Machine Learning Identification of Neutrophil Extracellular Trap-Related Genes as Potential Biomarkers and Therapeutic Targets for Bronchopulmonary Dysplasia
Abstract
1. Introduction
2. Results
2.1. Determination and Enrichment Analysis of DEGs
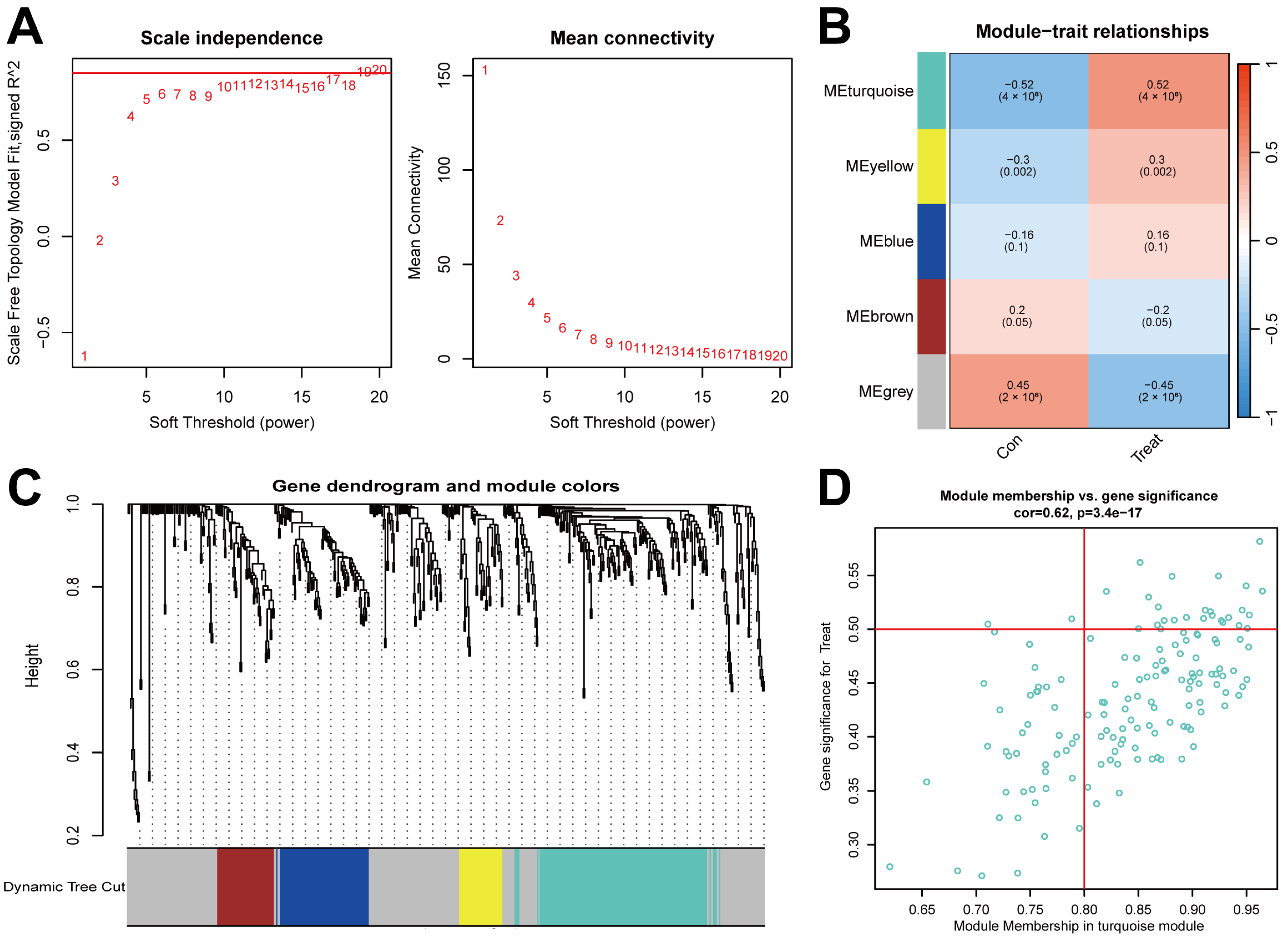
2.2. Gene Module Identification and Co-Expression Network Formation
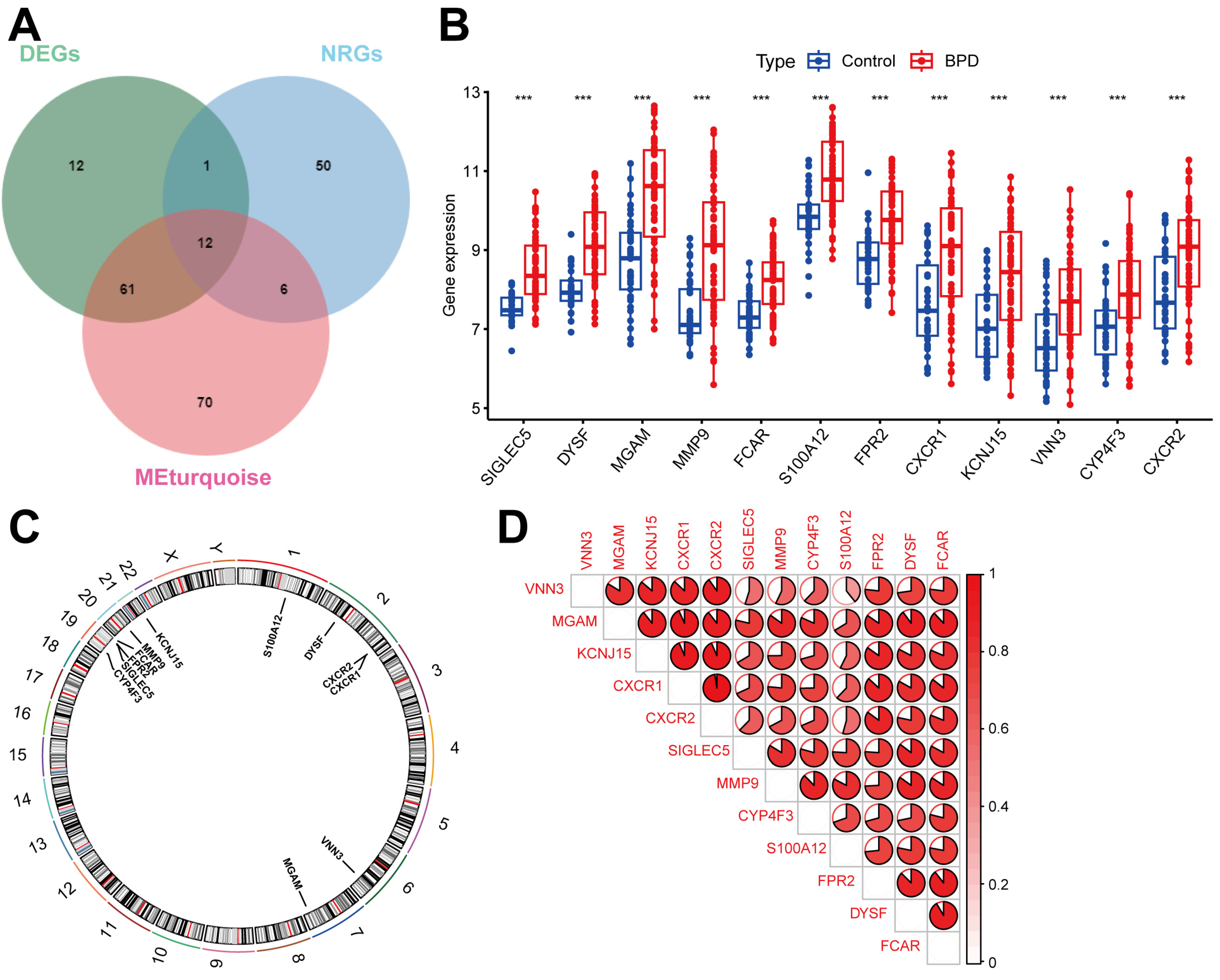
2.3. Identifying DENRGs in BPD
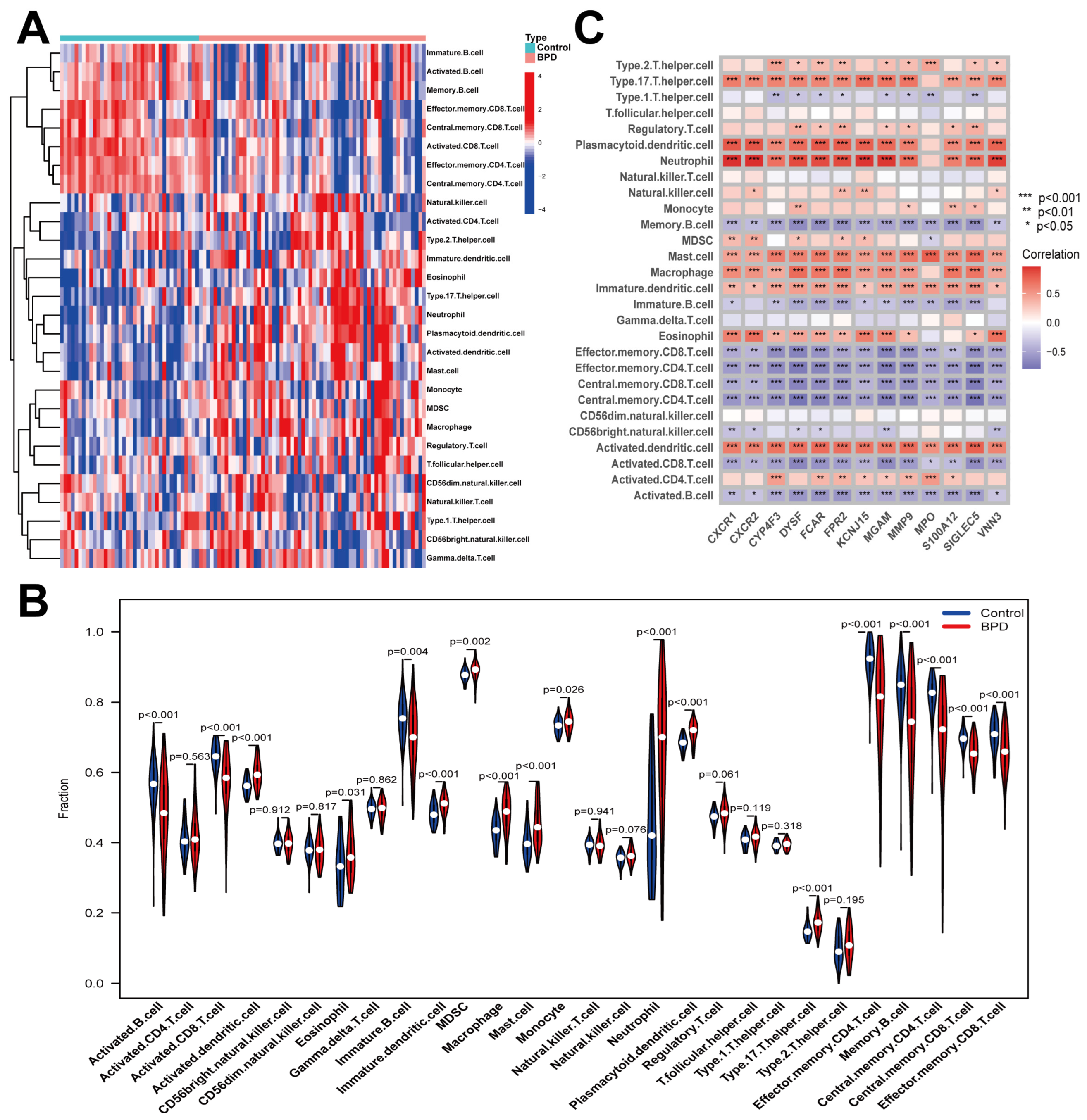
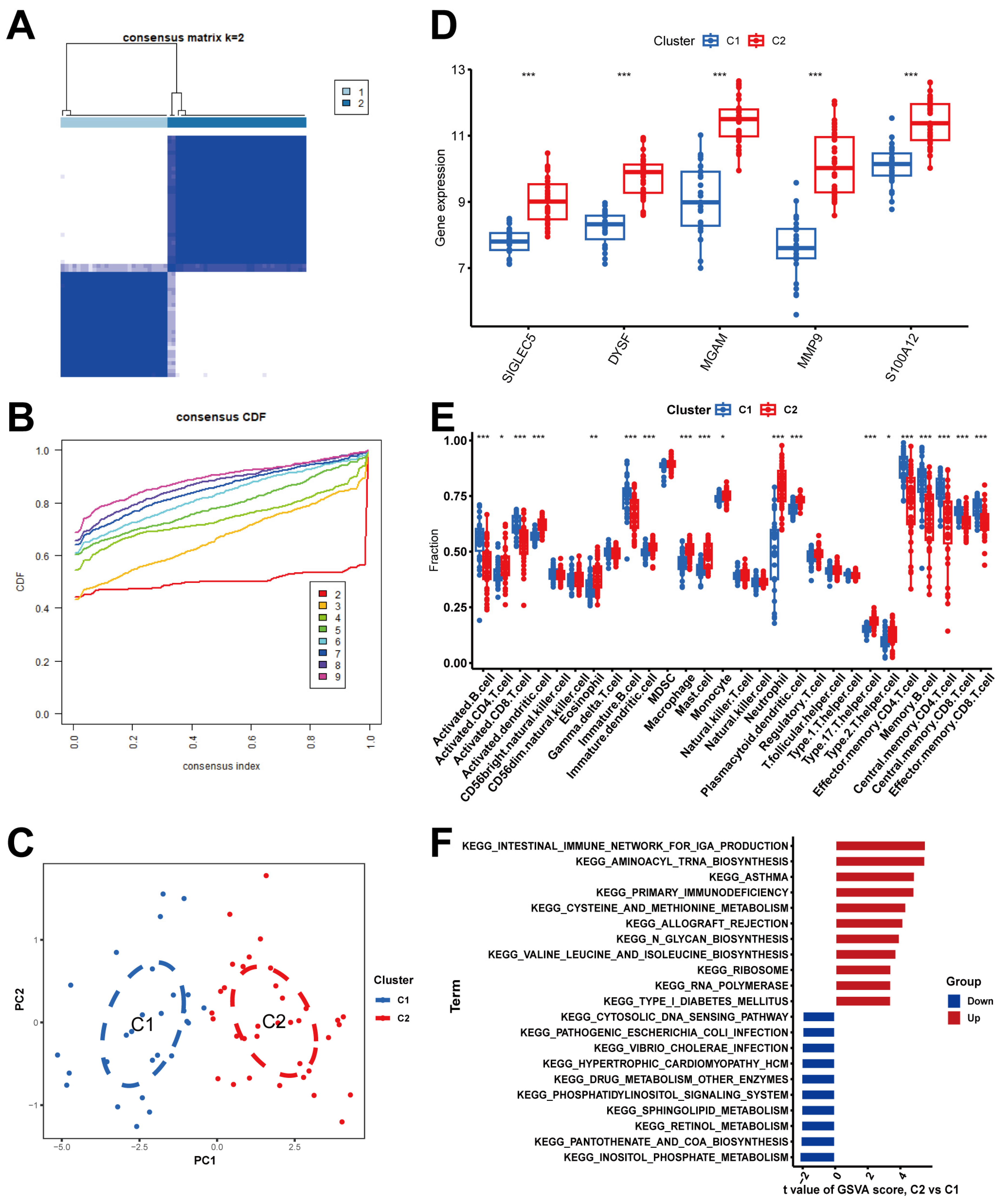
2.4. Immune Cell Infiltration in BPD
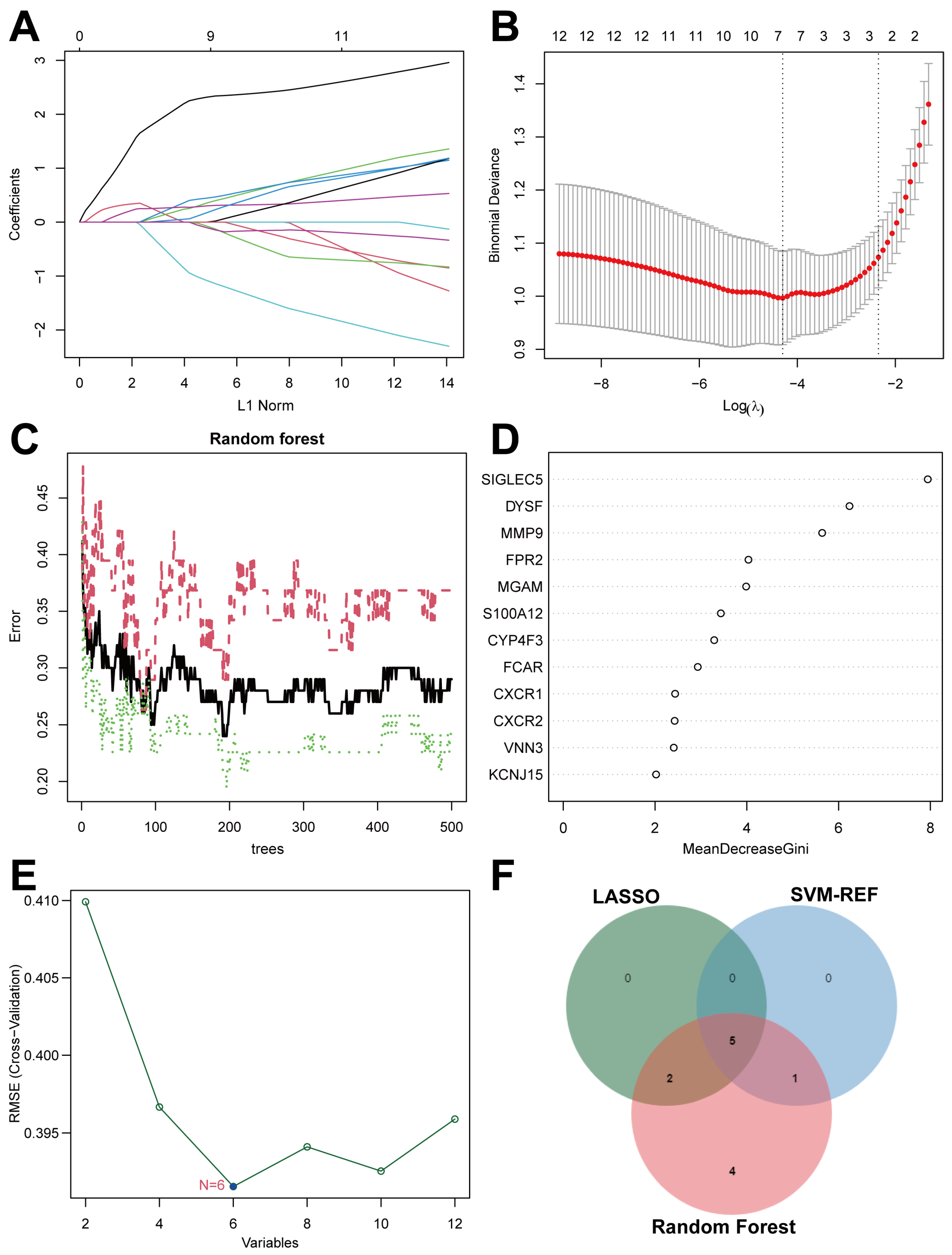
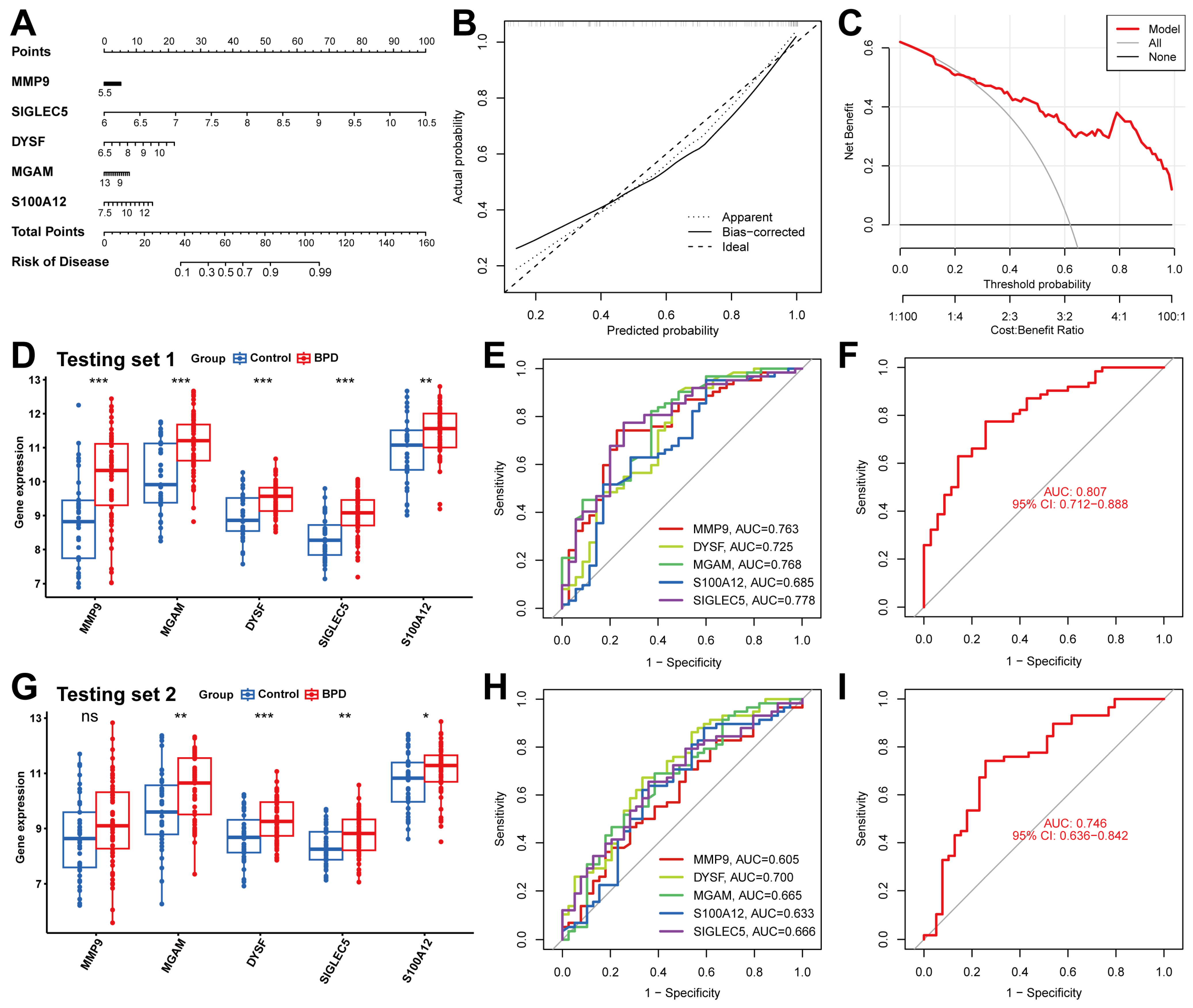
2.5. Screen Key Biomarkers Machine Learning Models
2.6. Validation of the Diagnostic Capabilities of Five Biomarkers
2.7. Determination of NRG Molecular Clusters in BPD
2.8. Drug–Gene Interactions for BPD Patients
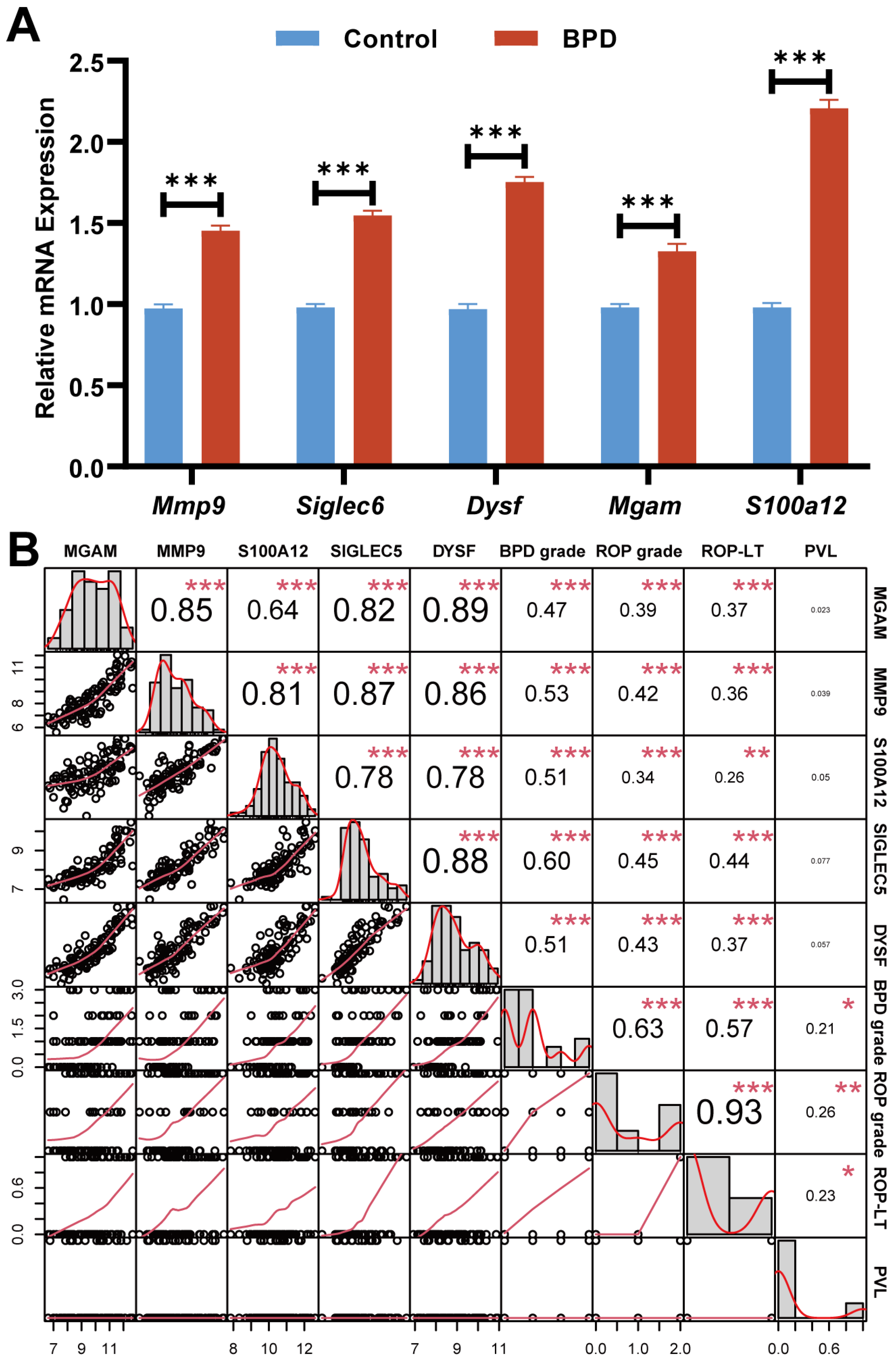
2.9. Relationship Between Biomarkers and Patients’ Clinical Characteristics
3. Discussion
4. Materials and Methods
4.1. Peripheral Blood Collection from Clinical BPD Patients
4.2. Gathering BPD Data from Gene Expression Omnibus (GEO) Database
4.3. Gene Expression and Functional Enrichment Analyses in BPD
4.4. Weighted Gene Co-Expression Network Analysis (WGCNA) in BPD
4.5. Immune Cells Infiltrating in BPD
4.6. Identifying Key Biomarkers in BPD
4.7. Building and Validating the Column Line Plot Model
4.8. Unsupervised Clustering and Gene Set Variation Analysis (GSVA) Analysis of BPD
4.9. Drug-Gene Interaction Insights
4.10. qRT-PCR in Peripheral Blood of BPD
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abiramalatha, T.; Ramaswamy, V.V.; Bandyopadhyay, T.; Somanath, S.H.; Shaik, N.B.; Pullattayil, A.K.; Weiner, G.M. Interventions to Prevent Bronchopulmonary Dysplasia in Preterm Neonates: An Umbrella Review of Systematic Reviews and Meta-analyses. JAMA Pediatr. 2022, 176, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, P.; Ho, J.J.; Davis, P.G. Prophylactic or very early initiation of continuous positive airway pressure (CPAP) for preterm infants. Cochrane Database Syst. Rev. 2021, 10, CD001243. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.W.; Cheong, J.L.; Hay, S.; Manley, B.J.; Halliday, H.L. Late (>/= 7 days) systemic postnatal corticosteroids for prevention of bronchopulmonary dysplasia in preterm infants. Cochrane Database Syst. Rev. 2021, 11, CD001145. [Google Scholar] [CrossRef]
- Gilfillan, M.; Bhandari, A.; Bhandari, V. Diagnosis and management of bronchopulmonary dysplasia. BMJ 2021, 375, n1974. [Google Scholar] [CrossRef]
- Wu, T.J.; Jing, X.; Teng, M.; Pritchard, K.A., Jr.; Day, B.W.; Naylor, S.; Teng, R.J. Role of Myeloperoxidase, Oxidative Stress, and Inflammation in Bronchopulmonary Dysplasia. Antioxidants 2024, 13, 889. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Zaramella, P.; Munari, F.; Stocchero, M.; Molon, B.; Nardo, D.; Priante, E.; Tosato, F.; Bonadies, L.; Viola, A.; Baraldi, E. Innate immunity ascertained from blood and tracheal aspirates of preterm newborn provides new clues for assessing bronchopulmonary dysplasia. PLoS ONE 2019, 14, e0221206. [Google Scholar] [CrossRef]
- Jin, R.; Xu, J.; Gao, Q.; Mao, X.; Yin, J.; Lu, K.; Guo, Y.; Zhang, M.; Cheng, R. IL-33-induced neutrophil extracellular traps degrade fibronectin in a murine model of bronchopulmonary dysplasia. Cell Death Discov. 2020, 6, 33. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, M.; Jiang, J.; Liu, W.; Zhao, W.; Li, F. Neutrophil extracellular traps promote bronchopulmonary dysplasia-like injury in neonatal mice via the WNT/beta-catenin pathway. Front. Cell Infect. Microbiol. 2023, 13, 1126516. [Google Scholar] [CrossRef]
- Fasoulakis, Z.; Koutras, A.; Ntounis, T.; Antsaklis, P.; Theodora, M.; Valsamaki, A.; Daskalakis, G.; Kontomanolis, E.N. Inflammatory Molecules Responsible for Length Shortening and Preterm Birth. Cells 2023, 12, 209. [Google Scholar] [CrossRef]
- Xing, D.; Wells, J.M.; Giordano, S.S.; Feng, W.; Gaggar, A.; Yan, J.; Hage, F.G.; Li, L.; Chen, Y.F.; Oparil, S. Induced pluripotent stem cell-derived endothelial cells attenuate lipopolysaccharide-induced acute lung injury. J. Appl. Physiol. (1985) 2019, 127, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Loering, S.; Cameron, G.J.M.; Starkey, M.R.; Hansbro, P.M. Lung development and emerging roles for type 2 immunity. J. Pathol. 2019, 247, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wu, X. Effect of early preventive use of caffeine citrate on prevention together with treatment of BPD within premature infants and its influence on inflammatory factors. Biotechnol. Genet. Eng. Rev. 2024, 40, 2730–2744. [Google Scholar] [CrossRef] [PubMed]
- Harijith, A.; Choo-Wing, R.; Cataltepe, S.; Yasumatsu, R.; Aghai, Z.H.; Janer, J.; Andersson, S.; Homer, R.J.; Bhandari, V. A role for matrix metalloproteinase 9 in IFNgamma-mediated injury in developing lungs: Relevance to bronchopulmonary dysplasia. Am. J. Respir. Cell Mol. Biol. 2011, 44, 621–630. [Google Scholar] [CrossRef]
- Zhang, Z.; Han, N.; Shen, Y. S100A12 promotes inflammation and cell apoptosis in sepsis-induced ARDS via activation of NLRP3 in fl ammasome signaling. Mol. Immunol. 2020, 122, 38–48. [Google Scholar] [CrossRef]
- Loughran-Fowlds, A.; Leach, S.; Lin, J.; Oei, J.; Henry, R.; Day, A.S.; Lui, K. Respiratory disease and early serum S100A12 changes in very premature infants. Acta Paediatr. 2011, 100, 1538–1543. [Google Scholar] [CrossRef]
- Ali, S.R.; Fong, J.J.; Carlin, A.F.; Busch, T.D.; Linden, R.; Angata, T.; Areschoug, T.; Parast, M.; Varki, N.; Murray, J.; et al. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J. Exp. Med. 2014, 211, 1231–1242. [Google Scholar] [CrossRef]
- Wielgat, P.; Mroz, R.M.; Stasiak-Barmuta, A.; Szepiel, P.; Chyczewska, E.; Braszko, J.J.; Holownia, A. Inhaled corticosteroids increase siglec-5/14 expression in sputum cells of COPD patients. Adv. Exp. Med. Biol. 2015, 839, 1–5. [Google Scholar] [CrossRef]
- Ballouhey, O.; Chapoton, M.; Alary, B.; Courrier, S.; Da Silva, N.; Krahn, M.; Levy, N.; Weisleder, N.; Bartoli, M. A Dysferlin Exon 32 Nonsense Mutant Mouse Model Shows Pathological Signs of Dysferlinopathy. Biomedicines 2023, 11, 1438. [Google Scholar] [CrossRef]
- Leung, C.; Shaheen, F.; Bernatchez, P.; Hackett, T.L. Expression of myoferlin in human airway epithelium and its role in cell adhesion and zonula occludens-1 expression. PLoS ONE 2012, 7, e40478. [Google Scholar] [CrossRef][Green Version]
- Tannous, S.; Stellbrinck, T.; Hoter, A.; Naim, H.Y. Interaction between the alpha-glucosidases, sucrase-isomaltase and maltase-glucoamylase, in human intestinal brush border membranes and its potential impact on disaccharide digestion. Front. Mol. Biosci. 2023, 10, 1160860. [Google Scholar] [CrossRef] [PubMed]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef]
- Jobe, A.H.; Bancalari, E. Bronchopulmonary dysplasia. Am. J. Respir. Crit. Care Med. 2001, 163, 1723–1729. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, L.; Dai, Q.; Shang, B.; Xiao, T.; Di, X.; Zhang, K.; Feng, L.; Shou, J.; Wang, Y. A signature for pan-cancer prognosis based on neutrophil extracellular traps. J. Immunother. Cancer. 2022, 10, e004210. [Google Scholar] [CrossRef]
- Zhou, Y.; Luo, Y.; Zeng, W.; Mao, L.; Le, F.; Lou, H.; Wang, L.; Mao, Y.; Jiang, Z.; Jin, F. FANCD2 as a ferroptosis-related target for recurrent implantation failure by integrated bioinformatics and Mendelian randomization analysis. J. Cell. Mol. Med. 2024, 28, e70119. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Zhang, T.; Wong, G. Gene expression data analysis using Hellinger correlation in weighted gene co-expression networks (WGCNA). Comput. Struct. Biotechnol. J. 2022, 20, 3851–3863. [Google Scholar] [CrossRef]
- Frost, H.R.; Amos, C.I. Gene set selection via LASSO penalized regression (SLPR). Nucleic Acids Res. 2017, 45, e114. [Google Scholar] [CrossRef] [PubMed]
- Mi, X.; Zou, B.; Zou, F.; Hu, J. Permutation-based identification of important biomarkers for complex diseases via machine learning models. Nat. Commun. 2021, 12, 3008. [Google Scholar] [PubMed]
- Kursa, M.B. Robustness of Random Forest-based gene selection methods. BMC Bioinform. 2014, 15, 8. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]
- David, C.C.; Jacobs, D.J. Principal component analysis: A method for determining the essential dynamics of proteins. Methods Mol. Biol. 2014, 1084, 193–226. [Google Scholar] [CrossRef] [PubMed]


| Primers | Sequence (5′->3′) | |
|---|---|---|
| MMP9 | Forward | 5′-TGTACCGCTATGGTTACACTCG-3′ |
| Reverse | 5′-GGCAGGGACAGTTGCTTCT-3′ | |
| Siglec-5 | Forward | 5′-TTCAGGAACGGCATAGCCCTA-3′ |
| Reverse | 5′-TACTCGACGAAGCTCCAAGAT-3′ | |
| DYSF | Forward | 5′-AAGAACAGCGTGAACCCTGTA-3′ |
| Reverse | 5′-CCTCTCGGAGTGGGACCTT-3′ | |
| MGAM | Forward | 5′-GCTCAGTGTTCTTCTGCTTGT-3′ |
| Reverse | 5′-CGTTGTCCTAGCATGTGTGGTA-3′ | |
| S100A12 | Forward | 5′-AGCATCTGGAGGGAATTGTCA-3′ |
| Reverse | 5′-GCAATGGCTACCAGGGATATGAA-3′ | |
| GAPDH | Forward | 5′-GCCTCAAAATCCTCTCGTTGTG-3′ |
| Reverse | 5′-GGAAGATGGTGATGGGATTTC-3′ | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Yan, B.; Jiang, Z.; Luo, Y. Machine Learning Identification of Neutrophil Extracellular Trap-Related Genes as Potential Biomarkers and Therapeutic Targets for Bronchopulmonary Dysplasia. Int. J. Mol. Sci. 2025, 26, 3230. https://doi.org/10.3390/ijms26073230
Zhang X, Yan B, Jiang Z, Luo Y. Machine Learning Identification of Neutrophil Extracellular Trap-Related Genes as Potential Biomarkers and Therapeutic Targets for Bronchopulmonary Dysplasia. International Journal of Molecular Sciences. 2025; 26(7):3230. https://doi.org/10.3390/ijms26073230
Chicago/Turabian StyleZhang, Xuandong, Bingqian Yan, Zhou Jiang, and Yujia Luo. 2025. "Machine Learning Identification of Neutrophil Extracellular Trap-Related Genes as Potential Biomarkers and Therapeutic Targets for Bronchopulmonary Dysplasia" International Journal of Molecular Sciences 26, no. 7: 3230. https://doi.org/10.3390/ijms26073230
APA StyleZhang, X., Yan, B., Jiang, Z., & Luo, Y. (2025). Machine Learning Identification of Neutrophil Extracellular Trap-Related Genes as Potential Biomarkers and Therapeutic Targets for Bronchopulmonary Dysplasia. International Journal of Molecular Sciences, 26(7), 3230. https://doi.org/10.3390/ijms26073230






