Abstract
Recent evidence indicates that FAM111B is significantly involved in the progression of various cancers. Nonetheless, the potential pan-cancer implications of FAM111B have not been systematically investigated. In this study, FAM111B’s expression and oncogenic potential were studied using TCGA and GTEx data via GEPIA2, TIMER2.0, and STRING tools. Pathway enrichment analyses with the GO, KEGG, Reactome, and WikiPathways databases were conducted to explore its role in cancer development. The results were validated via multiplex immunofluorescence assays of pancreatic cancer tissues, microarray assays of ovarian cancer tissues, and protein transcriptomics of ovarian cancer cells. The expression levels of FAM111B were elevated in most cancer types and were associated with poor prognostic outcomes. Mechanistically, FAM111B expression was positively correlated with the expression of genes involved in DNA homologous recombination repair and with the infiltration of Th2 CD4+ T cells. These observations were further substantiated in ovarian cancer cell lines and tissue specimens from pancreatic and ovarian cancers. FAM111B functions as a biomarker for the DNA repair pathway and Th2 CD4+ T-cell infiltration in human malignancies.
1. Introduction
The FAM111 trypsin-like peptidase B (FAM111B) gene, located on chromosome 11q12.1, encodes a protein with a trypsin-like peptidase domain for protein hydrolysis [1,2]. It is expressed in multiple human tissues, such as the liver, lung, and pancreas. In 2013, a mutation in FAM111B was linked to hereditary fibrosing poikiloderma with tendon contractures, myopathy, and pulmonary fibrosis (POIKTMP) [1]. Recent evidence demonstrates that FAM111B, which also has been referred to as a cancer-associated nucleoprotein, plays a crucial role in promoting various cancers, such as breast, lung, and pancreatic cancer (PC) [3,4,5,6,7,8,9]. In our previous study, we demonstrated that FAM111B is significantly overexpressed in ovarian cancer (OV) compared to adjacent non-cancerous tissues, and that its expression is linked to an aggressive pathology and poor prognosis [10]. FAM111B has also been shown to promote proliferation, migration, and metastasis in breast, lung, ovarian, and prostate cancer cells, and to accelerate epithelial–mesenchymal transition in breast cancer [3,11,12,13].
Recent research suggests that FAM111B promotes tumorigenesis by modulating DNA damage repair and altering the immune microenvironment. FAM111B has been demonstrated to break down DNA–protein crosslinks, which can interfere with DNA processes [14]. Gene set enrichment analysis (GSEA) further suggested that FAM111B is involved in nucleotide and base excision repair mechanisms in esophageal cancer and PC [13,15]. Additionally, research on lung adenocarcinoma and thyroid cancer has demonstrated that FAM111B is associated with immune-cell infiltration and positively correlates with immune checkpoints, including PDL-1 and CTLA-4 [16,17]. Our previous study of 141 OV cases confirmed the positive correlation between FAM111B and PDL-1 expression [10]. However, the broad application of these findings across cancer types has not been evaluated.
In this study, we conducted a pan-cancer analysis using TCGA data to investigate FAM111B’s role in DNA damage repair and the immune microenvironment across cancers. Our results were validated through immunofluorescence experiments and tissue microarrays of 83 PC and 125 OV samples, as well as protein transcriptomics of OV cells. This study represents the first comprehensive analysis of FAM111B’s role in facilitating DNA damage repair and modulating tumor immunity within a pan-cancer context.
2. Results
2.1. Elevated Expression of FAM111B Is Correlated with Aggressive Clinicopathological Features and Poor Prognosis in Pan-Cancers
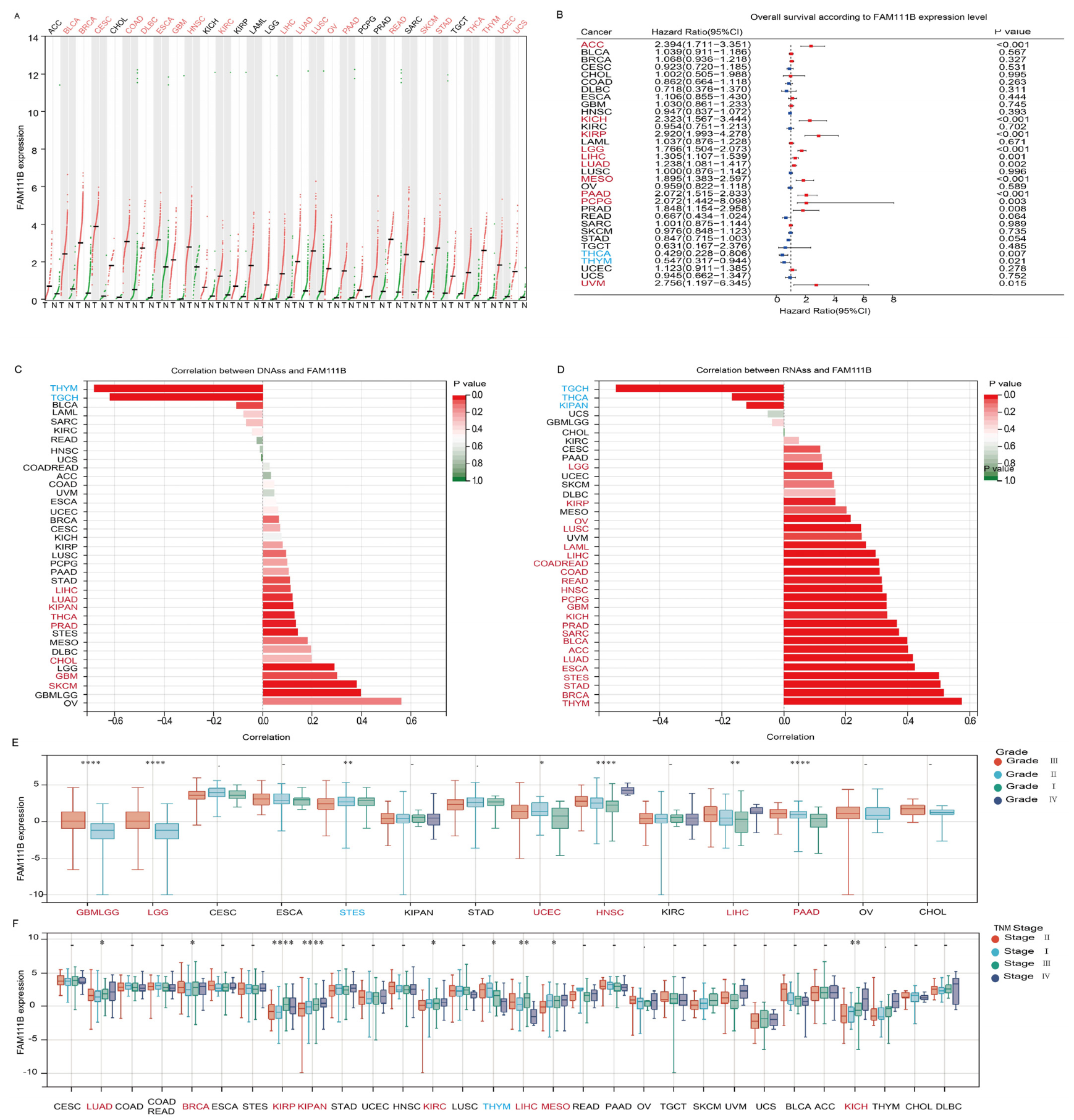
To evaluate pan-cancer FAM111B expression, we analyzed RNA-seq data from healthy controls and patients with 31 types of cancer from the TCGA and GTEx datasets. The results revealed that FAM111B mRNA expression is upregulated in 21 tumor types (Figure 1A). Prognostic analysis suggested that elevated FAM111B expression may function as an adverse prognostic biomarker in 10 cancer types, ACC, KICH, KIRP, LGG, LIHC, LUAD, MESO, PAAD, PCPG, and UVM (Figure 1B). Further analysis showed that FAM111B is significantly positively correlated with DNAss in 11 tumor types and RNAss in 26 tumor types, indicating an association with more aggressive tumor behavior (Figure 1C,D). Additionally, FAM111B expression correlated with higher pathological grades in six tumor types (GBMLGG, LGG, UCEC, HNSC, LIHC, PAAD) and higher TNM stages in eight tumor types (LUAD, BRCA, KIRP, KIRPN, KIRC, LIHC, MESO, KICH) (Figure 1E,F). These findings suggest that FAM111B is highly expressed in the majority of tumors, with correlations with aggressive features and poor prognosis in several cancer types.
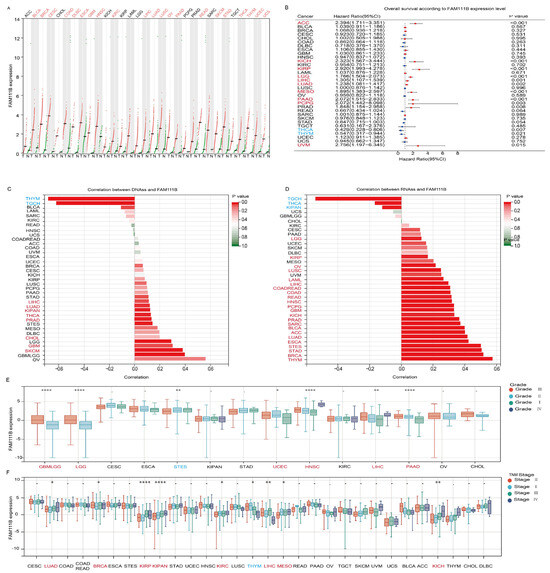
Figure 1.
Evaluation of the prognostic value and clinical significance of FAM111B. (A) Differential expression of FAM111B between tumor (T) and normal (N) tissue in 31 cancer types. MESO and UVM were excluded due to the absence of corresponding healthy control samples. (B) Survival analysis based on FAM111B expression. (C) Correlation between FAM111B expression and DNA expression-based stemness score (DNAss). (D) Correlation between FAM111B expression and RNA expression-based stemness score (RNAss). (E) Association between FAM111B expression and pathological grade. (F) Relationship between FAM111B expression and TNM stage. Data were obtained from TCGA and GTEx databases. Red font indicates cancers with elevated FAM111B expression and blue font indicates cancers with reduced FAM111B expression. Statistical significance was established at a threshold of p < 0.05, with the following designations: * for p < 0.05, ** for p < 0.01, and **** for p < 0.0001.
2.2. FAM111B Is Integral to the DNA Replication Process Across Various Cancer Types
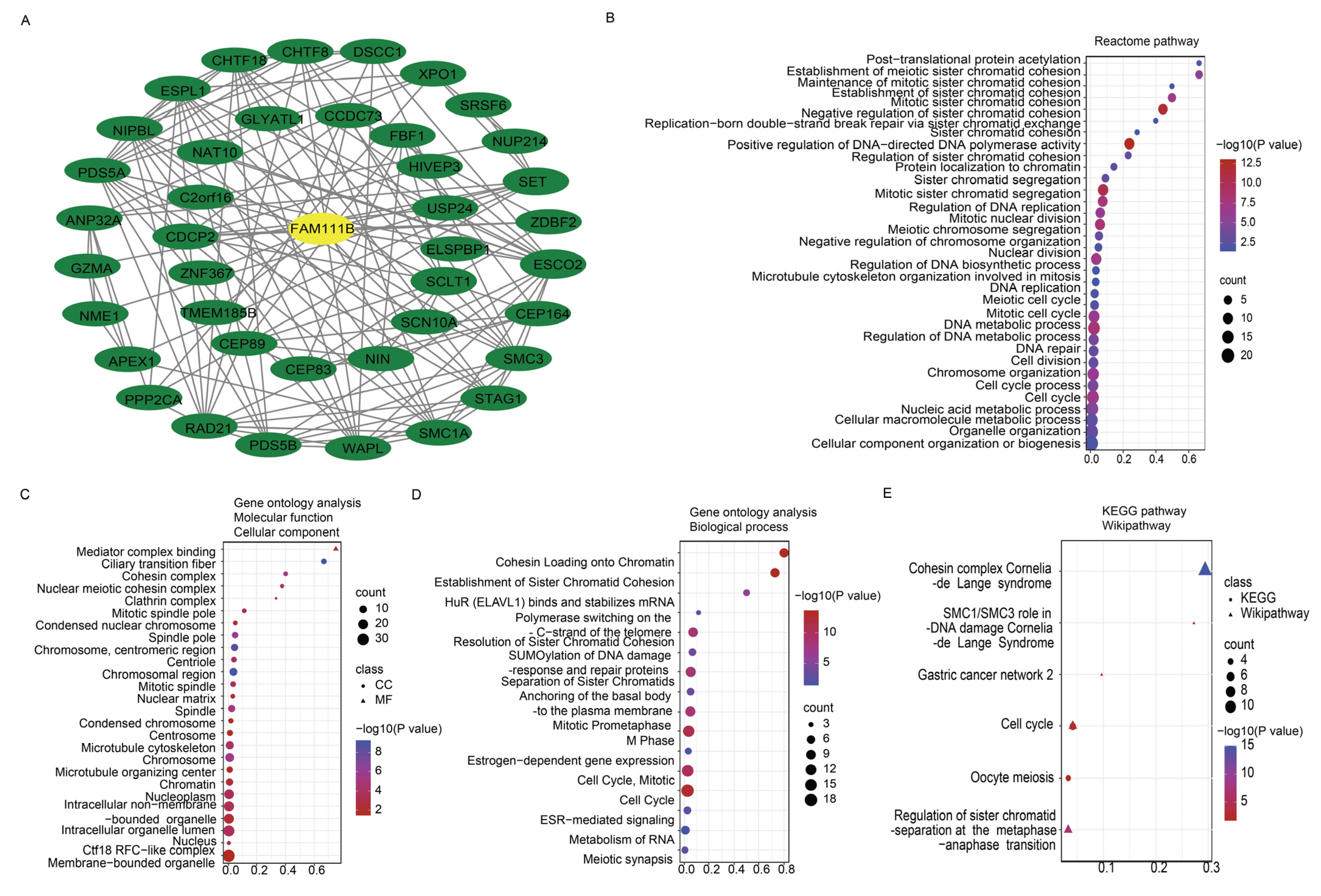
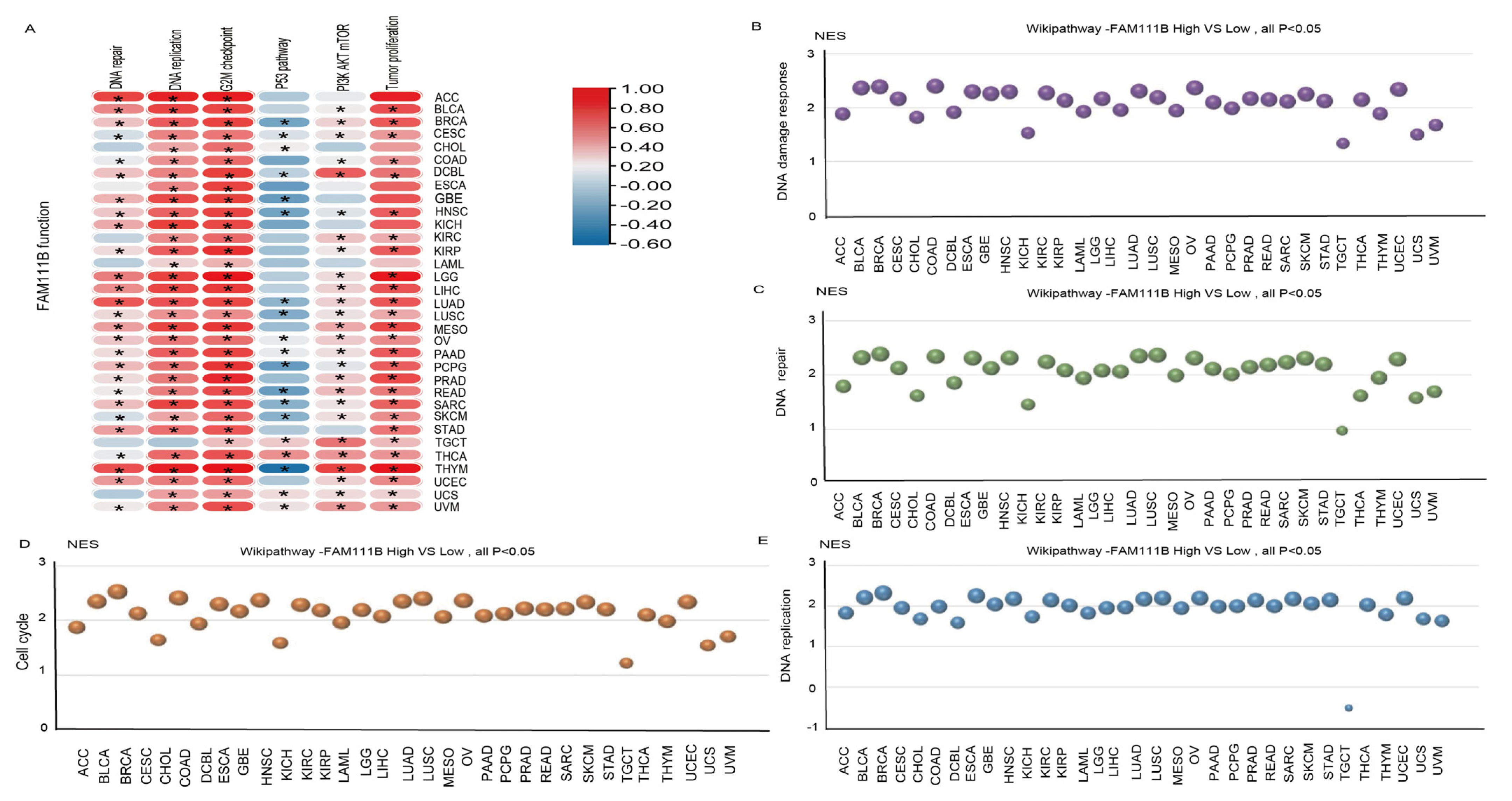
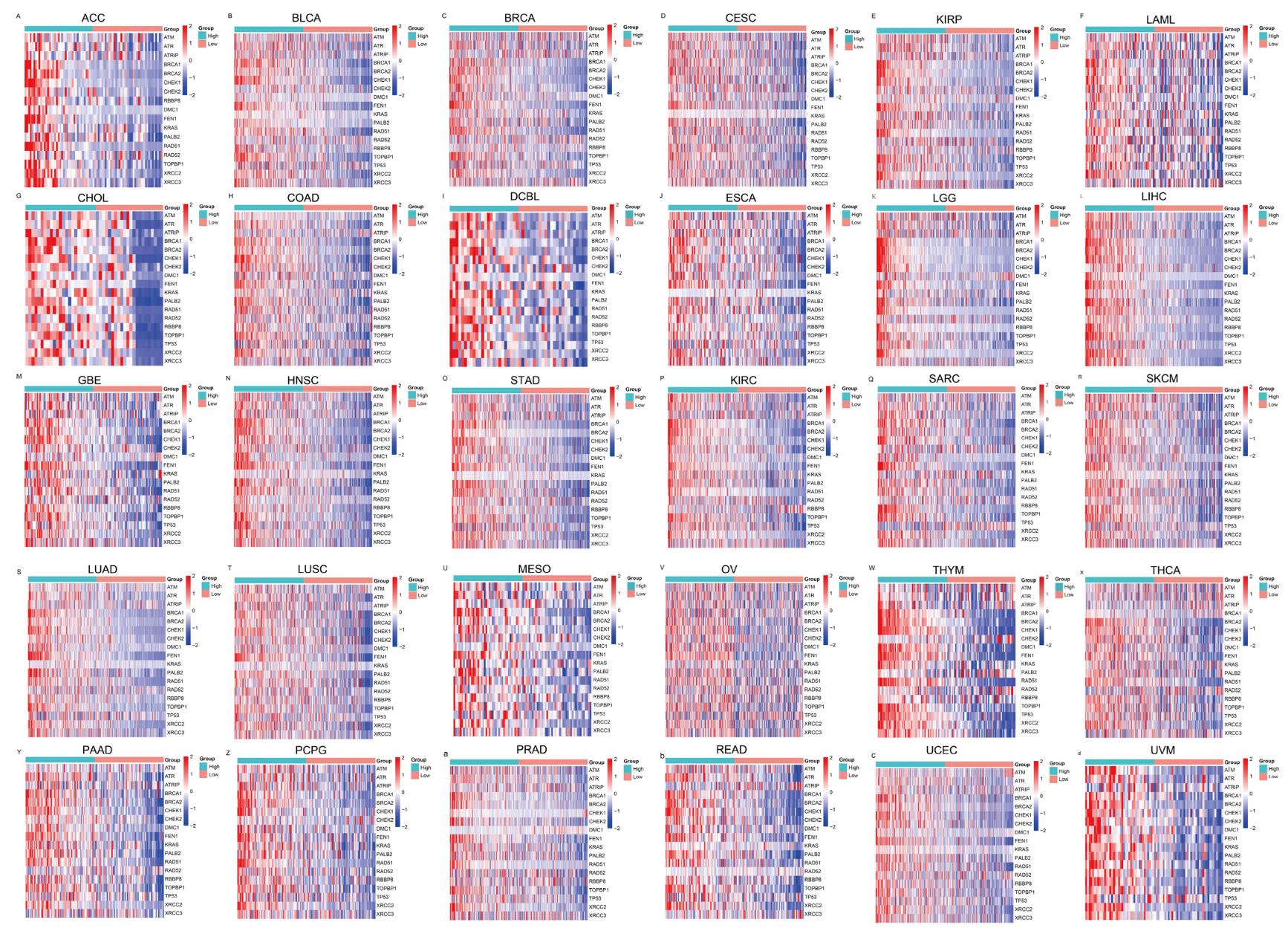
To further evaluate the pan-cancer role of FAM111B in the DNA replication process, we constructed a protein–protein interaction (PPI) network graph, which identified interactions between FAM111B and 40 other genes, including genes that encode (SMC3, SMC1A) and regulate (ESCO2) the cohesion complex (Figure 2A) [18], an integral component of the replication fork [19]. Functional enrichment analyses of the 40 FAM111B-interacting genes were conducted using GO, KEGG, Reactome, and WikiPathways. The findings indicate that FAM111B is crucial in genetic information processing, playing roles in DNA replication, DNA repair, sister chromatid segregation, and double-stranded DNA break repair, among others (Figure 2B–E). Using ssGSEA analysis of the TCGA database, we discovered that FAM111B is positively associated with DNA repair, replication, G2M checkpoint, P13K/AKT/mTOR, and tumor proliferation pathways in most cancers, and it is negatively associated with the P53 pathway (Figure 3A). For additional verification, we categorized the tumor samples into FAM111B high- and FAM111B low-expression groups and conducted GSEA analysis using the Wikipathway database. The results confirmed FAM111B’s significant role in DNA repair, the damage response, the cell cycle, and replication across TCGA tumors (Figure 3B–E, all p < 0.05). Co-expression heatmaps for homologous recombination-related genes and FAM111B (Figure 4) revealed a positive correlation in 30 of 33 TCGA tumors (all except KIHC, TGCT, and UCS). Therefore, we hypothesize that FAM111B plays a role in the DNA homologous recombination (HR) repair pathway.
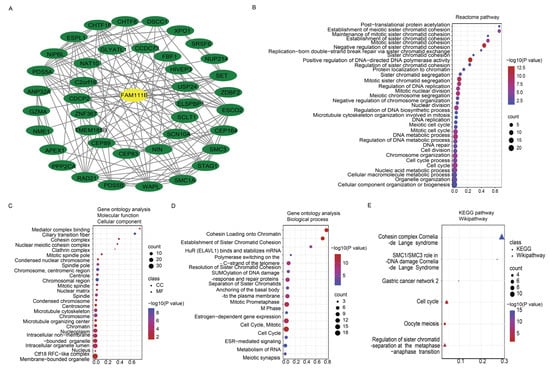
Figure 2.
Protein–protein interaction (PPI) network and enrichment analysis pertaining to FAM111B and co-expressed genes. (A) PPI network involving FAM111B and 40 associated genes. (B–E) Enrichment analysis of FAM111B and its related genes, utilizing the Reactome Pathway (panel B), Gene Ontology (GO) (panels C,D), Kyoto Encyclopedia of Genes and Genomes (KEGG), and WikiPathways (panel E) databases.
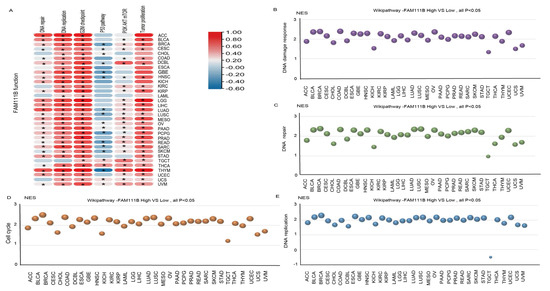
Figure 3.
Involvement of FAM111B in the DNA repair pathway. (A) The functional pathway of FAM111B according to ssGSEA analysis of TCGA data. (B–E) Roles of FAM111B in the DNA damage response, DNA repair, cell-cycle regulation, and DNA replication pathways. GSEA analysis of high and low FAM111B expression groups was performed using Wikipathway.
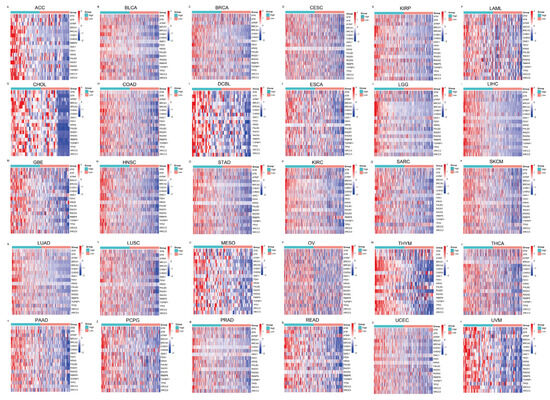
Figure 4.
Heatmaps showing the association between FAM111B and homologous recombination repair genes, as derived from the TCGA database. The relationships are depicted across multiple cancer types: (A–F) ACC, BLCA, BRCA, CESC, KIRP, and LAML; (G–L) CHOL, COAD, DCBL, ESCA, LGG, and LIHC. (M–R) GBE, HNSC, STAD, KIRC, SARC, and SKCM. (S–X) LUAD, LUSC, MESO, OV, THYM, and THCA. (Y–d) PAAD, PCPG, PRAD, READ, UCEC, and UVM.
2.3. FAM111B Plays a Crucial Role in Facilitating DNA Damage Repair Across Various Cancer Types
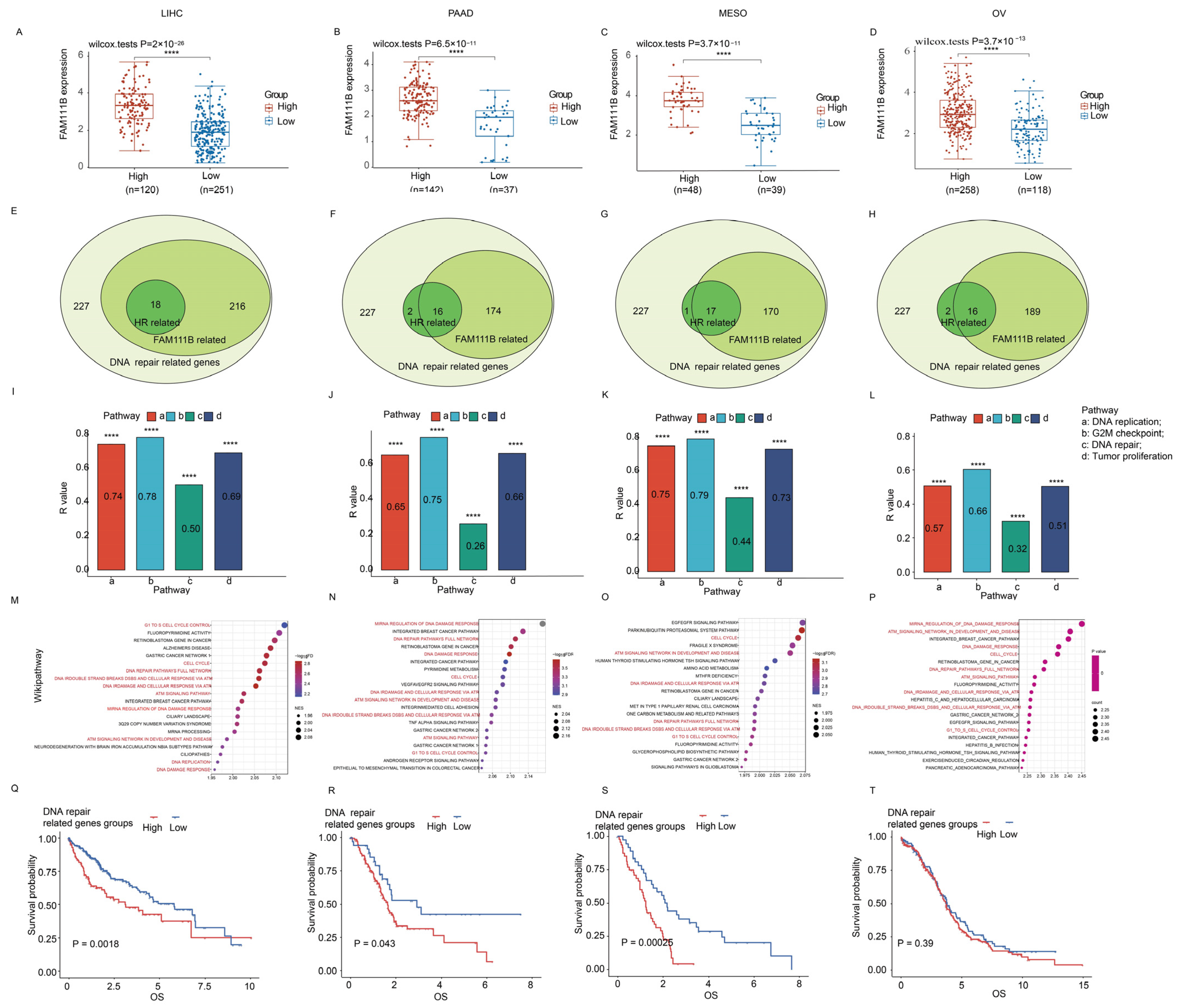
For additional insight into the role of FAM111B in DNA damage repair, we conducted the focused transcriptomic analysis of 227 DNA repair genes in LIHC, PAAD, MESO, and OV patients in the TCGA database. The patients were grouped into high and low DNA repair gene expression clusters (k = 2). Our analysis showed that FAM111B was significantly upregulated in the high-expression cluster compared to the low-expression cluster across all four tumor types (Figure 5A–D; all p < 0.05). Furthermore, correlation analyses indicated that 170–216 of the 227 DNA damage repair genes and 16–18 HR-related genes were positively correlated with FAM111B expression (Figure 5E–H). Additionally, ssGSEA analysis suggested that FAM111B positively influences DNA replication, G2M checkpoint control, DNA repair, and tumor proliferation pathways in LIHC, PAAD, MESO, and OV (Figure 5I–L, all p < 0.05), while GSEA analysis revealed FAM111B’s active role in DNA damage response pathways, including G1 cell-cycle control, the complete DNA repair network, DNA double-strand breaks, and ATR-mediated responses (Figure 5M–P; all p < 0.05). For additional prognostic insight, we performed a survival analysis of the high and low DNA repair gene expression groups. The results showed significantly shorter overall survival times in the high-expression groups for LIHC, PAAD, and MESO (p < 0.05), but not for OV (p > 0.05) (Figure 5Q–T). These findings indicate that FAM111B is closely linked to and regulates genes involved in DNA damage repair.
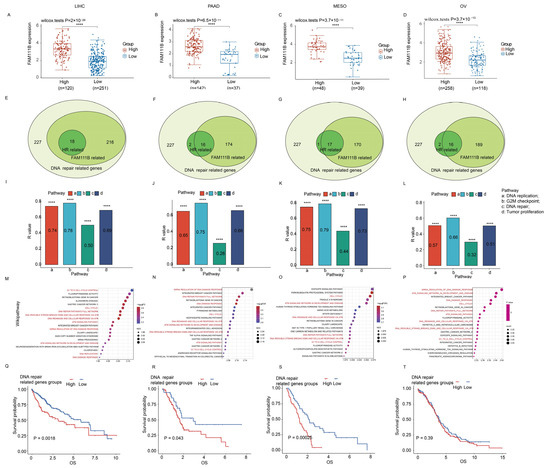
Figure 5.
FAM111B’s role in DNA repair across LIHC, PAAD, MESO, and OV. (A–D) FAM111B expression in high/low DNA repair gene groups. (E–H) Venn diagram analysis of FAM111B expression among 227 DNA repair–related genes (light green). The numbers of genes with high FAM111B expression (medium green) and those that are homologous recombination (HR) repair genes (dark green) are shown. (I–L) ssGSEA enrichment of FAM111B expression in four cancer-associated pathways. (M–P) GSEA enrichment of FAM111B in the Wikipathway database. Pathways associated with DNA damage response are indicated in red. (G–T) Kaplan–Meier analysis of DNA repair gene expression in LIHC, PAAD, MESO, and OV. Statistical significance was established at a threshold of p < 0.05, with the following designations: **** for p < 0.0001.
2.4. Multiplex Immunofluorescence Analysis Demonstrates That Pan-Cancer FAM111B Expression Is Positively Correlated with DNA Damage Repair Pathways and BRCA1 Expression
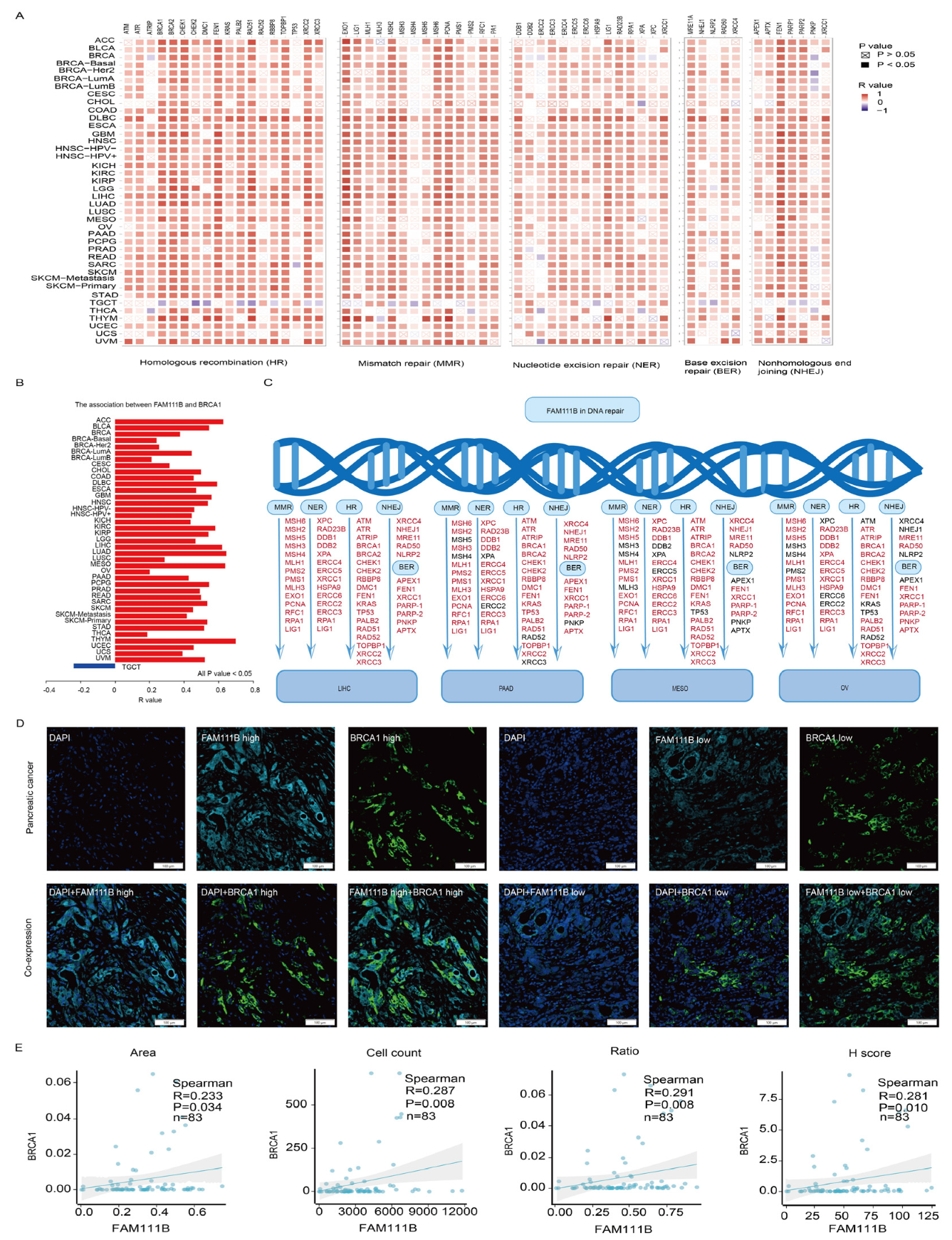
To further clarify FAM111B’s role in DNA damage repair, we examined its relationship with key proteins involved in various DNA repair mechanisms, including HR, mismatch repair (MMR), nucleotide excision repair (NER), base excision repair (BER), and nonhomologous end joining (NHEJ), which demonstrated positive correlations across multiple cancers (Figure 6A). Moreover, FAM111B and BRCA1 were positively correlated in 39 tumors; the exception was a negative correlation in TGCT (Figure 6B, all p < 0.05). In LIHC, PAAD, MESO, and OV, FAM111B also was positively correlated with HR, MMR, NER, BER, and NHEJ repair proteins (Figure 6C). To confirm the correlation between FAM111B and BRCA1, we performed a multiplex immunofluorescence analysis of 83 PC samples (Figure 6D). The results verify positive correlation between FAM111B and BRCA1 across the area, cell count, ratio, and H-score metrics (Figure 6E, all p < 0.05), further supporting the role of FAM111B in HR.
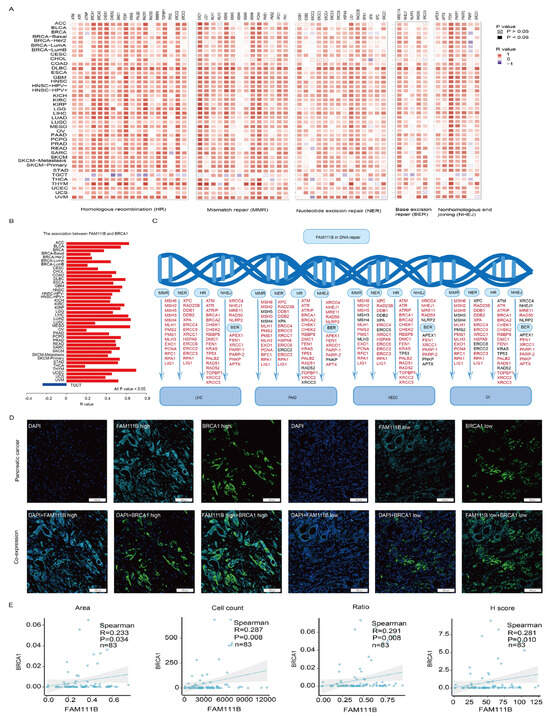
Figure 6.
The positive correlation between FAM111B and genes involved in DNA repair. (A) The positive correlation between FAM111B and sets of genes associated with specific DNA damage processes. Correlations were evaluated according to tumor type using data in TCGA and GTEx databases. (B) The correlation between FAM111B and BRCA1 across various cancer types. (C) The correlation between FAM111B and DNA repair genes specifically within LIHC, PAAD, MESO, and OV. (D) Representative images of FAM111B and BRCA1 immunofluorescence staining in PC and adjacent tissues. (E) The correlation between FAM111B and BRCA1 across the area (image section), cell count (number of positive cells), H-score (immunofluorescence assessment), and ratio (number of positive cells divided by the total cells). The H-score was calculated as ((intensity of 1 + cell positive rate) × 1 + (intensity of 2 + cell positive rate) × 2 + (intensity of 3 + cell positive rate) × 3) × 100.
2.5. FAM111B Knockdown Modulates DNA Repair Processes
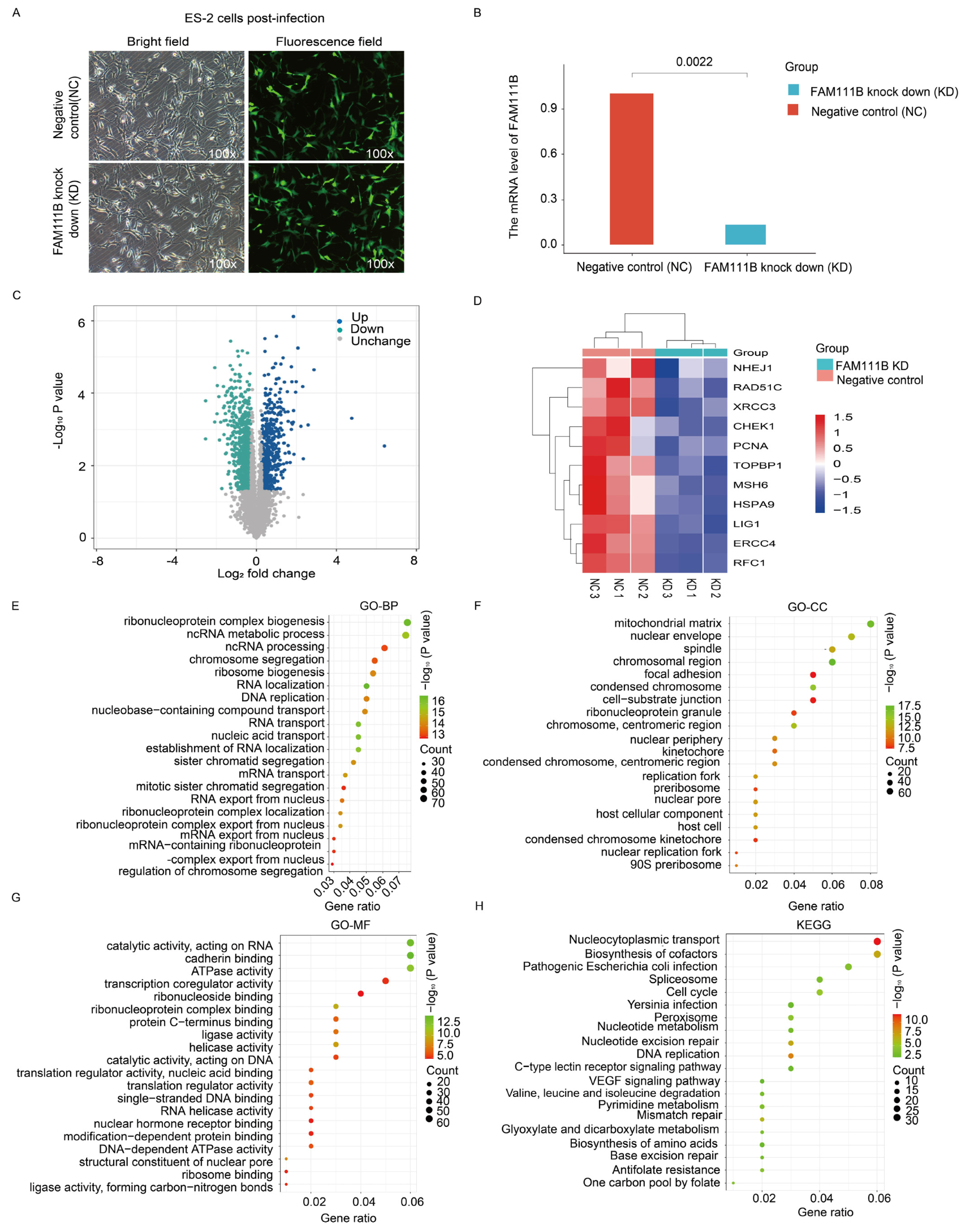
To verify the role of FAM111B in DNA repair pathways, we transfected ES-2 ovarian cancer cells with FAM111B siRNA lentivirus to silence FAM111B expression (Figure 7A). The qPCR results indicated an 86.63% reduction in FAM111B expression in the knockdown (KD) group compared to the negative control group (NC) (Figure 7B). Furthermore, a volcano plot analysis revealed 535 up-regulated and 864 down-regulated genes between the FAM111B-KD and NC cells (Figure 7C). Notably, heat maps showed a reduced expression of HR-related genes, including NHEJ1, RAD51C, and XRCC3, in the FAM111B-KD group (Figure 7D; all p < 0.05). Enrichment analysis of the 864 down-regulated genes, using the GO database, confirmed that FAM111B knockdown modulates genetic information processing activities, such as ncRNA metabolic processes and chromosome segregation (Figure 7E–G). Furthermore, KEGG analysis indicated that FAM111B knockdown modulates DNA repair mechanisms, including MMR and BER (Figure 7H). Therefore, our data confirm the close association of FAM111B with DNA damage repair in OV.
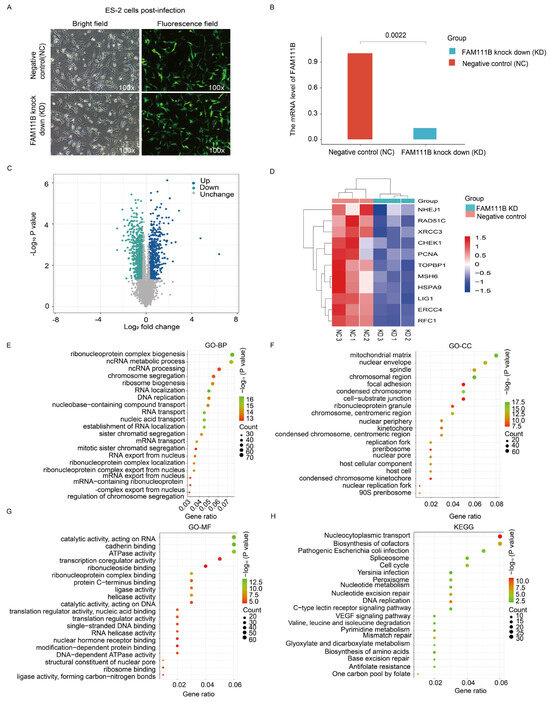
Figure 7.
Transcriptomic analysis of FAM111B knockdown cells. (A) Fluorescence image of ES-2 cells infected with a negative control siRNA (NC) or FAM111B knockdown siRNA (KD). (B) qPCR detection of FAM111B levels in infected cells. (C) Volcano plot of gene expression changes in NC versus KD cells. (D) Heat map of differentially expressed DNA repair genes. (E–H) Enrichment analysis of differentially expressed genes in GO (BP, biological processes; CC, cellular component; MF, molecular function) and KEGG databases.
2.6. FAM111B Modulates the Anti-Tumor Immune Microenvironment Across Various Cancer Types
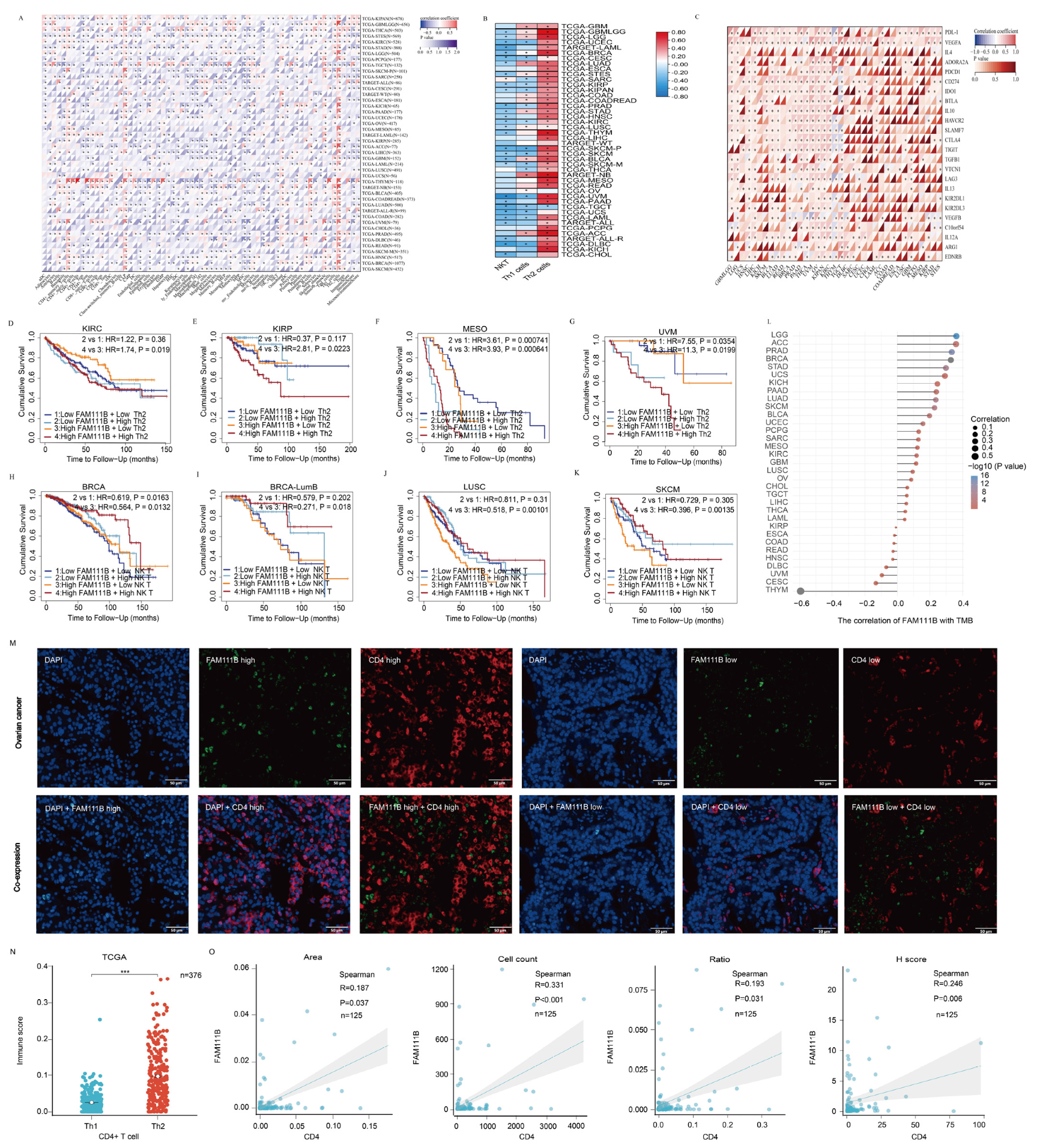
To investigate FAM111B’s role in the tumor immune microenvironment, we examined its correlation with the infiltration of 64 immune cell types using TCGA data (Figure 8A). FAM111B was negatively correlated with NK T cells and Th1 CD4+ T lymphocytes but positively correlated with Th2 CD4+ T lymphocytes in most tumors (Figure 8B). In addition, FAM111B showed a positive correlation with PDL-1, CTLA-4, and other immunosuppressive checkpoints in most tumors (Figure 8C). We evaluated the prognostic value of Th2 and NK T cells alongside FAM111B expression, which indicated that Th2 enrichment predicted a worse prognosis in KIRC, KIRP, MESO, and UVM, while a reduced number of NK T cells indicated a worse prognosis in BRCA, LUSC, and SKCM, particularly in the high FAM111B expression group (Figure 8D–K). Finally, we investigated the association between FAM111B expression and the tumor mutation burden (TMB). The results identify a positive correlation with LGG, ACC, PRAD, and several other tumor types (Figure 8L). Consequently, we hypothesize that elevated FAM111B expression may modulate the anti-tumor immune microenvironment, thereby facilitating tumor malignancy progression. To further explore the relationship between FAM111B and the anti-tumor immune microenvironment, we conducted multiplex immunofluorescence experiments using tissue microarrays from 125 OV patients (Figure 8M). FAM111B was positively correlated with CD4+ T cells in terms of the area, cell count, ratio, and H-score (Figure 8N, p < 0.05). Moreover, the analysis of 376 OV cases in the TCGA database suggested that Th2 CD4+ T cells were found to be significantly more prevalent than Th1 CD4+ T cells in OV, indicating a Th2 dominance among CD4+ T cells (Figure 8O, p < 0.05). These findings suggest that FAM111B may exert a chemotactic effect on CD4+ T cells in OV, with a possible preference for Th2 CD4+ T cells.
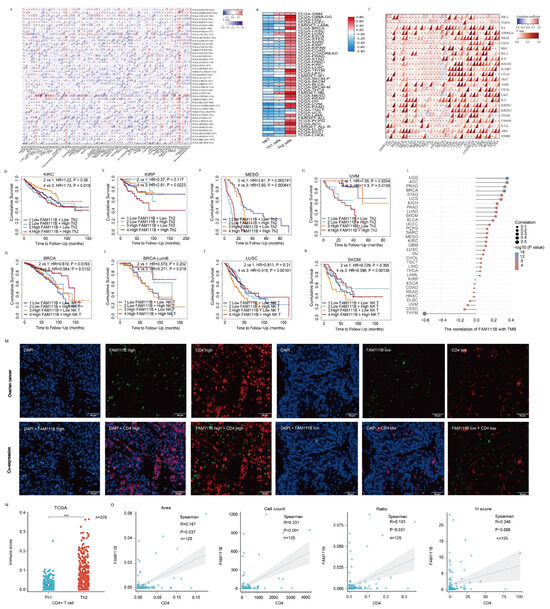
Figure 8.
The role of FAM111B in the tumor immune microenvironment. (A) FAM111B’s association with immune cell marker expression in different types of cancer. (B) Correlation between FAM111B expression and NK T, Th1, and Th2 cell marker enrichment. (C) Correlation between FAM111B expression and immune checkpoint markers. (D–G) The prognostic impact of FAM111B and Th2 cells in specific cancer types was evaluated according to Kaplan-Meier survival analysis of high/low FAM111B and CD4 expression groups. (H–K) The prognostic impact of FAM111B and NK T cells in specific cancer types was evaluated according to Kaplan-Meier survival analysis of high/low FAM111B and NK marker expression groups. (L) The association between FAM111B expression and the tumor mutation burden (TMB). (M) Representative immunofluorescence staining of FAM111B and CD4 expression for high- and low-expression samples. (N) Th1 and Th2 CD4+ T−cell infiltration scores for OV samples from the TCGA database. (O) FAM111B correlation with CD4 in OV samples according to the area, cell count, H-score, and ratio. *** for p < 0.001.
3. Discussion
In our pan-cancer analysis, we demonstrated that FAM111B is linked to malignancy and poor prognosis, and that alterations in DNA repair and the immune microenvironment underlie the severity of diseases. FAM111B’s role in DNA repair was verified in LIHC, PAAD, MESO, and OV. Multiplex immunofluorescence assays showed a positive correlation between FAM111B and the DNA repair protein BRCA1 in PC tissues, while transcriptomic analysis verified FAM111B’s role in DNA repair in OV cells. Furthermore, immunoassays revealed FAM111B’s negative correlation with Th1 CD4+ T cells and positive correlation with Th2 CD4+ T cells, which was verified by multiplex immunofluorescence assays of OV tissues. Thus, our results suggest that FAM111B aids DNA repair and influences Th1/Th2 differentiation in CD4+ T cells, potentially contributing to cancer development across various types.
Evidence has increasingly supported the involvement of FAM111B in the damage response and repair of DNA. For example, proteomic transcriptome analysis demonstrated FAM111B’s role in DNA repair pathways in ESCA cells [15]. Furthermore, GSEA analysis demonstrated that FAM111B is significantly enriched in NER and BER in PC [13]. However, this study is the first to evaluate the role of FAM111B in mediating DNA repair mechanisms across cancers. We evaluated its expression patterns in 33 cancer types and comprehensively assessed the FAM111B-associated pathways, which suggested mechanisms that are generalizable to many different cancer types. Notably, our findings revealed a negative association between FAM111B expression and the P53 pathway. The P53 signaling pathway, when phosphorylated and activated, triggers stress responses like DNA damage [20]. Research shows that the expression of FAM111B is increased during the DNA damage response and is linked to the activated P53 pathway in LUAD [11]. In OV, FAM111B correlates positively with phosphorylated AKT protein, which negatively influenced the P53 pathway [12]. FAM111B has also been reported to interact with CAPNS1, which indirectly regulates FANCD2, a key DNA damage response protein [21,22]. Studies suggest that FAM111B, like its homolog FAM111A, aids in removing DNA protein crosslinks during replication, enhancing DNA repair [2]. Therefore, these results support our findings suggesting that FAM111B influences the DNA damage and repair in multiple types of cancer.
Our research indicates that FAM111B is positively associated with DNA damage repair genes and plays a significant role in the occurrence of poor prognosis. Previous studies have shown that, in breast cancer, the analysis of eight DNA repair-related genes, including RPA3 and XRCC4, among others, reveals that patients exhibiting high expression levels of these genes tend to have poorer prognoses [23]. Similarly, elevated Rad51 expression is correlated with adverse outcomes in OV [24]. Active DNA damage repair mechanisms enable cancer cells to survive under conditions of DNA damage and may introduce mutations during the repair process, potentially activating oncogenes or inactivating tumor suppressor genes and thereby accelerating tumor progression [25,26]. Moreover, the accumulation of such mutations provides tumor cells with a growth advantage, driving their clonal evolution and enhancing their invasive and metastatic potential [27].
The targeting of key proteins, such as PARP, in DNA damage repair has been demonstrated to cause “synthetic lethality” by exploiting HR deficiencies, leading to tumor cell death [28]. Furthermore, mutations in HR-related genes, such as BRCA1/2, have been demonstrated to cause HR deficiency, thus increasing the effectiveness of PARP inhibitors [29]. In this study, we demonstrated that FAM111B expression correlates with 17 HR repair proteins, including ATM, ATR, and BRCA1/2, in 30 out of 33 of the studied cancers. The positive correlation between FAM111B and BRCA1 was verified by multiplex immunofluorescence assays of 83 PC samples. Based on these findings, the inhibition of FAM111B may induce HR deficiency and increase PARP inhibitor sensitivity. Research indicates that the knockout of FAM111A, a structurally and functionally similar protein, boosts PARP inhibitor effectiveness [30]. Given that FAM111B shares a 43% sequence similarity and structural domains with FAM111A, it may perform a similar role [1,2,31]. Moreover, our research showed that FAM111B deficiency may lead to HR deficiency by targeting multiple DNA damage and repair pathways. Therefore, the inhibition of FAM111B in patients with high FAM111B expression could potentially provide an approach for improving their treatment by disrupting the DNA damage response. FAM111B inhibitors are not yet available for clinical examination; however, the HERB database predicts that 12 natural products, including oleum anisistellati, fructus cinnamomi cassiae, and cassia bark oil, are potential FAM111B inhibitors [32]. Therefore, our findings could provide insights for the development of natural remedies for cancer treatment.
Our research also demonstrated that FAM111B is negatively correlated with Th1 CD4+ T cells and positively correlated with Th2 CD4+ T cells across various cancers, indicating its role in shifting the balance of these cell populations. The functions of Th1 and Th2 populations are distinctly different, with Th1 cells boosting anti-tumor immunity and Th2 cells aiding in tumor immune evasion [33], although both cell types originate from CD4+ T cells and inhibit each other to maintain immune system balance [34]. In the tumor microenvironment, a shift from Th1 to Th2 immune responses has been shown to influence cancer progression [35]; increased Th2 and decreased Th1 levels are linked to poor prognosis [36,37]. Moreover, studies have shown that FAM111B affects the T-cell balance in thyroid cancer and LUAD [16,17]. Therefore, our findings are consistent with the established role of FAM111B in modulating the immune environment to promote cancer progression.
Our study is constrained by several limitations. In the experimental validation phase, we exclusively utilized tissue microarrays of PC and OV cells to investigate the role of FAM111B in DNA repair. However, further validation across a broader range of cancer types is warranted. Additionally, within the tumor microenvironment, we observed a correlation between FAM111B and CD4+ T cells in the OV tissue microarrays; however, our analysis did not distinguish between specific subtypes, such as Th1 and Th2 cells. This limitation will be addressed in future research efforts.
4. Materials and Methods
4.1. Patient Cohorts
In this study, pan-cancer analysis data were sourced from the comprehensive data on various cancer types available in The Cancer Genome Atlas (TCGA) database, which includes 33 distinct cancer types and encompasses a total of 10,228 patients. Based on the TCGA database, BRCA can be subdivided into four subtypes: BRCA-Basal, BRCA-Her2, BRCA-LumA, and BRCA-LumB; HNSC can be divided into two subtypes: HNSC-HPV (−) and HNSC-HPV (+); and SKCM can be divided into two subtypes: SKCM-Metastasis and SKCM-Primary. Therefore, the 33 types of tumors in the TCGA database can be further divided into 41 subtypes, which were used to conduct a correlation analysis between FAM111B and DNA repair proteins or tumor stemness scores. For expression controls, the Genotype-Tissue Expression (GTEx) database (https://www.gtexportal.org/home/) (accessed on 21 September 2023), containing genomic information from 838 healthy donors, was utilized. Furthermore, for comprehensive immune analysis, we expanded our analysis to the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) database (https://ocg.cancer.gov/programs/target) (accessed on 21 September 2023) to incorporate four additional cancer types: osteosarcoma, acute lymphoblastic leukemia, neuroblastoma, and high-risk Wilms tumor.
4.2. Differential Expression and Prognostic Evaluation of FAM111B in Pan-Cancers
To analyze FAM111B expression and prognosis, transcription data from TCGA and GTEx were downloaded. Tumor versus normal tissue expression differences were evaluated with ANOVA using GEPIA2. FAM111B’s prognostic value was assessed using a Cox proportional hazards model via TIMER2.0. We retrieved the TCGA pan-cancer tumor stemness scores from the UCSC (https://xenabrowser.net/) (accessed on 21 September 2023) database, excluding cancer types with fewer than three samples, and ultimately acquired expression data for 37 cancer types to explore FAM111B’s link with tumor stemness. The DNA expression-based stemness score (DNAss) and RNA expression-based stemness score (RNAss) were derived for Pearson analysis using previous methylation signatures [18]. FAM111B expression across various pathological grades and TNM stages was analyzed using R software (v3.6.4) with unpaired Student’s t-test or ANOVA.
4.3. Functional Enrichment Analysis of FAM111B in Various Cancers
Using the STRING online tool, a PPI network was created for FAM111B and 40 related genes. These genes were analyzed using GO, KEGG, Reactome, and Wikipathway. FAM111B-related pathways were further examined using ssGSEA and the “GSVA” package in R, and analyses were based on TCGA data. GSEA with Wikipathway analysis was conducted to validate FAM111B’s pathways across 33 tumors. Heatmaps from an online platform (https://www.bioinformatics.com.cn) (accessed on 15 August 2023) were generated to visualize co-expression patterns of homologous recombination-related genes in groups with high (50–100%) and low (0–50%) FAM111B expression.
4.4. Correlation Analysis of FAM111B Within the DNA Repair Pathway in Four Types of Cancers
The “ConsensusClusterPlus” R package was used to analyze 227 DNA damage repair genes in LIHC, PAAD, MESO, and OV, and patients were categorized into high- and low-expression groups. Kaplan–Meier survival and differential expression analyses were performed between these groups, and Spearman analysis was employed to evaluate the correlation between FAM111B and the 227 genes. GSEA and ssGSEA were employed to investigate FAM111B’s functional pathways, while Spearman correlation analysis with TCGA DNA repair proteins was used to examine its role in DNA repair through TIMER2.0.
4.5. Immunoassay Investigation of the Role of FAM111B in the Immune Microenvironment
Gene expression profiles for each cancer type were extracted from the TCGA dataset, and immune cell infiltration scores were calculated using the IOBR R package with the xCell algorithm [38]. The prognostic significance of FAM111B’s correlation with Th2 and NK T-cell infiltration was analyzed using TIMER 2.0 and KM mapper (https://kmplot.com/analysis/) (accessed on 18 October 2023). Correlation data between FAM111B and the tumor mutation burden (TMB) were derived using TCGA transcriptome data.
4.6. Tissue Microarrays
Tissue microarrays from Shanghai Outdo Biotechnology Co. Ltd. included samples from 83 PC and 125 OV patients, and all clinical samples were clustered with patients’ informed consent. The PC microarrays (HPanA180Su10) included 78 pancreatic ductal adenocarcinoma cases and 5 cases of other subtypes, including adeno-squamous carcinoma, ampullary adenocarcinoma, high-grade intraductal papillary mucinous neoplasm with pancreatic ductal adenocarcinoma, and adenocarcinoma. The OV tissue microarrays (panel HOvaC151Su01) comprised 65 serous adenocarcinoma cases, 30 mucinous adenocarcinoma cases, 13 endometrioid adenocarcinoma cases, and 17 cases of various other subtypes, including clear cell carcinoma, serous-mucinous adenocarcinoma, dysgerminoma, squamous cell carcinoma, borderline mucinous tumors, adenocarcinoma, malignant Brenner tumors, and yolk sac tumors.
4.7. Multiplex Immunohistochemistry
Tissue samples were dewaxed in xylene, rehydrated with 10% formalin, and subjected to antigen retrieval in a microwave. Antibodies against FAM111B (Novus, NBP1-86645, Chesterfield, Missouri, USA), BRCA1 (Novus, NB100-404, Missouri, USA), or CD4 (ZSGB, ZM0418, Beijing, China) were applied, and fluorescent signals were stained and amplified using the PANO 7-plex IHC kit (Panovue, Beijing, China). After staining, DAPI solution was applied, and the slides were sealed with mounting medium. Section scanning was conducted using the Olympus VS200 MTL (Olympus, Hamburg, Germany) and UPLXAPO20X lenses. QuPath v0.3.0 software was used for analyzing multi-color fluorescence images.
4.8. Cell Culture and FAM111B Gene Knockdown
The ES-2 ovarian cancer cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in McCoy’s 5A medium with 10% FBS (Ausbian, Cat. No. A11-102) at 37 °C in 5% CO2. At 80% confluence, a 105 cells/mL suspension of FAM111B-targeting shRNA lentivirus (sense sequence: 5′-GCCTGCCTAGTGATTCTCATT-3′, anti-sense sequence: 5′-AATGAGAATCACTAGGCAGGC-3′) was applied. As a negative control, a nonsense shRNA lentivirus (target sequence 5′-TTCTCCGAACGTGTCACGT-3′) was applied. After 72 h, the expressed on the reporter gene was assessed by fluorescence microscopy to evaluate infection levels. An infection efficiency of 80% or higher was deemed satisfactory.
4.9. Real-Time Quantitative PCR
After centrifugation of cultured cells, the supernatants were removed, and 1 mL of Trizol (Promega, 3111-100, Fitchburg, Wisconsin, USA) per sample was added for cell lysis and RNA extraction. The cDNA was synthesized using the M-MLV Kit (Promega, M1705, Wisconsin, USA), and PCR was performed with SYBR Master Mix. RNA levels were measured using the 2−ΔΔCT method. Primers from Guangzhou Ribobio Co., Ltd. (Guangzhou, China) included GAPDH forward (5′-TGACTTCAACAGCGACACCCA-3′) and reverse (5′-CACCCTGTTGCTGTAGCCAAA-3′); and FAM111B forward (5′-GAGTTCTGCCCTACTCCTGAC-3′) and reverse (5′-AAATCTGTCGCCATAGTCCTG-3′).
4.10. Protein Extraction and Transcriptomic Analysis
Proteins were extracted from cells using the SDT lysis method and then quantified using the BCA Protein Assay Kit (P0012, Beyotime, Shanghai, China). UA buffer (8 M Urea, 150 mM Tris-HCl pH 8.5) was added for peptide quantification using the FASP enzymatic digestion method. Mass spectrometry analysis was conducted on the Orbitrap Exploris 480 in positive ion mode, with a parent ion scan range of 350–1200 m/z and a first-level resolution of 120,000. The AGC target was set at 300%, with a first-level IT cap of 50 ms. Mass spectrometry data were collected, processed, and analyzed using data-dependent acquisition and the UniProt protein sequence database (http://www.uniprot.org) (accessed on 15 February 2023). The “enhanced volcano” R package was used to create a volcano plot to visualize gene expression differences between the FAM111B KD and NC groups. The downregulated genes of the FAM111B KD and NC groups were input into the GO and KEGG databases for functional enrichment analysis.
4.11. Statistical Analysis
Statistical analysis was performed using R v4.0.3 and ggplot2 v3.3.3 for graphing. Correlations were assessed with Spearman and Pearson analyses, and group differences were evaluated using the unpaired Student’s t-test or ANOVA. Statistical significance was established at a threshold of p < 0.05, with the following designations: * for p < 0.05, ** for p < 0.01, *** for p < 0.001, and **** for p < 0.0001.
5. Conclusions
In summary, this study represents the first demonstration that FAM111B facilitates cancer progression by modulating DNA repair processes, particularly HR repair in various cancers. This mechanism has potential therapeutic implications for augmenting the effectiveness of PARP inhibitors through “synthetic lethality”. Furthermore, we demonstrated that FAM111B promotes the transition from Th1 to Th2 cells, thereby suggesting a mechanism for contributing to tumor immune evasion. These findings provide new insights into the oncogenic mechanisms associated with FAM111B expression.
Author Contributions
F.W. and W.L.: writing—original draft preparation, formal analysis, methodology, visualization, software, investigation. T.Z.: formal analysis, visualization. X.Y.: writing—review and editing, investigation. L.Z.: writing—review and editing, conceptualization, funding acquisition, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 81803009), the Natural Science Foundation of Hubei Province (Grant No. 2024AFB995), the Chinese Society of Clinical Oncology (CSCO) Research Fund (Grant Nos. Y-L2018-003, Y-sy2018-039), the CSCO-BMS Immune-Oncology Research Fund (Grant No. Y-BMS 2019-011), and the Chen Xiaoping Foundation for the Development of Science and Technology of Hubei Province (Grant No. CXPJJH122006-1027).
Institutional Review Board Statement
The use of TMA samples was approved by the Ethics Committee of Shanghai Outdo Biotech Co., Ltd. (Approval number: YBM-05-02; date of approval: 11 January 2019).
Informed Consent Statement
The informed consent was obtained from all patients.
Data Availability Statement
The public datasets utilized in this study are accessible through online database. Publicly available data were obtained from the TCGA websites (https://portal.gdc.cancer.gov/ or https://xenabrowser.net/datapages/) (accessed on 21 September 2023), GTEx website (https://www.gtexportal.org/home/) (accessed on 21 September 2023), and TARGET database (https://ocg.cancer.gov/programs/target) (accessed on 21 September 2023).
Conflicts of Interest
The authors state that they have no competing interests.
Abbreviations
| FAM111B | the FAM111 trypsin like peptidase B |
| POIKTMP | hereditary fibrosing poikiloderma with tendon contractures, myopathy, and pulmonary fibrosis; |
| PC | pancreatic cancer |
| OV | ovarian cancer |
| DPC | DNA-protein crosslinking complexes |
| TCGA | the cancer genome atlas database |
| mIF | multiple immunofluorescence |
| GTEx | the genotype-tissue expression |
| DNAss | DNA expression-based stemness scores |
| RNAss | RNA expression-based stemness scores |
| TMB | tumor mutation burden; |
| HR | homologous recombination |
| MMR | mismatch repair; |
| NER | nucleotide excision repair; |
| BER | base excision repair; |
| NHEJ | non-homologous end joining repair; |
| PPI | protein-protein interaction; |
| GO | gene ontology; |
| KEGG | kyoto encyclopedia of genes and genomes; |
| GSEA | gene set enrichment; |
| CAPNS1 | calpain small subunit 1; |
| DDR | DNA damage repair; |
| HRD | HR deficiency; |
| TME | tumor microenvironment; |
| LAML | acute myeloid leukemia; |
| ACC | adrenocortical carcinoma; |
| BLCA | bladder urothelial carcinoma; |
| LGG | brain lower grade glioma; |
| BRCA | breast invasive carcinoma; |
| CESC | cervical squamous cell carcinoma and endocervical adenocarcinoma; |
| CHOL | cholangiocarcinoma; |
| COAD | colon adenocarcinoma; |
| GBM | glioblastoma multiforme; |
| HNSC | head and neck squamous cell carcinoma; |
| KICH | kidney chromophobe; |
| KIRC | kidney renal clear cell carcinoma; |
| KIRP | kidney renal papillary cell carcinoma; |
| LIHC | liver hepatocellular carcinoma; |
| LUAD | lung adenocarcinoma; |
| LUSC | lung squamous cell carcinoma; |
| DLBC | lymphoid neoplasm diffuse large B-cell lymphoma; |
| MESO | mesothelioma; |
| PAAD | pancreatic adenocarcinoma; |
| PCPG | pheochromocytoma and paraganglioma; |
| PRAD | prostate adenocarcinoma; |
| READ | rectum adenocarcinoma; |
| SARC | sarcoma |
| SKCM | skin cutaneous melanoma |
| STAD | stomach adenocarcinoma |
| TGCT | testicular germ cell tumors; |
| THYM | thymoma; |
| THCA | thyroid carcinoma; |
| UCS | uterine carcinosarcoma; |
| UCEC | uterine corpus endometrial carcinoma; |
| UVM | uveal melanoma. |
References
- Hoffmann, S.; Pentakota, S.; Mund, A.; Haahr, P.; Coscia, F.; Gallo, M.; Mann, M.; Taylor, N.M.; Mailand, N. FAM111 protease activity undermines cellular fitness and is amplified by gain-of-function mutations in human disease. EMBO Rep. 2020, 21, e50662. [Google Scholar]
- Mercier, S.; Küry, S.; Shaboodien, G.; Houniet, D.T.; Khumalo, N.P.; Bou-Hanna, C.; Bodak, N.; Cormier-Daire, V.; David, A.; Faivre, L.; et al. Mutations in FAM111B cause hereditary fibrosing poikiloderma with tendon contracture, myopathy, and pulmonary fibrosis. Am. J. Hum. Genet. 2013, 93, 1100–1107. [Google Scholar] [CrossRef]
- Akamatsu, S.; Takata, R.; Haiman, C.A.; Takahashi, A.; Inoue, T.; Kubo, M.; Furihata, M.; Kamatani, N.; Inazawa, J.; Chen, G.K.; et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat. Genet. 2012, 44, 426–429, s1. [Google Scholar] [PubMed]
- Kawasaki, K.; Nojima, S.; Hijiki, S.; Tahara, S.; Ohshima, K.; Matsui, T.; Hori, Y.; Kurashige, M.; Umeda, D.; Kiyokawa, H.; et al. FAM111B enhances proliferation of KRAS-driven lung adenocarcinoma by degrading p16. Cancer Sci. 2020, 111, 2635–2646. [Google Scholar] [PubMed]
- Li, W.; Hu, S.; Han, Z.; Jiang, X. YY1-Induced Transcriptional Activation of FAM111B Contributes to the Malignancy of Breast Cancer. Clin. Breast Cancer. 2022, 22, e417–e425. [Google Scholar]
- Mercier, S.; Küry, S.; Nahon, S.; Salort-Campana, E.; Barbarot, S.; Bézieau, S. FAM111B Mutation Is Associated with Pancreatic Cancer Predisposition. Pancreas 2019, 48, e41–e42. [Google Scholar] [PubMed]
- Gupta, S.; Silveira, D.A.; Hashimoto, R.F. A Boolean model of the oncogene role of FAM111B in lung adenocarcinoma. Comput Biol Chem. 2023, 106, 107926. [Google Scholar]
- Wu, H.; Liang, C. Pan-Cancer Analysis of the Tumorigenic Effect and Prognostic Diagnostic Value of FAM111B in Human Carcinomas. Int. J. Gen. Med. 2023, 16, 1845–1865. [Google Scholar]
- Zhu, X.; Xue, C.; Kang, X.; Jia, X.; Wang, L.; Younis, M.H.; Liu, D.; Huo, N.; Han, Y.; Chen, Z.; et al. DNMT3B-mediated FAM111B methylation promotes papillary thyroid tumor glycolysis, growth and metastasis. Int. J. Biol. Sci. 2022, 18, 4372–4387. [Google Scholar]
- Wei, F.; Yu, G.; Si, C.; Chao, T.; Xiong, H.; Zhang, L. High FAM111B expression predicts aggressive clinicopathologic features and poor prognosis in ovarian cancer. Transl. Oncol. 2023, 32, 101659. [Google Scholar]
- Gong, Q.; Dong, Q.; Zhong, B.; Zhang, T.; Cao, D.; Zhang, Y.; Ma, D.; Cai, X.; Li, Z. Clinicopathological features, prognostic significance, and associated tumor cell functions of family with sequence similarity 111 member B in pancreatic adenocarcinoma. J. Clin. Lab. Anal. 2022, 36, e24784. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, K.; Huang, J.; Sun, Q.; Shao, C.; Luo, J.; Xu, L.; Shen, Y.; Ren, B. FAM111B, a direct target of p53, promotes the malignant process of lung adenocarcinoma. Onco Targets Ther. 2019, 12, 2829–2842. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gu, Y.; Ni, H.; Quan, Q.; Guo, L. Silencing of FAM111B inhibits tumor growth and promotes apoptosis by decreasing AKT activity in ovarian cancer. Exp. Biol. Med. 2023, 248, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Arowolo, A.; Malebana, M.; Sunda, F.; Rhoda, C. Proposed Cellular Function of the Human FAM111B Protein and Dysregulation in Fibrosis and Cancer. Front. Oncol. 2022, 12, 932167. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Chen, J.; Liu, P.; Xiao, X. Overexpressed FAM111B degrades GSDMA to promote esophageal cancer tumorigenesis and cisplatin resistance. Cell Oncol. 2024, 47, 343–359. [Google Scholar] [CrossRef]
- Hong, K.; Cen, K.; Chen, Q.; Dai, Y.; Mai, Y.; Guo, Y. Identification and validation of a novel senescence-related biomarker for thyroid cancer to predict the prognosis and immunotherapy. Front. Immunol. 2023, 14, 1128390. [Google Scholar]
- Lao, Y.; Li, T.; Xie, X.; Chen, K.; Li, M.; Huang, L. MiR-195-3p is a Novel Prognostic Biomarker Associated with Immune Infiltrates of Lung Adenocarcinoma. Int. J. Gen. Med. 2022, 15, 191–203. [Google Scholar] [CrossRef]
- Banerji, R.; Skibbens, R.V.; Iovine, M.K. Cohesin mediates Esco2-dependent transcriptional regulation in a zebrafish regenerating fin model of Roberts Syndrome. Biol. Open 2017, 6, 1802–1813. [Google Scholar] [CrossRef]
- Murayama, Y.; Endo, S.; Kurokawa, Y.; Kurita, A.; Iwasaki, S.; Araki, H. Coordination of cohesin and DNA replication observed with purified proteins. Nature 2024, 626, 653–660. [Google Scholar] [CrossRef]
- Hassin, O.; Oren, M. Drugging p53 in cancer: One protein, many targets. Nat. Rev. Drug Discov. 2023, 22, 127–144. [Google Scholar] [CrossRef]
- Alcón, P.; Kaczmarczyk, A.P.; Ray, K.K.; Liolios, T.; Guilbaud, G.; Sijacki, T.; Shen, Y.; McLaughlin, S.H.; Sale, J.E.; Knipscheer, P.; et al. FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions. Nature 2024, 632, 1165–1173. [Google Scholar] [PubMed]
- Roversi, G.; Colombo, E.A.; Magnani, I.; Gervasini, C.; Maggiore, G.; Paradisi, M.; Larizza, L. Spontaneous chromosomal instability in peripheral blood lymphocytes from two molecularly confirmed Italian patients with Hereditary Fibrosis Poikiloderma: Insights into cancer predisposition. Genet. Mol. Biol. 2021, 44, e20200332. [Google Scholar]
- Zhang, D.; Yang, S.; Li, Y.; Yao, J.; Ruan, J.; Zheng, Y.; Deng, Y.; Li, N.; Wei, B.; Wu, Y.; et al. Prediction of Overall Survival Among Female Patients With Breast Cancer Using a Prognostic Signature Based on 8 DNA Repair-Related Genes. JAMA Netw. Open. 2020, 3, e2014622. [Google Scholar]
- Hoppe, M.M.; Jaynes, P.; Wardyn, J.D.; Upadhyayula, S.S.; Tan, T.Z.; Lie, S.; Lim, D.G.Z.; Pang, B.N.K.; Lim, S.; Yeong, J.P.S.; et al. Quantitative imaging of RAD51 expression as a marker of platinum resistance in ovarian cancer. EMBO Mol. Med. 2021, 13, e13366. [Google Scholar] [PubMed]
- Achar, Y.J.; Foiani, M. An Error-Prone Polymerase in the Fight against Cancer. Cell 2019, 176, 1241–1243. [Google Scholar]
- Lodovichi, S.; Cervelli, T.; Pellicioli, A.; Galli, A. Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach. Int. J. Mol. Sci. 2020, 21, 6684. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar]
- Li, S.; Wang, L.; Wang, Y.; Zhang, C.; Hong, Z.; Han, Z. The synthetic lethality of targeting cell cycle checkpoints and PARPs in cancer treatment. J. Hematol. Oncol. 2022, 15, 147. [Google Scholar] [CrossRef]
- Murai, J.; Pommier, Y. BRCAness, Homologous Recombination Deficiencies, and Synthetic Lethality. Cancer Res. 2023, 83, 1173–1174. [Google Scholar]
- Kojima, Y.; Machida, Y.; Palani, S.; Caulfield, T.R.; Radisky, E.S.; Kaufmann, S.H.; Machida, Y.J. FAM111A protects replication forks from protein obstacles via its trypsin-like domain. Nat. Commun. 2020, 11, 1318. [Google Scholar] [CrossRef]
- Fine, D.A.; Rozenblatt-Rosen, O.; Padi, M.; Korkhin, A.; James, R.L.; Adelmant, G.; Yoon, R.; Guo, L.; Berrios, C.; Zhang, Y.; et al. Identification of FAM111A as an SV40 host range restriction and adenovirus helper factor. PLoS Pathog. 2012, 8, e1002949. [Google Scholar]
- Li, W.; Feng, S.S.; Wu, H.; Deng, J.; Zhou, W.Y.; Jia, M.X.; Shi, Y.; Ma, L.; Zeng, X.X.; Zuberi, Z.; et al. Comprehensive Analysis of CDK1-Associated ceRNA Network Revealing the Key Pathways LINC00460/LINC00525-Hsa-Mir-338-FAM111/ZWINT as Prognostic Biomarkers in Lung Adenocarcinoma Combined with Experiments. Cells 2022, 11, 1220. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, H.; Tracy, S.I.; Farrar, M.A. Cytotoxic CD4 T cells in the mucosa and in cancer. Front. Immunol. 2023, 14, 1233261. [Google Scholar]
- Basu, A.; Ramamoorthi, G.; Albert, G.; Gallen, C.; Beyer, A.; Snyder, C.; Koski, G.; Disis, M.L.; Czerniecki, B.J.; Kodumudi, K. Differentiation and Regulation of T(H) Cells: A Balancing Act for Cancer Immunotherapy. Front. Immunol. 2021, 12, 669474. [Google Scholar]
- Lee, Y.H.; Tsai, K.W.; Lu, K.C.; Shih, L.J.; Hu, W.C. Cancer as a Dysfunctional Immune Disorder: Pro-Tumor TH1-like Immune Response and Anti-Tumor THαβ Immune Response Based on the Complete Updated Framework of Host Immunological Pathways. Biomedicines 2022, 10, 2497. [Google Scholar] [CrossRef]
- Hu, X.; Luo, B.; Wu, Q.; Chen, Q.; Lu, P.; Huang, J.; Liang, X.; Ling, C.; Li, Y. Effects of Dezocine and Sufentanil on Th1/Th2 Balance in Breast Cancer Patients Undergoing Surgery. Drug Des. Devel Ther. 2021, 15, 4925–4938. [Google Scholar]
- Shang, Q.; Yu, X.; Sun, Q.; Li, H.; Sun, C.; Liu, L. Polysaccharides regulate Th1/Th2 balance: A new strategy for tumor immunotherapy. Biomed. Pharmacother. 2024, 170, 115976. [Google Scholar]
- Zeng, D.; Ye, Z.; Shen, R.; Yu, G.; Wu, J.; Xiong, Y.; Zhou, R.; Qiu, W.; Huang, N.; Sun, L.; et al. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Front. Immunol. 2021, 12, 687975. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).