Phenotypic, Physiological, and Transcriptomic Analyses Reveal Different Responses to Salt Stress in Cultivated Red Lettuce and Wild Lettuce Seedlings
Abstract
1. Introduction
2. Results
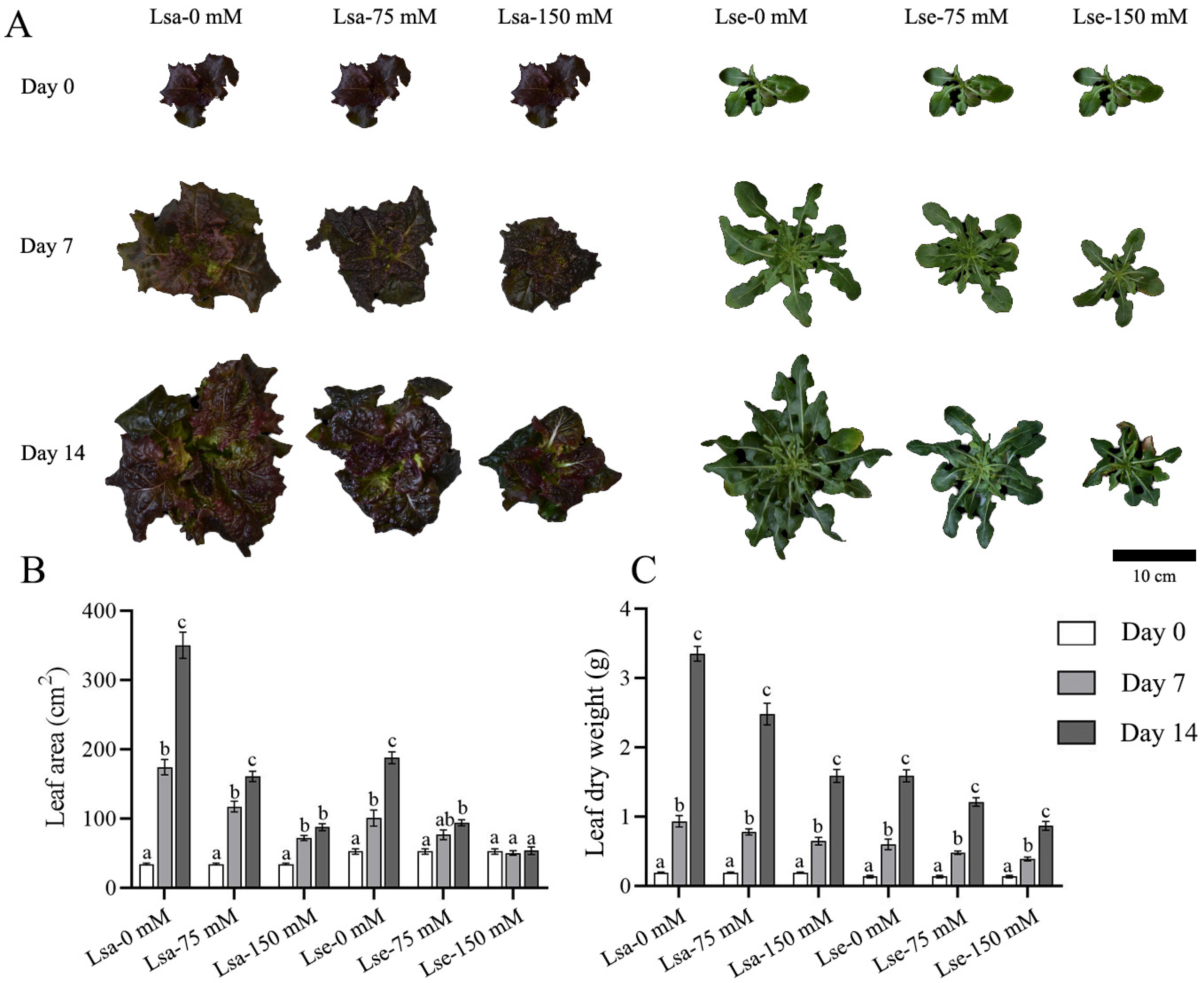
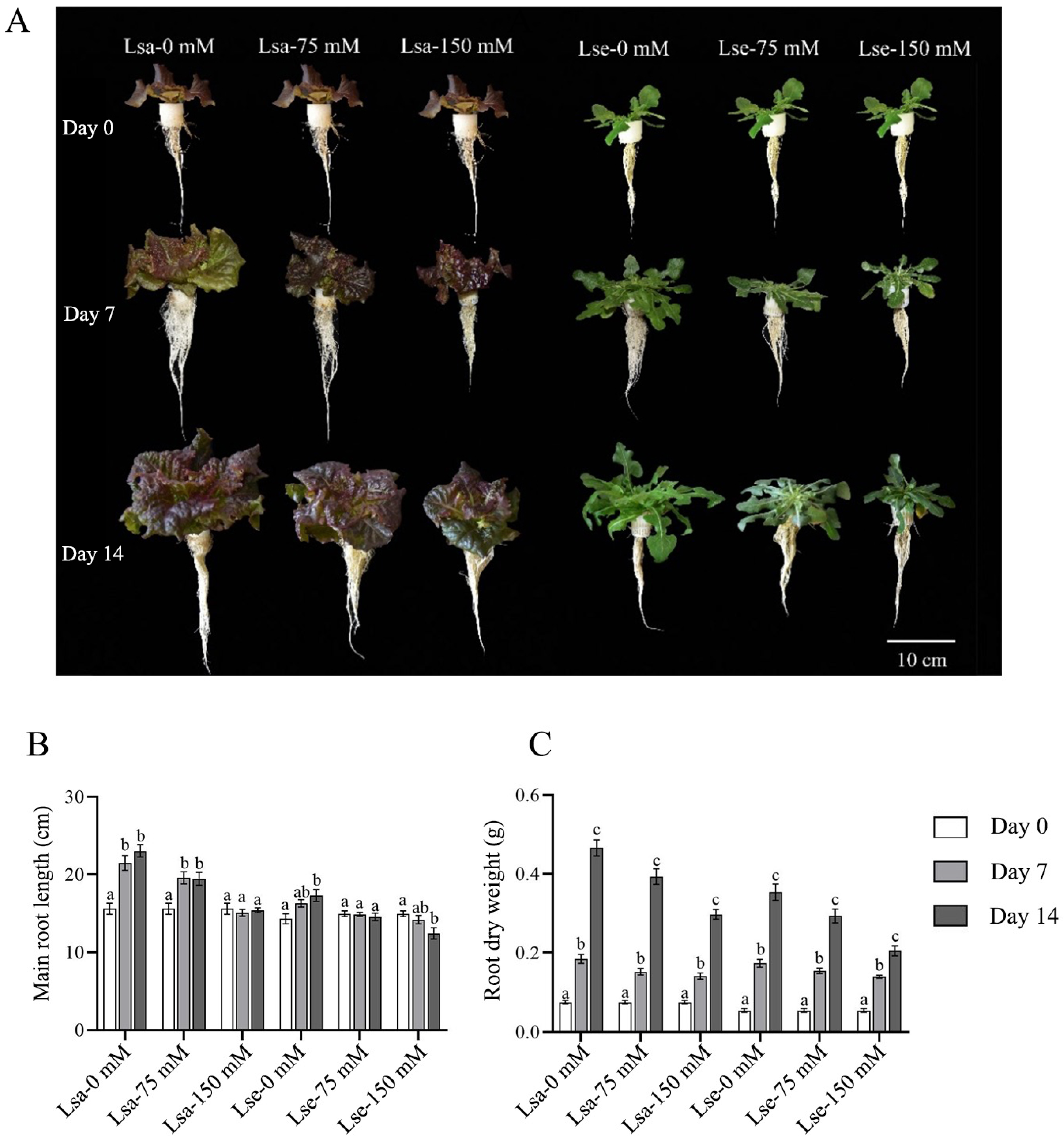
2.1. Variations in the Phenotypic Traits of Lettuce During Salt Stress Treatment
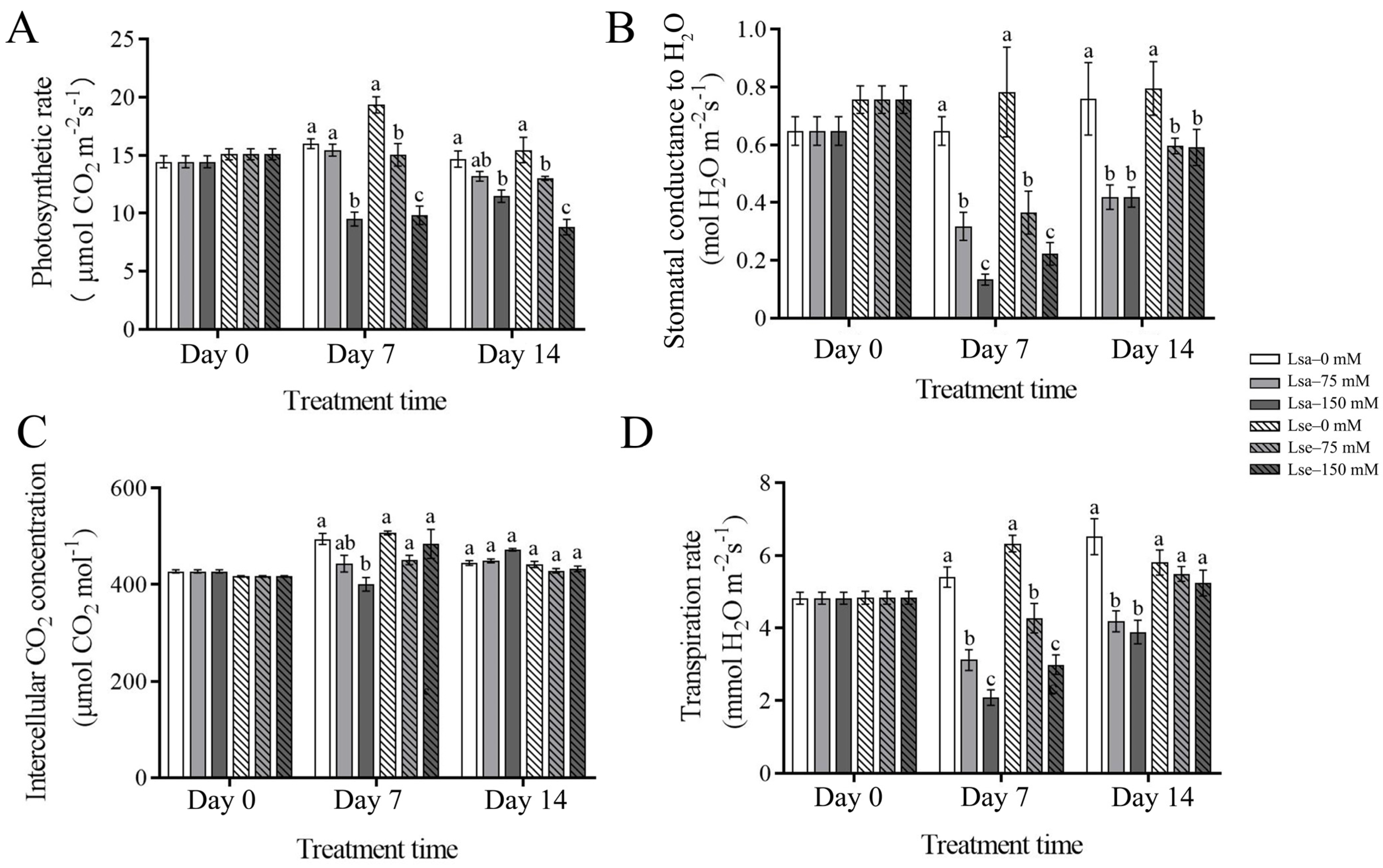
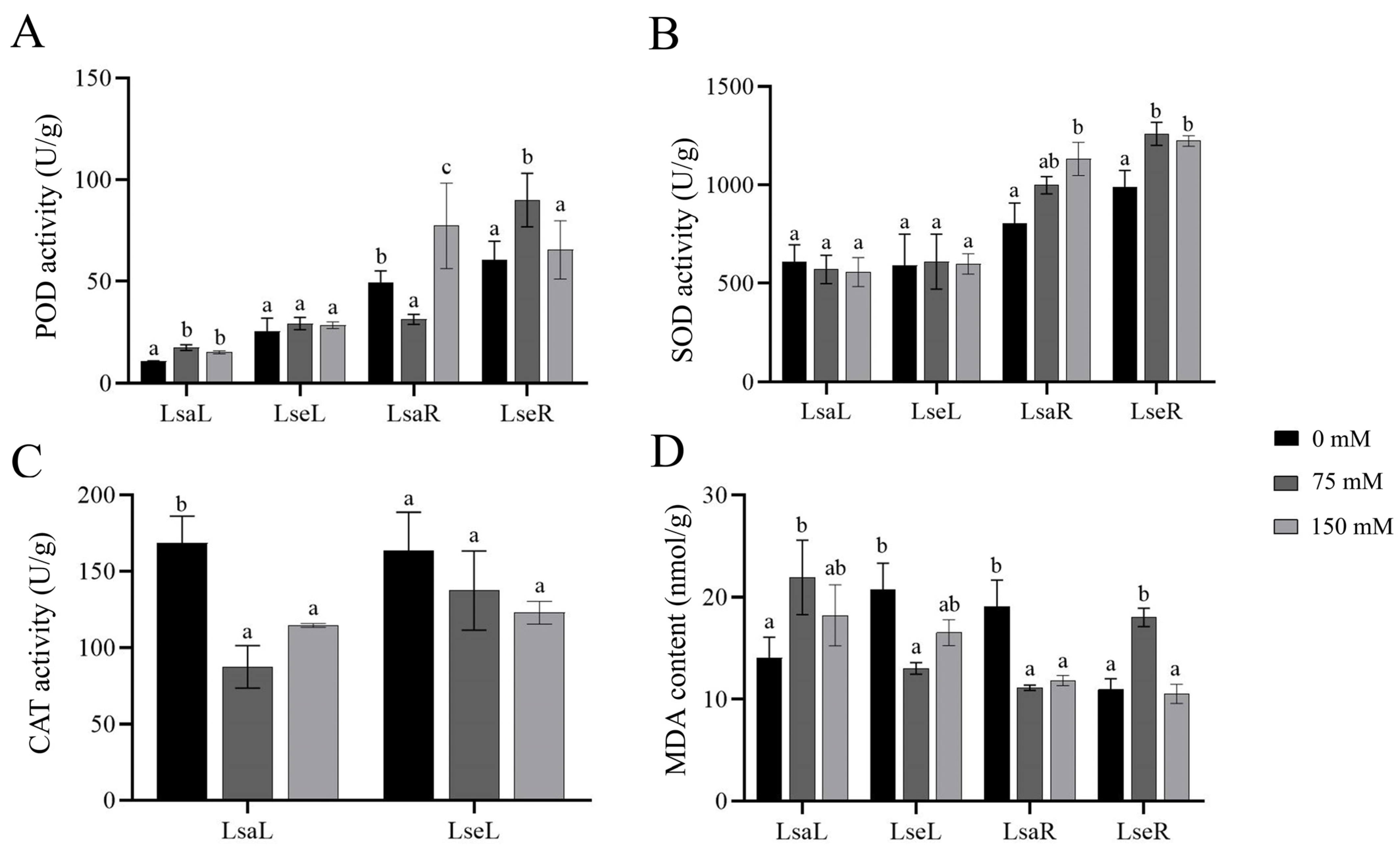
2.2. Physiological Changes in the Photosynthetic Traits, Antioxidant Enzymatic Activity, and Proline and Soluble Sugar Contents of Wild and Cultivated Lettuces Under the Salt Treatments
2.3. The Identification of Differentially Expressed Genes in Lettuce Under Normal and Salt Treatment Conditions
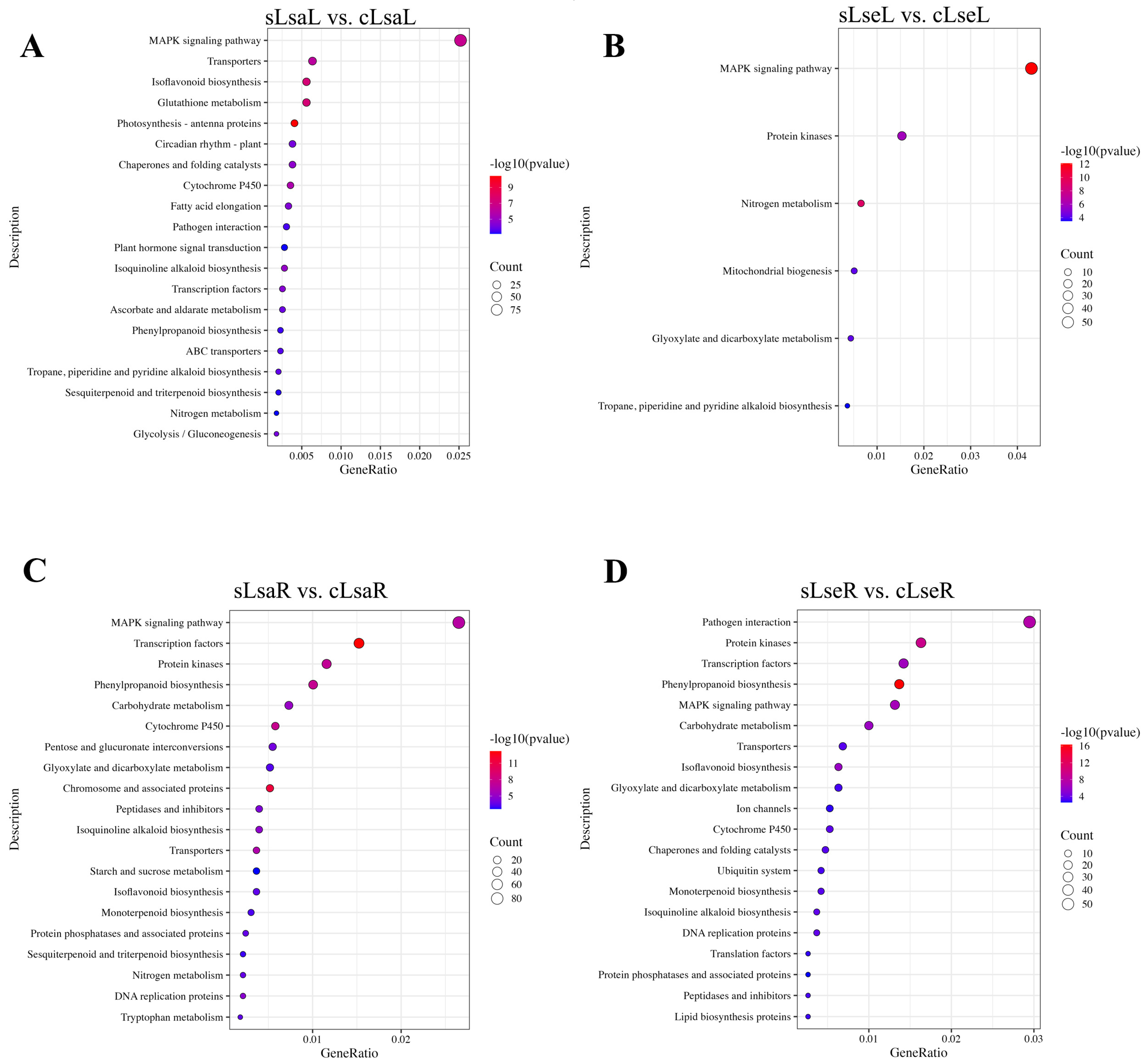
2.4. Functional Annotation of the DEGs
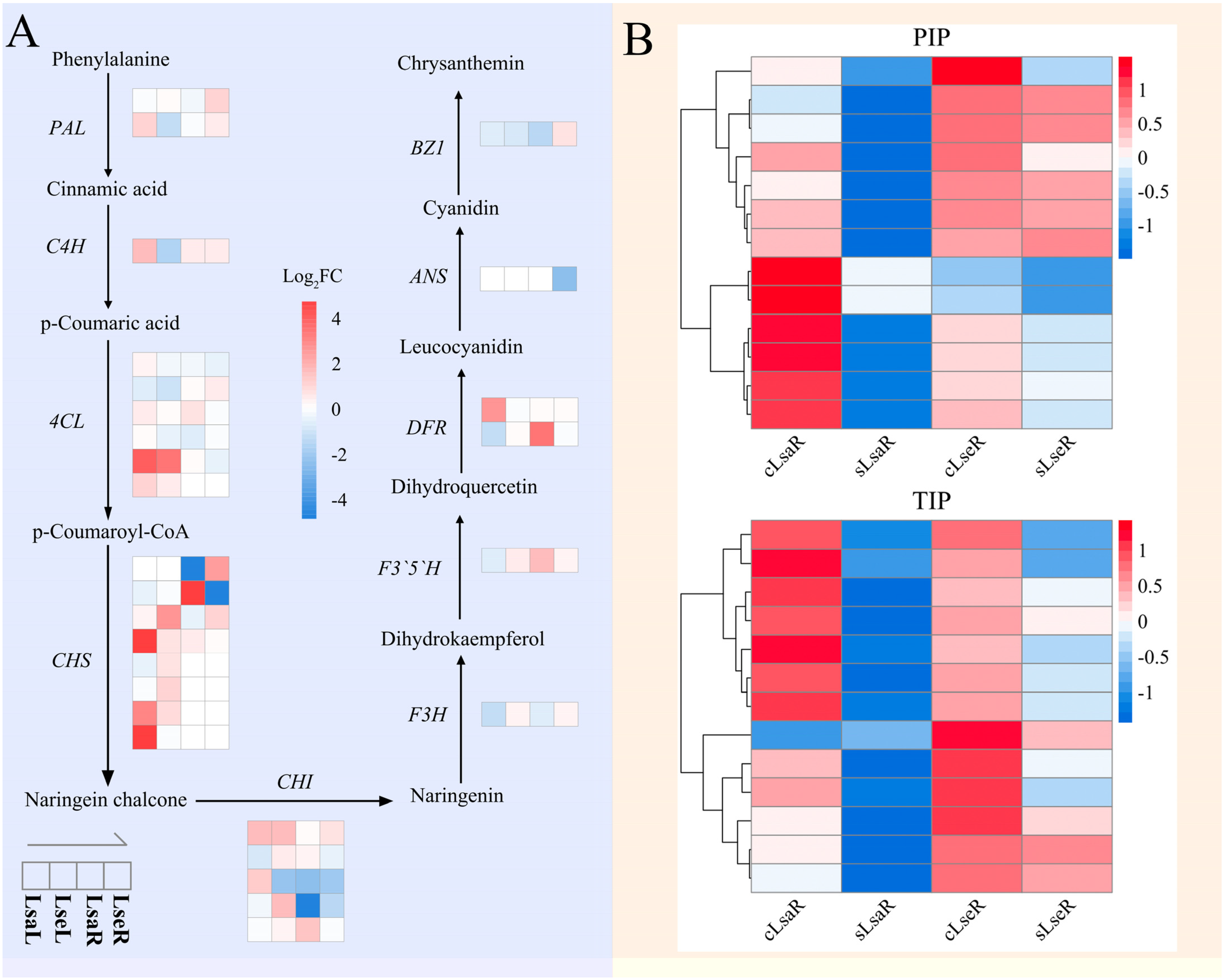
2.5. The Key DEGs Involved in Salt Stress
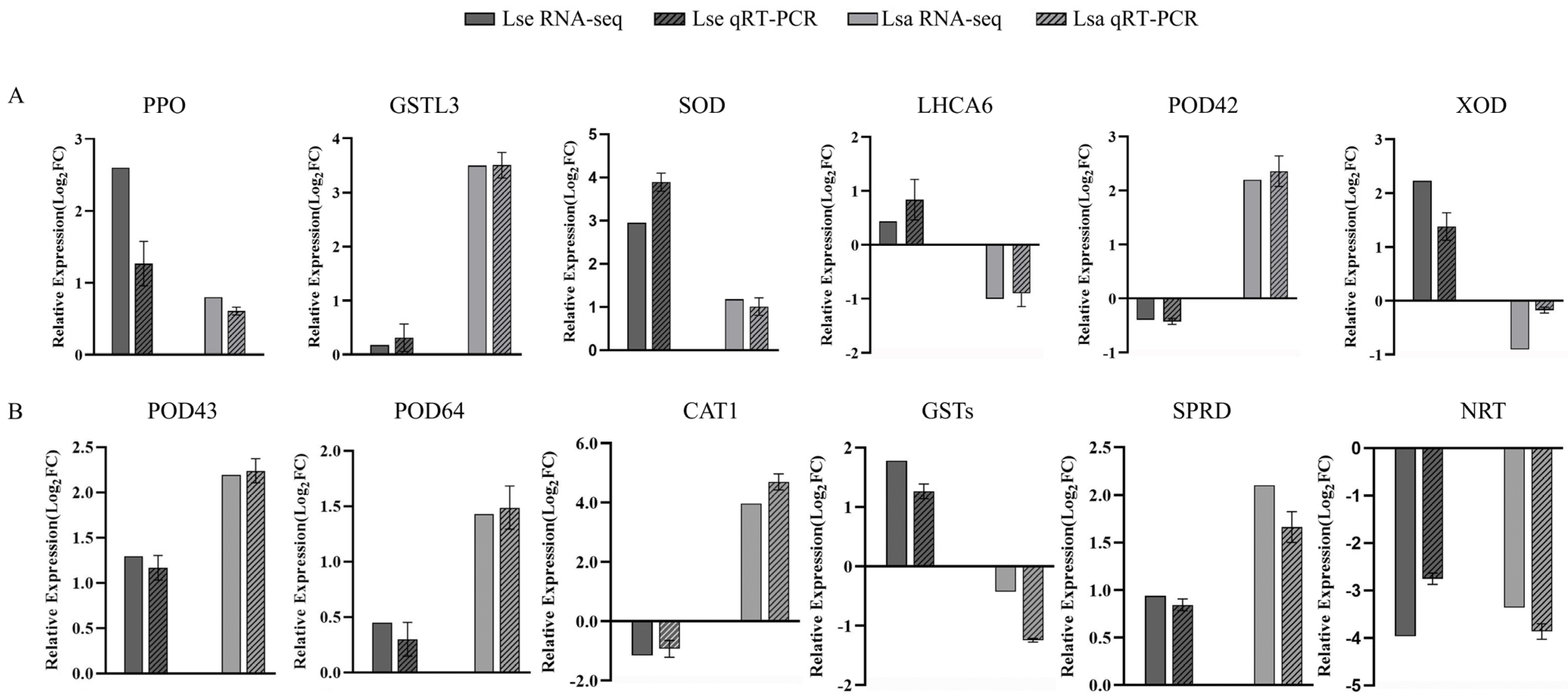
2.6. qRT-PCR Validation of the DEGs
3. Discussion
4. Materials and Methods
4.1. The Plant Materials and Salt Stress Treatments
4.2. Measurements of Their Phenotypic and Photosynthetic Traits, Antioxidant Enzymatic Activity, and Proline and Soluble Sugar Contents
4.3. RNA Extraction, Library Preparation, and RNA Sequencing
4.4. Quality Control and Transcriptome Assembly
4.5. The Identification of Differential Expression Genes and the Functional Enrichment Analysis
4.6. Quantitative Real-Time PCR Validation
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Lsa | L. sativa ‘Lollo Rosso’ |
| Lse | L. serriola |
| LsaL | L. sativa ‘Lollo Rosso’ leaf |
| LsaR | L. sativa ‘Lollo Rosso’ root |
| LseL | L. serriola leaf |
| LseR | L. serriola root |
| LA | Leaf area |
| LDW | Leaf dry weight |
| MRL | Main root length |
| RDW | Root dry weight |
References
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, B. Protection of Halophytes and Their Uses for Cultivation of Saline-Alkali Soil in China. Biology 2021, 10, 353. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef]
- Berthomieu, P.; Conéjéro, G.; Nublat, A.; Brackenbury, W.J.; Lambert, C.; Savio, C.; Uozumi, N.; Oiki, S.; Yamada, K.; Cellier, F.; et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003, 22, 2004–2014. [Google Scholar]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Khare, T.; Sharma, M.; Wani, S.H. ROS-Induced Signaling and Gene Expression in Crops Under Salinity Stress; Springer: Singapore, 2017; pp. 159–184. [Google Scholar]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Cao, X.; Khan, M.A.R. Proline, a multifaceted signalling molecule in plant responses to abiotic stress: Understanding the physiological mechanisms. Plant Biol. J. 2022, 24, 227–239. [Google Scholar] [CrossRef]
- Khan, A.; Kamran, M.; Imran, M.; Al-Harrasi, A.; Al-Rawahi, A.; Al-Amri, I.; Lee, I.-J.; Khan, A.L. Silicon and salicylic acid confer high-pH stress tolerance in tomato seedlings. Sci. Rep. 2019, 9, 19788. [Google Scholar] [CrossRef]
- Niu, M.; Xie, J.; Chen, C.; Cao, H.; Sun, J.; Kong, Q.; Shabala, S.; Shabala, L.; Huang, Y.; Bie, Z. An early ABA-induced stomatal closure, Na+ sequestration in leaf vein and K+ retention in mesophyll confer salt tissue tolerance in Cucurbita species. J. Exp. Bot. 2018, 69, 4945–4960. [Google Scholar] [CrossRef]
- Valenzuela, C.E.; Acevedo-Acevedo, O.; Miranda, G.S.; Vergara-Barros, P.; Holuigue, L.; Figueroa, C.R.; Figueroa, P.M. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4209–4220. [Google Scholar] [CrossRef]
- Chu, R.; Xu, X.; Lu, Z.; Ma, Y.; Cheng, H.; Zhu, S.; Bakker, F.T.; Schranz, M.E.; Wei, Z. Plastome-based phylogeny and biogeography of Lactuca L. (Asteraceae) support revised lettuce gene pool categories. Front. Plant Sci. 2022, 13, 978417. [Google Scholar] [CrossRef] [PubMed]
- Baslam, M.; Pascual, I.; Sánchez-Díaz, M.; Erro, J.; García-Mina, J.M.; Goicoechea, N. Improvement of Nutritional Quality of Greenhouse-Grown Lettuce by Arbuscular Mycorrhizal Fungi Is Conditioned by the Source of Phosphorus Nutrition. J. Agric. Food Chem. 2011, 59, 11129–11140. [Google Scholar] [CrossRef]
- Camejo, D.; Frutos, A.; Mestre, T.C.; Del Carmen Piñero, M.; Rivero, R.M.; Martínez, V. Artificial light impacts the physical and nutritional quality of lettuce plants. Hortic. Environ. Biotechnol. 2020, 61, 69–82. [Google Scholar] [CrossRef]
- Adhikari, N.D.; Simko, I.; Mou, B. Phenomic and Physiological Analysis of Salinity Effects on Lettuce. Sensors 2019, 19, 4814. [Google Scholar] [CrossRef]
- Kalhor, M.S.; Aliniaeifard, S.; Seif, M.; Asayesh, E.J.; Bernard, F.; Hassani, B.; Li, T. Title: Enhanced salt tolerance and photosynthetic performance: Implication of ɤ-amino butyric acid application in salt-exposed lettuce (Lactuca sativa L.) plants. Plant Physiol. Biochem. 2018, 130, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Miras-Moreno, B.; Yildiztugay, E.; Ozfidan-Konakci, C.; Arikan, B.; Elbasan, F.; Ak, G.; Rouphael, Y.; Zengin, G.; Lucini, L. Metabolomics and Physiological Insights into the Ability of Exogenously Applied Chlorogenic Acid and Hesperidin to Modulate Salt Stress in Lettuce Distinctively. Molecules 2021, 26, 6291. [Google Scholar] [CrossRef]
- van Treuren, R.; Coquin, P.; Lohwasser, U. Genetic resources collections of leafy vegetables (lettuce, spinach, chicory, artichoke, asparagus, lamb’s lettuce, rhubarb and rocket salad): Composition and gaps. Genet. Resour. Crop Evol. 2012, 59, 981–997. [Google Scholar] [CrossRef]
- Wei, Z.; Julkowska, M.M.; Laloë, J.-O.; Hartman, Y.; de Boer, G.-J.; Michelmore, R.W.; van Tienderen, P.H.; Testerink, C.; Schranz, M.E. A mixed-model QTL analysis for salt tolerance in seedlings of crop-wild hybrids of lettuce. Mol. Breed. 2014, 34, 1389–1400. [Google Scholar] [CrossRef]
- Chalecka, M.; Kazberuk, A.; Palka, J.; Surazynski, A. P5C as an Interface of Proline Interconvertible Amino Acids and Its Role in Regulation of Cell Survival and Apoptosis. Int. J. Mol. Sci. 2021, 22, 11763. [Google Scholar] [CrossRef]
- Basilio Guimarães, R.F.; Nascimento, R.D.; Melo, D.F.D.; Ramos, J.G.; Pereira, M.D.O.; Cardoso, J.A.F.; De Lima, S.C. Production of Hydroponic Lettuce under Different Salt Levels of Nutritive Solution. J. Agric. Sci. 2017, 9, 242. [Google Scholar] [CrossRef][Green Version]
- Shin, Y.K.; Bhandari, S.R.; Jo, J.S.; Song, J.W.; Cho, M.C.; Yang, E.Y.; Lee, J.G. Response to Salt Stress in Lettuce: Changes in Chlorophyll Fluorescence Parameters, Phytochemical Contents, and Antioxidant Activities. Agronomy 2020, 10, 1627. [Google Scholar] [CrossRef]
- Blankenship, R.E. Early Evolution of Photosynthesis. Plant Physiol. 2010, 154, 434–438. [Google Scholar] [PubMed]
- Hasanuzzaman, M.; Raihan, M.d.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of Reactive Oxygen Species and Antioxidant Defense in Plants under Salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Meng, C.; Zhang, K.; Wang, K.; Fan, H.; Li, Y. Study on Physiological Mechanism of Using Cottonseed Meal to Improve Salt–Alkali Tolerance of Cotton. J. Plant Growth Regul. 2021, 40, 126–136. [Google Scholar] [CrossRef]
- Gharaghanipor, N.; Arzani, A.; Rahimmalek, M.; Ravash, R. Physiological and Transcriptome Indicators of Salt Tolerance in Wild and Cultivated Barley. Front. Plant Sci. 2022, 13, 819282. [Google Scholar]
- Zeng, Y.; Li, L.; Yang, R.; Yi, X.; Zhang, B. Contribution and distribution of inorganic ions and organic compounds to the osmotic adjustment in Halostachys caspica response to salt stress. Sci. Rep. 2015, 5, 13639. [Google Scholar]
- Saddhe, A.A.; Manuka, R.; Penna, S. Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol. Plant. 2021, 171, 739–755. [Google Scholar]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar]
- Kumar, K.; Raina, S.K.; Sultan, S.M. Arabidopsis MAPK signaling pathways and their cross talks in abiotic stress response. J. Plant Biochem. Biotechnol. 2020, 29, 700–714. [Google Scholar]
- Jia, M.; Luo, N.; Meng, X.; Song, X.; Jing, Y.; Kou, L.; Liu, G.; Huang, X.; Wang, Y.; Li, J.; et al. OsMPK4 promotes phosphorylation and degradation of IPA1 in response to salt stress to confer salt tolerance in rice. J. Genet. Genom. 2022, 49, 766–775. [Google Scholar]
- Sun, Q.; Liu, X.; Kitagawa, Y.; Calamita, G.; Ding, X. Plant aquaporins: Their roles beyond water transport. Crop J. 2024, 12, 641–655. [Google Scholar] [CrossRef]
- Byrt, C.S.; Zhao, M.; Kourghi, M.; Bose, J.; Henderson, S.W.; Qiu, J.; Gilliham, M.; Schultz, C.; Schwarz, M.; Ramesh, S.A.; et al. Non-selective cation channel activity of aquaporin AtPIP2;1 regulated by Ca2+ and pH. Plant Cell Environ. 2017, 40, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Hooijmaijers, C.; Rhee, J.Y.; Kwak, K.J.; Chung, G.C.; Horie, T.; Katsuhara, M.; Kang, H. Hydrogen peroxide permeability of plasma membrane aquaporins of Arabidopsis thaliana. J. Plant Res. 2012, 125, 147–153. [Google Scholar] [CrossRef]
- Kumari, A.; Bhatla, S.C. Regulation of salt-stressed sunflower (Helianthus annuus) seedling’s water status by the coordinated action of Na+/K+ accumulation, nitric oxide, and aquaporin expression. Funct. Plant Biol. 2021, 48, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gil, M.I.; Yang, Q.; Tomas-Barberan, F.A. Bioactive compounds in lettuce: Highlighting the benefits to human health and impacts of preharvest and postharvest practices. Compr. Rev. Food Sci. Food Saf. 2021, 21, 4–45. [Google Scholar] [CrossRef]
- Shi, M.; Gu, J.; Wu, H.; Rauf, A.; Emran, T.B.; Khan, Z.; Mitra, S.; Aljohani, A.S.M.; Alhumaydhi, F.A.; Al-Awthan, Y.S.; et al. Phytochemicals, Nutrition, Metabolism, Bioavailability, and Health Benefits in Lettuce—A Comprehensive Review. Antioxid. 2022, 11, 1158. [Google Scholar] [CrossRef]
- Souza, A.S.N.; Schmidt, H.O.; Pagno, C.; Rodrigues, E.; Silva, M.; Flores, S.H.; Rios, A.O. Influence of cultivar and season on carotenoids and phenolic compounds from red lettuce influence of cultivar and season on lettuce. Food Res. Int. 2022, 155, 111110. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Tak, Y.; Kaur, M.; Gautam, C.; Kumar, R.; Tilgam, J.; Natta, S. Phenolic biosynthesis and metabolic pathways to alleviate stresses in plants. In Plant Phenolics in Abiotic Stress Management; Springer Nature: Singapore, 2023; pp. 63–87. [Google Scholar]
- Marchiosi, R.; dos Santos, W.D.; Constantin, R.P.; de Lima, R.B.; Soares, A.R.; Finger-Teixeira, A.; Mota, T.R.; de Oliveira, D.M.; Foletto-Felipe, M.d.P.; Abrahão, J.; et al. Biosynthesis and metabolic actions of simple phenolic acids in plants. Phytochem. Rev. 2020, 19, 865–906. [Google Scholar] [CrossRef]
- Ramaroson, M.-L.; Koutouan, C.; Helesbeux, J.-J.; Le Clerc, V.; Hamama, L.; Geoffriau, E.; Briard, M. Role of Phenylpropanoids and Flavonoids in Plant Resistance to Pests and Diseases. Molecules 2022, 27, 8371. [Google Scholar] [CrossRef]
- Ren, H.; Yang, W.; Jing, W.; Shahid, M.O.; Liu, Y.; Qiu, X.; Choisy, P.; Xu, T.; Ma, N.; Gao, J.; et al. Multi-omics analysis reveals key regulatory defense pathways and genes involved in salt tolerance of rose plants. Hortic. Res. 2024, 11, uhae068. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Tohge, T.; Winkler, J.B.; Albert, A.; Schäffner, A.R.; Fernie, A.R.; Zuther, E.; Hincha, D.K. Natural Variation among Arabidopsis Accessions in the Regulation of Flavonoid Metabolism and Stress Gene Expression by Combined UV Radiation and Cold. Plant Cell Physiol. 2021, 62, 502–514. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhou, Y.; Liu, X.; Meng, F.; Xu, C.; Chen, M. Integrated transcriptomic and metabolomic analyses uncover the key pathways of Limonium bicolor in response to salt stress. Plant Biotechnol. J. 2025, 23, 715–730. [Google Scholar] [CrossRef]
- Medina-Lozano, I.; Bertolín, J.R.; Díaz, A. Impact of drought stress on vitamin C and anthocyanin content in cultivated lettuces (Lactuca sativa L.) and wild relatives (Lactuca spp.). Front. Plant Sci. 2024, 15, 1369658. [Google Scholar] [CrossRef]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant cytochrome P450 plasticity and evolution. Mol. Plant 2021, 14, 1244–1265. [Google Scholar] [CrossRef]
- De Bruyn, C.; Ruttink, T.; Lacchini, E.; Rombauts, S.; Haegeman, A.; De Keyser, E.; Van Poucke, C.; Desmet, S.; Jacobs, T.B.; Eeckhaut, T.; et al. Identification and characterization of CYP71 subclade cytochrome P450 enzymes involved in the biosynthesis of bitterness compounds in Cichorium intybus. Front. Plant Sci. 2023, 14, 1200253. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Zhou, H.; Ashworth, K.; Dodd, I.C.; Foyer, C. Exogenous monoterpenes mitigate H2O2-induced lipid damage but do not attenuate photosynthetic decline during water deficit in tomato. J. Exp. Bot. 2023, 74, 5327–5340. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [PubMed]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS. Comput. Biol. 2022, 18, e1009730. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 2014, 15, 550. [Google Scholar]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Lian, J.; Hong, C.; Sun, S.; Hao, J.; Huang, S.; Wang, J.; Guan, Y.; Lu, Z.; Wang, Z.; et al. Phenotypic, Physiological, and Transcriptomic Analyses Reveal Different Responses to Salt Stress in Cultivated Red Lettuce and Wild Lettuce Seedlings. Int. J. Mol. Sci. 2025, 26, 3425. https://doi.org/10.3390/ijms26073425
Chen W, Lian J, Hong C, Sun S, Hao J, Huang S, Wang J, Guan Y, Lu Z, Wang Z, et al. Phenotypic, Physiological, and Transcriptomic Analyses Reveal Different Responses to Salt Stress in Cultivated Red Lettuce and Wild Lettuce Seedlings. International Journal of Molecular Sciences. 2025; 26(7):3425. https://doi.org/10.3390/ijms26073425
Chicago/Turabian StyleChen, Wei, Jiahao Lian, Caiyun Hong, Shuguang Sun, Jia Hao, Shengqi Huang, Jialin Wang, Yue Guan, Zhenwei Lu, Zhenlong Wang, and et al. 2025. "Phenotypic, Physiological, and Transcriptomic Analyses Reveal Different Responses to Salt Stress in Cultivated Red Lettuce and Wild Lettuce Seedlings" International Journal of Molecular Sciences 26, no. 7: 3425. https://doi.org/10.3390/ijms26073425
APA StyleChen, W., Lian, J., Hong, C., Sun, S., Hao, J., Huang, S., Wang, J., Guan, Y., Lu, Z., Wang, Z., Zhu, S., & Wei, Z. (2025). Phenotypic, Physiological, and Transcriptomic Analyses Reveal Different Responses to Salt Stress in Cultivated Red Lettuce and Wild Lettuce Seedlings. International Journal of Molecular Sciences, 26(7), 3425. https://doi.org/10.3390/ijms26073425




