Inc.: St. Louis, MO, USA, 2017; pp. 2–43.e19. [Google Scholar] [CrossRef]
- Buck, M.D.; O’Sullivan, D.; Klein Geltink, R.I.; Curtis, J.D.; Chang, C.H.; Sanin, D.E.; Qiu, J.; Kretz, O.; Braas, D.; van der Windt, G.J.; et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 2016, 166, 63–76. [Google Scholar] [CrossRef]
- Russell, B.E.; Whaley, K.G.; Bove, K.E.; Labilloy, A.; Lombardo, R.C.; Hopkin, R.J.; Leslie, N.D.; Prada, C.; Assouline, Z.; Barcia, G.; et al. Expanding and Underscoring the Hepato-Encephalopathic Phenotype of QIL1/MIC13. Hepatology 2019, 70, 1066–1070. [Google Scholar] [CrossRef]
- Zeharia, A.; Friedman, J.R.; Tobar, A.; Saada, A.; Konen, O.; Fellig, Y.; Shaag, A.; Nunnari, J.; Elpeleg, O. Mitochondrial hepato-encephalopathy due to deficiency of QIL1/MIC13 (C19orf70), a MICOS complex subunit. Eur. J. Hum. Genet. 2016, 24, 1778–1782. [Google Scholar] [CrossRef]
- Shimura, M.; Kuranobu, N.; Ogawa-Tominaga, M.; Akiyama, N.; Sugiyama, Y.; Ebihara, T.; Fushimi, T.; Ichimoto, K.; Matsunaga, A.; Tsuruoka, T.; et al. Clinical and molecular basis of hepatocerebral mitochondrial DNA depletion syndrome in Japan: Evaluation of outcomes after liver transplantation. Orphanet J. Rare Dis. 2020, 15, 169. [Google Scholar] [CrossRef]
- Peifer-Weiss, L.; Kurban, M.; David, C.; Lubeck, M.; Kondadi, A.K.; Nemer, G.; Reichert, A.S.; Anand, R. A X-linked nonsense APOO/MIC26 variant causes a lethal mitochondrial disease with progeria-like phenotypes. Clin. Genet. 2023, 104, 659–668. [Google Scholar] [CrossRef]
- Beninca, C.; Zanette, V.; Brischigliaro, M.; Johnson, M.; Reyes, A.; Valle, D.A.D.; Robinson, A.J.; Degiorgi, A.; Yeates, A.; Telles, B.A.; et al. Mutation in the MICOS subunit gene APOO (MIC26) associated with an X-linked recessive mitochondrial myopathy, lactic acidosis, cognitive impairment and autistic features. J. Med. Genet. 2021, 58, 155–167. [Google Scholar] [CrossRef]
- Ikeda, A.; Imai, Y.; Hattori, N. Neurodegeneration-associated mitochondrial proteins, CHCHD2 and CHCHD10-what distinguishes the two? Front. Cell Dev. Biol. 2022, 10, 996061. [Google Scholar] [CrossRef] [PubMed]
- Shammas, M.K.; Huang, T.H.; Narendra, D.P. CHCHD2 and CHCHD10-related neurodegeneration: Molecular pathogenesis and the path to precision therapy. Biochem. Soc. Trans. 2023, 51, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Funayama, M.; Ohe, K.; Amo, T.; Furuya, N.; Yamaguchi, J.; Saiki, S.; Li, Y.; Ogaki, K.; Ando, M.; Yoshino, H.; et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson’s disease: A genome-wide linkage and sequencing study. Lancet Neurol. 2015, 14, 274–282. [Google Scholar] [CrossRef]
- Li, Q.; Madden, J.A.; Lin, J.; Shi, J.; Rosen, S.M.; Schmitz-Abe, K.; Agrawal, P.B. Reanalysis of Exome Data Identifies Novel SLC25A46 Variants Associated with Leigh Syndrome. J. Pers. Med. 2021, 11, 1277. [Google Scholar] [CrossRef]
- Wan, J.; Steffen, J.; Yourshaw, M.; Mamsa, H.; Andersen, E.; Rudnik-Schoneborn, S.; Pope, K.; Howell, K.B.; McLean, C.A.; Kornberg, A.J.; et al. Loss of function of SLC25A46 causes lethal congenital pontocerebellar hypoplasia. Brain 2016, 139, 2877–2890. [Google Scholar] [CrossRef]
- Desai, R.; Frazier, A.E.; Durigon, R.; Patel, H.; Jones, A.W.; Dalla Rosa, I.; Lake, N.J.; Compton, A.G.; Mountford, H.S.; Tucker, E.J.; et al. ATAD3 gene cluster deletions cause cerebellar dysfunction associated with altered mitochondrial DNA and cholesterol metabolism. Brain 2017, 140, 1595–1610. [Google Scholar] [CrossRef]
- Peralta, S.; Gonzalez-Quintana, A.; Ybarra, M.; Delmiro, A.; Perez-Perez, R.; Docampo, J.; Arenas, J.; Blazquez, A.; Ugalde, C.; Martin, M.A. Novel ATAD3A recessive mutation associated to fatal cerebellar hypoplasia with multiorgan involvement and mitochondrial structural abnormalities. Mol. Genet. Metab. 2019, 128, 452–462. [Google Scholar] [CrossRef]
- Ishihara, T.; Ban-Ishihara, R.; Ota, A.; Ishihara, N. Mitochondrial nucleoid trafficking regulated by the inner-membrane AAA-ATPase ATAD3A modulates respiratory complex formation. Proc. Natl. Acad. Sci. USA 2022, 119, e2210730119. [Google Scholar] [CrossRef]
- Qin, J.; Guo, Y.; Xue, B.; Shi, P.; Chen, Y.; Su, Q.P.; Hao, H.; Zhao, S.; Wu, C.; Yu, L.; et al. ER-mitochondria contacts promote mtDNA nucleoids active transportation via mitochondrial dynamic tubulation. Nat. Commun. 2020, 11, 4471. [Google Scholar] [CrossRef]
- Lepelley, A.; Della Mina, E.; Van Nieuwenhove, E.; Waumans, L.; Fraitag, S.; Rice, G.I.; Dhir, A.; Fremond, M.L.; Rodero, M.P.; Seabra, L.; et al. Enhanced cGAS-STING-dependent interferon signaling associated with mutations in ATAD3A. J. Exp. Med. 2021, 218, e20201560. [Google Scholar] [CrossRef]
- Hirono, K.; Ichida, F.; Nishio, N.; Ogawa-Tominaga, M.; Fushimi, T.; Feichtinger, R.G.; Mayr, J.A.; Kohda, M.; Kishita, Y.; Okazaki, Y.; et al. Mitochondrial complex deficiency by novel compound heterozygous TMEM70 variants and correlation with developmental delay, undescended testicle, and left ventricular noncompaction in a Japanese patient: A case report. Clin. Case Rep. 2019, 7, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Kovalcikova, J.; Vrbacky, M.; Pecina, P.; Tauchmannova, K.; Nuskova, H.; Kaplanova, V.; Brazdova, A.; Alan, L.; Elias, J.; Cunatova, K.; et al. TMEM70 facilitates biogenesis of mammalian ATP synthase by promoting subunit c incorporation into the rotor structure of the enzyme. FASEB J. 2019, 33, 14103–14117. [Google Scholar] [CrossRef] [PubMed]
- Wortmann, S.B.; Zietkiewicz, S.; Kousi, M.; Szklarczyk, R.; Haack, T.B.; Gersting, S.W.; Muntau, A.C.; Rakovic, A.; Renkema, G.H.; Rodenburg, R.J.; et al. CLPB mutations cause 3-methylglutaconic aciduria, progressive brain atrophy, intellectual disability, congenital neutropenia, cataracts, movement disorder. Am. J. Hum. Genet. 2015, 96, 245–257. [Google Scholar] [CrossRef]
- Bonjour, K.; Palazzi, C.; Silva, T.P.; Malta, K.K.; Neves, V.H.; Oliveira-Barros, E.G.; Neves, I.; Kersten, V.A.; Fortuna, B.T.; Samarasinghe, A.E.; et al. Mitochondrial Population in Mouse Eosinophils: Ultrastructural Dynamics in Cell Differentiation and Inflammatory Diseases. Front. Cell Dev. Biol. 2022, 10, 836755. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, K.; Budin, I. Cardiolipin remodeling maintains the inner mitochondrial membrane in cells with saturated lipidomes. J. Lipid Res. 2024, 65, 100601. [Google Scholar] [CrossRef]
- Paradies, G.; Paradies, V.; Ruggiero, F.M.; Petrosillo, G. Role of Cardiolipin in Mitochondrial Function and Dynamics in Health and Disease: Molecular and Pharmacological Aspects. Cells 2019, 8, 728. [Google Scholar] [CrossRef]
- Ades, L.C.; Gedeon, A.K.; Wilson, M.J.; Latham, M.; Partington, M.W.; Mulley, J.C.; Nelson, J.; Lui, K.; Sillence, D.O. Barth syndrome: Clinical features and confirmation of gene localisation to distal Xq28. Am. J. Med. Genet. 1993, 45, 327–334. [Google Scholar] [CrossRef]
- Barth, P.G.; Scholte, H.R.; Berden, J.A.; Van der Klei-Van Moorsel, J.M.; Luyt-Houwen, I.E.; Van’t Veer-Korthof, E.T.; Van der Harten, J.J.; Sobotka-Plojhar, M.A. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci. 1983, 62, 327–355. [Google Scholar] [CrossRef]
- Byeon, S.K.; Ramarajan, M.G.; Madugundu, A.K.; Oglesbee, D.; Vernon, H.J.; Pandey, A. High-resolution mass spectrometric analysis of cardiolipin profiles in Barth syndrome. Mitochondrion 2021, 60, 27–32. [Google Scholar] [CrossRef]
- Clarke, S.L.; Bowron, A.; Gonzalez, I.L.; Groves, S.J.; Newbury-Ecob, R.; Clayton, N.; Martin, R.P.; Tsai-Goodman, B.; Garratt, V.; Ashworth, M.; et al. Barth syndrome. Orphanet J. Rare Dis. 2013, 8, 23. [Google Scholar] [CrossRef]
- Liang, Z.; Ralph-Epps, T.; Schmidtke, M.W.; Kumar, V.; Greenberg, M.L. Decreased pyruvate dehydrogenase activity in Tafazzin-deficient cells is caused by dysregulation of pyruvate dehydrogenase phosphatase 1 (PDP1). J. Biol. Chem. 2024, 300, 105697. [Google Scholar] [CrossRef] [PubMed]
- Vo, L.; Schmidtke, M.W.; Da Rosa-Junior, N.T.; Ren, M.; Schlame, M.; Greenberg, M.L. Cardiolipin metabolism regulates expression of muscle transcription factor MyoD1 and muscle development. J. Biol. Chem. 2023, 299, 102978. [Google Scholar] [CrossRef] [PubMed]
- Tapscott, S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005, 132, 2685–2695. [Google Scholar] [CrossRef]
- Xu, Y.; Kelley, R.I.; Blanck, T.J.; Schlame, M. Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 2003, 278, 51380–51385. [Google Scholar] [CrossRef] [PubMed]
- Maguire, J.J.; Tyurina, Y.Y.; Mohammadyani, D.; Kapralov, A.A.; Anthonymuthu, T.S.; Qu, F.; Amoscato, A.A.; Sparvero, L.J.; Tyurin, V.A.; Planas-Iglesias, J.; et al. Known unknowns of cardiolipin signaling: The best is yet to come. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 8–24. [Google Scholar] [CrossRef]
- Spiekerkoetter, U. Mitochondrial fatty acid oxidation disorders: Clinical presentation of long-chain fatty acid oxidation defects before and after newborn screening. J. Inherit. Metab. Dis. 2010, 33, 527–532. [Google Scholar] [CrossRef]
- Orrenius, S.; Zhivotovsky, B. Cardiolipin oxidation sets cytochrome c free. Nat. Chem. Biol. 2005, 1, 188–189. [Google Scholar] [CrossRef]
- Morita, S.Y.; Terada, T. Enzymatic measurement of phosphatidylglycerol and cardiolipin in cultured cells and mitochondria. Sci. Rep. 2015, 5, 11737. [Google Scholar] [CrossRef]
- Andrews, W.T.; Donahue, D.; Holmes, A.; Balsara, R.; Castellino, F.J.; Hummon, A.B. In situ metabolite and lipid analysis of GluN2D-/- and wild-type mice after ischemic stroke using MALDI MSI. Anal. Bioanal. Chem. 2020, 412, 6275–6285. [Google Scholar] [CrossRef]
- Mayr, J.A.; Haack, T.B.; Graf, E.; Zimmermann, F.A.; Wieland, T.; Haberberger, B.; Superti-Furga, A.; Kirschner, J.; Steinmann, B.; Baumgartner, M.R.; et al. Lack of the mitochondrial protein acylglycerol kinase causes Sengers syndrome. Am. J. Hum. Genet. 2012, 90, 314–320. [Google Scholar] [CrossRef]
- Richter-Dennerlein, R.; Korwitz, A.; Haag, M.; Tatsuta, T.; Dargazanli, S.; Baker, M.; Decker, T.; Lamkemeyer, T.; Rugarli, E.I.; Langer, T. DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metab. 2014, 20, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Corrado, M.; Edwards-Hicks, J.; Villa, M.; Flachsmann, L.J.; Sanin, D.E.; Jacobs, M.; Baixauli, F.; Stanczak, M.; Anderson, E.; Azuma, M.; et al. Dynamic Cardiolipin Synthesis Is Required for CD8+ T Cell Immunity. Cell Metab. 2020, 32, 981–995.e7. [Google Scholar] [CrossRef] [PubMed]
- Bailis, W.; Shyer, J.A.; Zhao, J.; Canaveras, J.C.G.; Al Khazal, F.J.; Qu, R.; Steach, H.R.; Bielecki, P.; Khan, O.; Jackson, R.; et al. Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature 2019, 571, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Lakhal-Littleton, S. Mechanisms of cardiac iron homeostasis and their importance to heart function. Free Radic. Biol. Med. 2019, 133, 234–237. [Google Scholar] [CrossRef]
- Gao, J.; Zhou, Q.; Wu, D.; Chen, L. Mitochondrial iron metabolism and its role in diseases. Clin. Chim. Acta 2021, 513, 6–12. [Google Scholar] [CrossRef]
- Lill, R.; Muhlenhoff, U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008, 77, 669–700. [Google Scholar] [CrossRef]
- Poulos, T.L. Heme enzyme structure and function. Chem. Rev. 2014, 114, 3919–3962. [Google Scholar] [CrossRef]
- Maio, N.; Rouault, T.A. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta 2015, 1853, 1493–1512. [Google Scholar] [CrossRef]
- Paterek, A.; Mackiewicz, U.; Maczewski, M. Iron and the heart: A paradigm shift from systemic to cardiomyocyte abnormalities. J. Cell. Physiol. 2019, 234, 21613–21629. [Google Scholar] [CrossRef]
- Zhang, H.; Jamieson, K.L.; Grenier, J.; Nikhanj, A.; Tang, J.; Nguyen, Q.; Wang, S.; Thompson, R.B.; Seubert, J.M.; Oudit, G.Y. Myocardial iron depletion exacerbates end-stage heart failure by promoting adverse remodeling and worsening mitochondrial function. Eur. Heart J. 2020, 41, ehaa946.1144. [Google Scholar] [CrossRef]
- Dong, F.; Zhang, X.; Culver, B.; Chew, H.G., Jr.; Kelley, R.O.; Ren, J. Dietary iron deficiency induces ventricular dilation, mitochondrial ultrastructural aberrations and cytochrome c release: Involvement of nitric oxide synthase and protein tyrosine nitration. Clin. Sci. 2005, 109, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, V.G.; Tongers, J.; Bode, J.; Berliner, D.; Widder, J.D.; Escher, F.; Mutsenko, V.; Chung, B.; Rostami, F.; Guba-Quint, A.; et al. Cardiac iron concentration in relation to systemic iron status and disease severity in non-ischaemic heart failure with reduced ejection fraction. Eur. J. Heart Fail. 2020, 22, 2038–2046. [Google Scholar] [CrossRef] [PubMed]
- Li, G.H.; Couto, G.; Chen, Y.; Sun, M.; Shi, Y.; Heng, Y.; Dawood, F.; Liu, Y.; Zong, Y.; Khaper, N.; et al. The Critical Role of Autophagy in Iron-Overload Cardiomyopathy: A Model of Diastolic Heart Failure Due to Oxidative Stress. Can. J. Cardiol. 2013, 29, S117–S118. [Google Scholar] [CrossRef]
- Tang, D.; Kroemer, G. Ferroptosis. Curr. Biol. 2020, 30, R1292–R1297. [Google Scholar] [CrossRef]
- Okonko, D.O.; Mandal, A.K.; Missouris, C.G.; Poole-Wilson, P.A. Disordered iron homeostasis in chronic heart failure: Prevalence, predictors, and relation to anemia, exercise capacity, and survival. J. Am. Coll. Cardiol. 2011, 58, 1241–1251. [Google Scholar] [CrossRef]
- Girelli, D.; Ugolini, S.; Busti, F.; Marchi, G.; Castagna, A. Modern iron replacement therapy: Clinical and pathophysiological insights. Int. J. Hematol. 2018, 107, 16–30. [Google Scholar] [CrossRef]
- Seguin, A.; Ward, D.M. Mitochondrial ABC Transporters and Iron Metabolism. J. Clin. Exp. Pathol. 2018, 8, 338. [Google Scholar] [CrossRef]
- Solbach, T.F.; Konig, J.; Fromm, M.F.; Zolk, O. ATP-binding cassette transporters in the heart. Trends Cardiovasc. Med. 2006, 16, 7–15. [Google Scholar] [CrossRef]
- Couture, L.; Nash, J.A.; Turgeon, J. The ATP-binding cassette transporters and their implication in drug disposition: A special look at the heart. Pharmacol. Rev. 2006, 58, 244–258. [Google Scholar] [CrossRef]
- Schaedler, T.A.; Faust, B.; Shintre, C.A.; Carpenter, E.P.; Srinivasan, V.; van Veen, H.W.; Balk, J. Structures and functions of mitochondrial ABC transporters. Biochem. Soc. Trans. 2015, 43, 943–951. [Google Scholar] [CrossRef]
- Liesa, M.; Qiu, W.; Shirihai, O.S. Mitochondrial ABC transporters function: The role of ABCB10 (ABC-me) as a novel player in cellular handling of reactive oxygen species. Biochim. Biophys. Acta 2012, 1823, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Moitra, K.; Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Hum. Mutat. 2022, 43, 1162–1182. [Google Scholar] [CrossRef]
- Krishnamurthy, P.C.; Du, G.; Fukuda, Y.; Sun, D.; Sampath, J.; Mercer, K.E.; Wang, J.; Sosa-Pineda, B.; Murti, K.G.; Schuetz, J.D. Identification of a mammalian mitochondrial porphyrin transporter. Nature 2006, 443, 586–589. [Google Scholar] [CrossRef]
- Song, G.; Zhang, S.; Tian, M.; Zhang, L.; Guo, R.; Zhuo, W.; Yang, M. Molecular insights into the human ABCB6 transporter. Cell Discov. 2021, 7, 55. [Google Scholar] [CrossRef]
- Murphy, A.J.; Sarrazy, V.; Wang, N.; Bijl, N.; Abramowicz, S.; Westerterp, M.; Welch, C.B.; Schuetz, J.D.; Yvan-Charvet, L. Deficiency of ATP-binding cassette transporter B6 in megakaryocyte progenitors accelerates atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 751–758. [Google Scholar] [CrossRef]
- Aivado, M.; Gattermann, N.; Rong, A.; Giagounidis, A.A.; Prall, W.C.; Czibere, A.; Hildebrandt, B.; Haas, R.; Bottomley, S.S. X-linked sideroblastic anemia associated with a novel ALAS2 mutation and unfortunate skewed X-chromosome inactivation patterns. Blood Cells Mol. Dis. 2006, 37, 40–45. [Google Scholar] [CrossRef]
- Kumar, V.; Aneesh, K.A.; Sanawar, R.; Jaleel, A.; Santhosh Kumar, T.R.; Kartha, C.C. Chronic Pressure Overload Results in Deficiency of Mitochondrial Membrane Transporter ABCB7 Which Contributes to Iron Overload, Mitochondrial Dysfunction, Metabolic Shift and Worsens Cardiac Function. Sci. Rep. 2019, 9, 13170. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, J.K.; Kim, H. ABCB7 simultaneously regulates apoptotic and non-apoptotic cell death by modulating mitochondrial ROS and HIF1alpha-driven NFkappaB signaling. Oncogene 2020, 39, 1969–1982. [Google Scholar] [CrossRef]
- Ardehali, H.; O’Rourke, B.; Marban, E. Cardioprotective role of the mitochondrial ATP-binding cassette protein 1. Circ. Res. 2005, 97, 740–742. [Google Scholar] [CrossRef]
- Paggio, A.; Checchetto, V.; Campo, A.; Menabo, R.; Di Marco, G.; Di Lisa, F.; Szabo, I.; Rizzuto, R.; De Stefani, D. Identification of an ATP-sensitive potassium channel in mitochondria. Nature 2019, 572, 609–613. [Google Scholar] [CrossRef]
- Ichikawa, Y.; Ghanefar, M.; Bayeva, M.; Wu, R.; Khechaduri, A.; Naga Prasad, S.V.; Mutharasan, R.K.; Naik, T.J.; Ardehali, H. Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J. Clin. Investig. 2014, 124, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Shirihai, O.S.; Gregory, T.; Yu, C.; Orkin, S.H.; Weiss, M.J. ABC-me: A novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 2000, 19, 2492–2502. [Google Scholar] [CrossRef] [PubMed]
- Hyde, B.B.; Liesa, M.; Elorza, A.A.; Qiu, W.; Haigh, S.E.; Richey, L.; Mikkola, H.K.; Schlaeger, T.M.; Shirihai, O.S. The mitochondrial transporter ABC-me (ABCB10), a downstream target of GATA-1, is essential for erythropoiesis in vivo. Cell Death Differ. 2012, 19, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Liesa, M.; Luptak, I.; Qin, F.; Hyde, B.B.; Sahin, E.; Siwik, D.A.; Zhu, Z.; Pimentel, D.R.; Xu, X.J.; Ruderman, N.B.; et al. Mitochondrial transporter ATP binding cassette mitochondrial erythroid is a novel gene required for cardiac recovery after ischemia/reperfusion. Circulation 2011, 124, 806–813. [Google Scholar] [CrossRef]
| Element | Mutated Gene | Mutation Type | Cardiomyopathy | References |
|---|---|---|---|---|
| Leucine tRNA, lysine tRNA, isoleucine tRNA, glycine tRNA | MT-TL1, MT-TK, MT-TI, and TRG | Point mutation | Hypertrophic cardiomyopathy, mitochondrial encephalopathy, lactic acidosis, stroke-like episodes, and myoclonus epilepsy with ragged-red fibers | [83] |
| Complex I, subunits 1–6 | ND 1–6 | mtDNA point mutation (m.3460G>A, m.11778G>A, and m.14484T>C) | Dilated cardiomyopathy and Leber hereditary optic neuropathy | [84] |
| Complex I, Cytochrome b | Cyt b | mtDNA point mutation (m.14757T>C) | Hypertrophic cardiomyopathy and macrocephaly–capillary malformation | [85] |
| Complex II subunits 1 and 2 | MC2DNI and MC2DNII | Point mutation, variations on the gene | Dilated cardiomyopathy, Leigh syndrome, and encephalopathy | [80,86] |
| Complex III, subunits 1 and 3 | COI and COIII | Point mutation | Dilated cardiomyopathy | [87] |
| Complex IV assembly factors | COX10, COX15, and TACO1 | Point mutation, variations on the gene | Cardiomyopathy, Leigh syndrome, and encephalopathy | [88,89,90] |
| Complex IV subunits | MTCO2, MTCO3, and NDUFA4 | Point mutation, variations on the gene | Hypertrophic cardiomyopathy and Leigh syndrome | [91] |
| Complex V subunits | MT-ATP6, MT-ATP8, ATP5I, ATP5MJ, ATP5IF1, and ATP6 | Point mutation, variations on the gene | Hypertrophic cardiomyopathy, Leigh syndrome, heart failure, and ischemic cardiomyopathy | [92,93,94] |
| Mitochondrial cardiolipin remodeling/maturation acyltransferase | TAZ (Tafazzin) | Point mutation, variations on the gene with missing base pairs | Barth syndrome | [92,93,94,95,96,97] |
| Heterotetrametric trifunctional mitochondrial protein (TFP) | HADHA (hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex α subunits) and HADHB (hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex β subunits) | Point mutation, variations on the gene with missing base pairs | Fatty acid β-oxidation diseases | [98] |
| Acylglycerol kinase | AGK | Variations on the gene with missing base pairs | Senger’s syndrome | [99] |
| Mitochondrial import inner membrane translocase subunit TIM14 | DNAJC19 | Point mutation variations on the gene with missing base pairs | Dilated cardiomyopathy with ataxia | [100] |
| ATP/ADP Antiporter (ANT1) | SLC25A4 | Variations on the gene with missing base pairs | Hypertrophic cardiomyopathy and dilated cardiomyopathy | [101,102] |
| Mitochondrial phosphate transporter (PiC) | SLC25A3 | Homozygous mutation c.158-9A>G | Progressive hypertrophic cardiomyopathy | [103] |
| Protein Name | Human Gene Name | Proposed MICOS Function | Other Names | References | |
|---|---|---|---|---|---|
| MIC60 subcomplex | Mic60 | IMMT | Core subunit required for normal cristae morphology | Mitofilin, Fcj1, Aim28, Fmp13 | [161,162,163,164,165] |
| Mic19 | CHCHD3 | Homologue of Mic25, responsible for MICOS–MIB stability through N-terminal myristoylation | MINOS3, AIM13 | [166,167] | |
| Mic25 | CHCHD6 | Mic19 homologue with a role in Mic60 stabilization and cristae morphology | CHCM1 | [168,169] | |
| Bridging | Mic13 | QIL1 | Bridges and stabilizes the Mic60 and Mic10 subcomplexes to form the mature MICOS | QIl1, C19orf70, MICOS13 | [170,171] |
| MIC10 subcomplex | Mic10 | MICOS10 | The core of the Mic10 subcomplex, oligomers with Mic60 upholding normal cristae morphology | Mio10, MINOS1, Mos1, Mcs10 | [161,162,172] |
| Mic26 | APOO | Paralogue of Mic27, stabilizing Mic10 subcomplex and cristae morphology | Mcs29, Mio27, Mos2 | [173] | |
| Mic27 | APOOL | Paralogue of Mic26, stabilizing Mic10 subcomplex and cristae morphology | Aim37, Mcs27, MOMA-1 | [174] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Carrada, M.; Vilchis-Landeros, M.M.; Vázquez-Meza, H.; Uribe-Ramírez, D.; Matuz-Mares, D. A New Perspective on the Role of Alterations in Mitochondrial Proteins Involved in ATP Synthesis and Mobilization in Cardiomyopathies. Int. J. Mol. Sci. 2025, 26, 2768. https://doi.org/10.3390/ijms26062768
Vázquez-Carrada M, Vilchis-Landeros MM, Vázquez-Meza H, Uribe-Ramírez D, Matuz-Mares D. A New Perspective on the Role of Alterations in Mitochondrial Proteins Involved in ATP Synthesis and Mobilization in Cardiomyopathies. International Journal of Molecular Sciences. 2025; 26(6):2768. https://doi.org/10.3390/ijms26062768
Chicago/Turabian StyleVázquez-Carrada, Melissa, María Magdalena Vilchis-Landeros, Héctor Vázquez-Meza, Daniel Uribe-Ramírez, and Deyamira Matuz-Mares. 2025. "A New Perspective on the Role of Alterations in Mitochondrial Proteins Involved in ATP Synthesis and Mobilization in Cardiomyopathies" International Journal of Molecular Sciences 26, no. 6: 2768. https://doi.org/10.3390/ijms26062768
APA StyleVázquez-Carrada, M., Vilchis-Landeros, M. M., Vázquez-Meza, H., Uribe-Ramírez, D., & Matuz-Mares, D. (2025). A New Perspective on the Role of Alterations in Mitochondrial Proteins Involved in ATP Synthesis and Mobilization in Cardiomyopathies. International Journal of Molecular Sciences, 26(6), 2768. https://doi.org/10.3390/ijms26062768








