Comparative Analysis of the Chloroplast Genomes of Cypripedium: Assessing the Roles of SSRs and TRs in the Non-Coding Regions of LSC in Shaping Chloroplast Genome Size
Abstract
1. Introduction
2. Results
2.1. Chloroplast Genomes of Cypripedium
2.2. Comparative Analysis of Chloroplast Genome Structure
2.3. Repeat Sequence Analysis
2.4. Gene Length Correlations
2.5. Phylogenetic Relationships
2.6. Phylogenetic Signal Analysis
3. Discussion
3.1. Characterization of Organellar Genomes
3.2. The Expansion and Contraction of Organellar Genomes
3.3. Phylogenetic Relationships in Organellar Genomes
4. Materials and Methods
4.1. Data Acquisition
4.2. Comparison of Chloroplast Genome Sequences
4.3. Analysis of Repeat Sequences
4.4. Correlation Analysis
4.5. Phylogenetic Analysis
4.6. Phylogenetic Signal Test
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LSC | Large single-copy region |
| SSC | Small single-copy region |
| IR | Inverted repeat |
| CDS | Coding sequence |
| NCDS | Non-coding sequence |
| SSRs | Simple sequence repeats |
| TRs | Tandem repeats (TRs) |
References
- Chase, M.W. Classification of Orchidaceae in the age of DNA data. Curtis’s Bot. Mag. 2005, 22, 2–7. [Google Scholar] [CrossRef]
- Szlachetko, D.L.; Górniak, M.; Kowalkowska, A.K.; Kolanowska, M.; Jurczak-Kurek, A.; Morales, F.A. The natural history of the genus Cypripedium (Orchidaceae). Plant Biosyst. 2020, 155, 772–796. [Google Scholar] [CrossRef]
- Chen, S.Q.; Liu, Z.J.; Chen, L.J.; Li, L.Q. The Genus Cypripedium in China; Science Press: Beijing, China, 2012; pp. 10–12. [Google Scholar]
- Liu, H.C.; Jacquemyn, H.; Chen, W.; Janssens, S.B.; He, X.Y.; Yu, S.; Huang, Y.Q. Niche evolution and historical biogeography of lady slipper orchids in North-American and Eurasia. J. Biogeogr. 2021, 48, 2727–2741. [Google Scholar] [CrossRef]
- Filippov, E.G.; Andronova, E.V. Genetic Differentiation in Plants of the Genus Cypripedium from Russia Inferred from Allozyme Data. Russ. J. Genet. 2011, 47, 538–545. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Yang, J.X.; Bai, M.Z.; Zhang, G.Q.; Liu, Z.J. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021, 21, 248. [Google Scholar] [CrossRef]
- Liao, M.; Zhang, J.Y.; Feng, Y.; Ren, Z.X.; Deng, H.N.; Xu, B. Phylogenomic insights into the historical biogeography, character-state evolution, and species diversification rates of Cypripedioideae (Orchidaceae). Mol. Phylogenet. Evol. 2024, 199, 108138. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Liao, M.; Cheng, Y.H.; Feng, Y.; Ju, W.B.; Deng, H.N.; Li, X.; Plenkovic-Moraj, A.; Xu, B. Comparative Chloroplast Genomics of Seven Endangered Cypripedium Species and Phylogenetic Relationships of Orchidaceae. Front. Plant Sci. 2022, 13, 911702. [Google Scholar] [CrossRef]
- Lagou, L.J.; Kadereit, G.; Morales-Briones, D.F. Phylogenomic analysis of target enrichment and transcriptome data uncovers rapid radiation and extensive hybridization in the slipper orchid genus Cypripedium. Ann. Bot. 2024, 134, 1229–1250. [Google Scholar] [CrossRef]
- Guo, F.C.; Yang, J.H.; Guo, Y.Y. The plastomes of Cypripedium (Orchidaceae: Cypripedioideae) exhibit atypical GC content and genome size based on different sequencing strategies. Gene 2025, 935, 149086. [Google Scholar] [CrossRef]
- Finkemeier, I.; Leister, D. Plant chloroplasts and other plastids. In Encyclopedia of Life Sciences (ELS); John Wiley & Sons, Ltd.: Chichester, UK, 2010. [Google Scholar] [CrossRef]
- Green, B.R. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011, 66, 34–44. [Google Scholar] [CrossRef]
- Mogensen, H.L. The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot. 1996, 83, 383–404. [Google Scholar] [CrossRef]
- Ravi, V.; Khurana, J.P.; Tyagi, A.K.; Khurana, P. An update on chloroplast genomes. Plant Syst. Evol. 2008, 271, 101–122. [Google Scholar] [CrossRef]
- Rogalski, M.; Vieira, L.D.; Fraga, H.P.; Guerra, M.P. Plastid genomics in horticultural species: Importance and applications for plant population genetics, evolution, and biotechnology. Front. Plant Sci. 2016, 6, 586. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Zhang, Y.Q.; Zhang, G.Q.; Huang, L.Q.; Liu, Z.J. Comparative transcriptomics provides insight into the molecular basis of species diversification of section Trigonopedia (Cypripedium) on the Qinghai-Tibetan Plateau. Sci. Rep. 2018, 8, 11640. [Google Scholar] [CrossRef]
- Li, Y.; Lu, X.; Li, S.; Sun, Y.; Shan, Y.; Wang, S.; Jiang, N.; Xiao, Y.; Wang, Q.; Yu, J.; et al. Unveiling a Meaningful Form of Cypripedium × ventricosum Sw. (Cypripedioideae, Orchidaceae) from Changbai Mountain, China: Insights from Morphological, Molecular, and Plastome Analyses. Plants 2025, 14, 772. [Google Scholar] [CrossRef]
- Jo, S.; Ochiai, M.; Furuta, K.; Yagi, K. Genetic analyses of genus Cypripedium found in northern Japanese Islands and related species endemic to northeast China. J. Jpn. Soc. Hortic. Sci. 2005, 74, 234–241. [Google Scholar] [CrossRef][Green Version]
- Li, J.H.; Liu, Z.J.; Salazar, G.A.; Bernhardt, P.; Perner, H.; Tomohisa, Y.; Jin, X.H.; Chung, S.W.; Luo, Y.B. Molecular phylogeny of Cypripedium (Orchidaceae: Cypripedioideae) inferred from multiple nuclear and chloroplast regions. Mol. Phylogenet. Evol. 2011, 61, 308–320. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Minasiewicz, J.; Znaniecka, J.M.; Górniak, M.; Kawinski, A. Spatial genetic structure of an endangered orchid Cypripedium calceolus (Orchidaceae) at a regional scale: Limited gene flow in a fragmented landscape. Conserv. Genet. 2018, 19, 1449–1460. [Google Scholar] [CrossRef]
- Liu, H.C.; Jacquemyn, H.; He, X.Y.; Chen, W.; Huang, Y.Q.; Yu, S.; Lu, Y.P.; Zhang, Y. The Impact of Human Pressure and Climate Change on the Habitat Availability and Protection of Cypripedium (Orchidaceae) in Northeast China. Pants 2021, 10, 84. [Google Scholar] [CrossRef]
- Li, L.; Wang, W.Y.; Zhang, G.Q.; Wu, K.L.; Fang, L.; Li, M.Z.; Liu, Z.J.; Zeng, S.J. Comparative analyses and phylogenetic relationships of thirteen Pholidota species (Orchidaceae) inferred from complete chloroplast genomes. BMC Plant Biol. 2023, 23, 269. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Duan, H.N.; Tao, K.F.; Luo, Y.; Li, Q.Q.; Li, L. Complete chloroplast genome structural characterization of two Phalaenopsis (Orchidaceae) species and comparative analysis with their alliance. BMC Genom. 2023, 24, 359. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Li, J.F.; Zhang, H.; Cai, B.H.; Gao, Z.H.; Qiao, Y.S.; Mi, L. The complete chloroplast genome sequence of strawberry (Fragaria × ananassa Duch.) and comparison with related species of Rosaceae. PeerJ 2017, 5, e3919. [Google Scholar] [CrossRef]
- Lan, T.Y.; Albert, V.A. Dynamic distribution patterns of ribosomal DNA and chromosomal evolution in Paphiopedilum.; a lady’s slipper orchid. BMC Plant Biol. 2011, 11, 126. [Google Scholar] [CrossRef]
- Bock, D.G.; Cai, Z.; Elphinstone, C.; González-Segovia, E.; Hirabayashi, K.; Huang, K.C.; Keais, G.L.; Kim, A.; Owens, G.L.; Rieseberg, L.H. Genomics of plant speciation. Plant Commun. 2023, 4, 5. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Joo, M.J.; Hong, J.R.; Kwak, M.; Kim, K.J. Plastome evolution and phylogeny of Orchidaceae, with 24 new sequences. Front. Plant Sci. 2020, 11, 22. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef]
- Weng, M.L.; Blazier, J.C.; Govindu, M.; Jansen, R.K. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 2014, 31, 645–659. [Google Scholar] [CrossRef]
- Cozzolino, S.; Cafasso, D.; Pellegrino, G.; Musacchio, A.; Widmer, A. Fine-scale phylogeographical analysis of Mediterranean Anacamptis palustris (Orchidaceae) populations based on chloroplast minisatellite and microsatellite variation. Mol. Ecol. 2003, 12, 2783–2792. [Google Scholar] [CrossRef]
- Ebert, D.; Hayes, C.; Peakall, R. Chloroplast simple sequence repeat markers for evolutionary studies in the sexually deceptive orchid genus Chiloglottis. Mol. Ecol. Resour. 2009, 9, 784–789. [Google Scholar] [CrossRef]
- Peakall, R.; Ebert, D.; Poldy, J.; Barrow, R.A.; Francke, W.; Bower, C.C.; Schiestl, F.P. Pollinator specificity, floral odour chemistry and the phylogeny of Australian sexually deceptive Chiloglottis orchids: Implications for pollinator-driven speciation. New Phytol. 2010, 188, 437–450. [Google Scholar] [CrossRef]
- Smidt, E.C.; Borba, E.L.; Gravendel, B.; Fischer, G.A.; van den Berg, C. Molecular phylogeny of the Neotropical sections of Bulbophyllum (Orchidaceae) using nuclear and plastid spacers. Taxon 2019, 60, 1050–1064. [Google Scholar] [CrossRef]
- Niu, Z.T.; Hou, Z.Y.; Wang, M.T.; Ye, M.R.; Zhang, B.H.; Xue, Q.Y.; Liu, W.; Ding, X.Y. A comparative plastomics approach reveals available molecular markers for the phylogeographic study of Dendrobium huoshanense, an endangered orchid with extremely small populations. Ecol. Evol. 2020, 10, 5332–5342. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.K.; Du, J.X.; Liu, Z.H.; Zuo, W.M.; Wang, Z.L.; Li, J.P.; Zeng, Y. Comparative and phylogenetic analyses of nine complete chloroplast genomes of Orchidaceae. Sci. Rep. 2023, 13, 21403. [Google Scholar] [CrossRef]
- Jiang, H.; Tian, J.; Yang, J.X.; Dong, X.; Zhong, Z.X.; Mwachala, G.; Zhang, C.F.; Hu, G.W.; Wang, Q.F. Comparative and phylogenetic analyses of six Kenya Polystachya (Orchidaceae) species based on the complete chloroplast genome sequences. BMC Plant Biol. 2022, 22, 177. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Hoffman, J.I.; Schielzeth, H. Comparative Analysis of Genomic Repeat Content in Gomphocerine Grasshoppers Reveals Expansion of Satellite DNA and Helitrons in Species with Unusually Large Genomes. Genome Biol. Evol. 2020, 12, 1180–1193. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.Y.; Ye, X.H.; Mei, Y.; He, K.; Li, F. Transposons and non-coding regions drive the intrafamily differences of genome size in insects. iScience 2022, 25, 104873. [Google Scholar] [CrossRef]
- Kashi, Y.; King, D.G. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006, 22, 253–259. [Google Scholar] [CrossRef]
- Abdullah, M.F.; Shahzadi, I.; Ali, Z.; Islam, M.; Naeem, M.; Mirza, B.; Lockhart, P.J.; Ahmed, I.; Waheed, M.T. Correlations among oligonucleotide repeats, nucleotide substitutions and insertion-deletion mutations in chloroplast genomes of plant family Malvaceae. J. Syst. Evol. 2021, 59, 388–402. [Google Scholar] [CrossRef]
- Wang, T.; Li, X.Z.; Tang, C.Y.; Cao, Z.F.; He, H.; Ma, X.P.; Li, Y.L.; De, K.J. Complete chloroplast genomes and phylogenetic relationships of Pedicularis chinensis and Pedicularis kansuensis. Sci. Rep. 2024, 14, 14357. [Google Scholar] [CrossRef]
- Brandham, P.E.; Pridgeon, A.M.; Cribb, P.J.; Chase, M.W.; Rasmussen, F.N. Cytogenetics. In Genera Orchidacearum, Volume 1: General introduction, Apostasioideae and Cypripedioideae; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Leitch, I.J.; Kahandawala, I.; Suda, J.; Fay, M.F. Genome size diversity in orchids: Consequences and evolution. Ann. Bot. 2009, 104, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. Plant genomes: Markers of evolutionary history and drivers of evolutionary change. Plants People Planet 2021, 3, 74–82. [Google Scholar] [CrossRef]
- Allen, J.F. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 10231–10238. [Google Scholar] [CrossRef]
- Klier, K.; Leoschke, M.J.; Wendel, J.F. Hybridization and Introgression in White and Yellow Ladyslipper Orchids (Cypripedium-candidum and C-pubescens). J. Hered. 1991, 82, 305–318. [Google Scholar] [CrossRef]
- Li, P.; Luo, Y.B. Reproductive biology of an endemic orchid Cypripedium smithii in China and reproductive isolation between C. smithii and C. tibeticum. Biodivers. Sci. 2009, 17, 406–413. [Google Scholar]
- Hu, S.J.; Hu, H.; Yan, N.; Huang, J.L.; Li, S.Y. Hybridization and asymmetric introgression between Cypripedium tibeticum and C. yunnanense in Shangrila County, Yunnan Province, China. Nord. J. Bot. 2011, 29, 625–631. [Google Scholar] [CrossRef]
- Tao, J.J.; Feng, C.; Ai, B.; Kang, M. Adaptive molecular evolution of the two-pore channel 1 gene TPC1 in the karst-adapted genus Primulina (Gesneriaceae). Ann. Bot. 2016, 118, 1257–1268. [Google Scholar] [CrossRef]
- Liu, Q.; Li, X.Y.; Li, M.Z.; Xu, W.K.; Schwarzacher, T.; Heslop-Harrison, J.S. Comparative chloroplast genome analyses of Avena: Insights into evolutionary dynamics and phylogeny. BMC Plant Biol. 2020, 20, 406. [Google Scholar] [CrossRef]
- Zhou, Y.Z.; Zheng, R.Y.; Peng, Y.K.; Chen, J.M.; Zhu, X.Y.; Xie, K.; Ahmad, S.; Chen, J.L.; Wang, F.; Shen, M.L.; et al. The first mitochondrial genome of Melastoma dodecandrum resolved structure evolution in Melastomataceae and micro inversions from inner horizontal gene transfer. Ind. Crops Prod. 2023, 205, 117390. [Google Scholar] [CrossRef]
- Li, H.E.; Guo, Q.Q.; Xu, L.; Gao, H.D.; Liu, L.; Zhou, X.Y. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 2023, 11, e15326. [Google Scholar] [CrossRef]
- Zheng, S.Y.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An Online Program for the Versatile Plotting of Organelle Genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenie. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Pennell, M.W.; Eastman, J.M.; Slater, G.J.; Brown, J.W.; Uyeda, J.C.; FitzJohn, R.G.; Alfaro, M.E.; Harmon, L.J. Geiger v2.0: An expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 2014, 30, 2216–2218. [Google Scholar] [CrossRef]
- Pagel, M. Inferring the historical patterns of biological evolution. Nature 1999, 401, 877–884. [Google Scholar] [CrossRef]
- Blomberg, S.P.; Garland, T.; Ives, A.R. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 2003, 57, 717–745. [Google Scholar] [CrossRef]
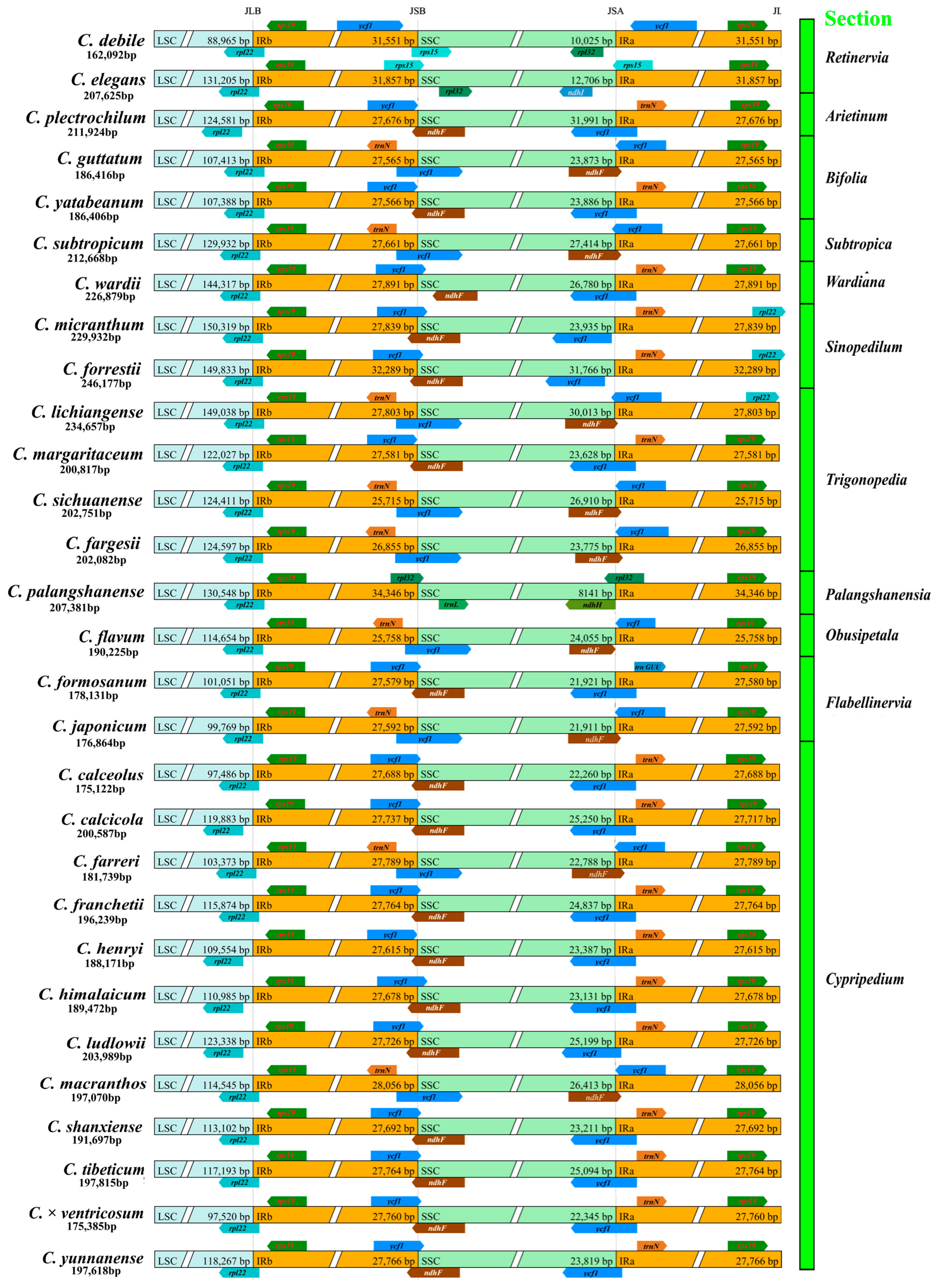
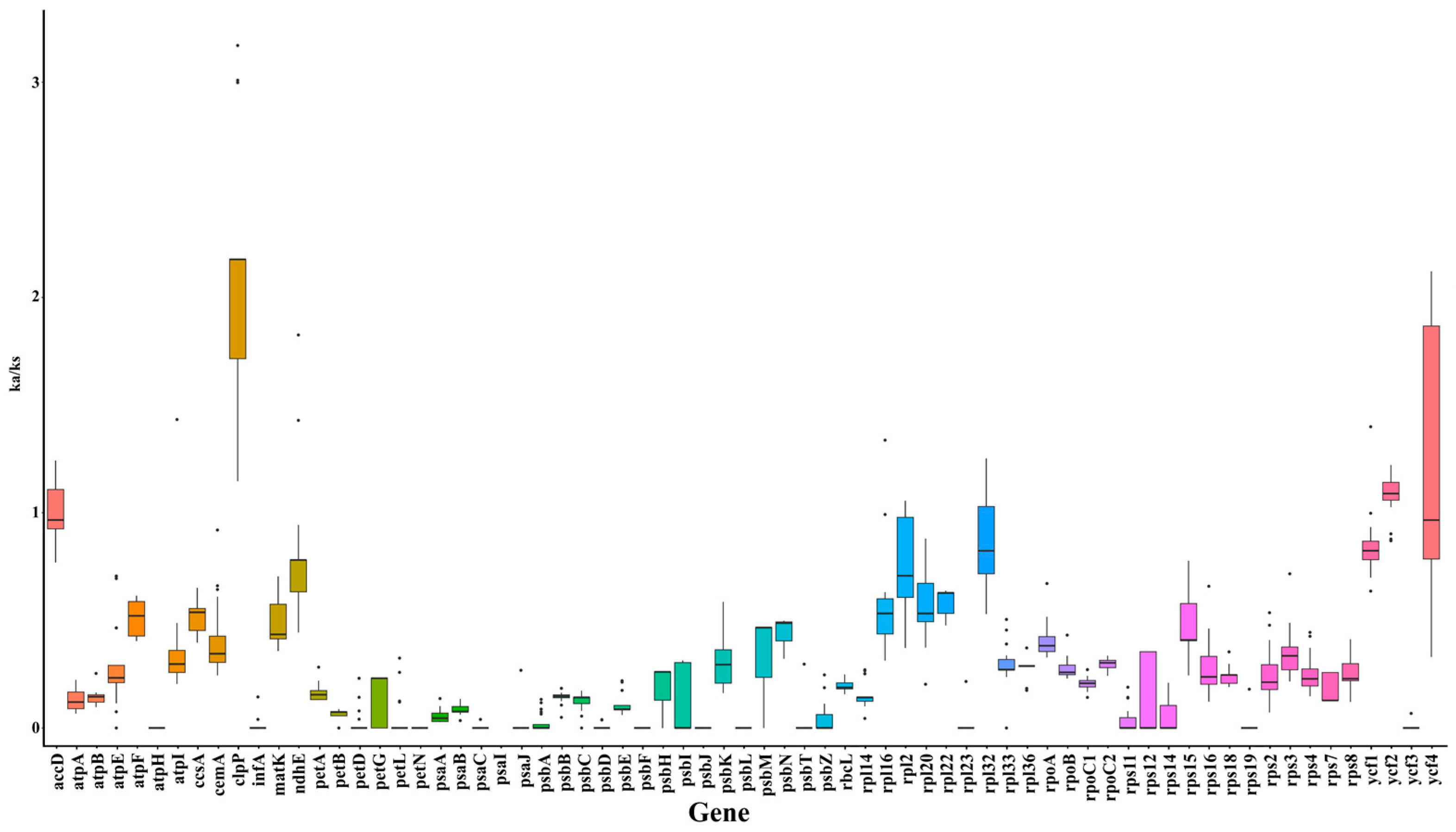


| Species | Total Length (bp) | GC Content (%) | Number of Genes | Protein Coding Genes | tRNA Genes | rRNA Genes | Pseudogenes | GenBank No. |
|---|---|---|---|---|---|---|---|---|
| C. debile | 162,092 | 35.4 | 128 | 76 | 8 | 38 | 6 | PP811663 |
| C. elegans | 207,625 | 27.7 | 127 | 81 | 8 | 38 | 2 | OR698927 |
| C. plectrochilum | 211,924 | 28.4 | 133 | 85 | 8 | 38 | 2 | PP811672 |
| C. guttatum | 186,416 | 32.4 | 133 | 86 | 8 | 38 | 1 | PP811666 |
| C. yatabeanum | 186,406 | 32.4 | 133 | 86 | 8 | 38 | 1 | PP811674 |
| C. subtropicum | 212,668 | 28.2 | 132 | 86 | 8 | 38 | 0 | MT937100 |
| C. wardii | 226,879 | 26.4 | 131 | 85 | 8 | 38 | 0 | OR698942 |
| C. micranthum | 229,932 | 26.7 | 133 | 88 | 8 | 37 | 0 | OR698938 |
| C. forrestii | 246,177 | 26.7 | 134 | 88 | 8 | 38 | 0 | OR698931 |
| C. lichiangense | 234,657 | 26 | 132 | 86 | 8 | 38 | 1 | NC_084419 |
| C. margaritaceum | 200,817 | 29.6 | 130 | 77 | 8 | 38 | 7 | PP811670 |
| C. sichuanense | 202,751 | 29.1 | 133 | 82 | 8 | 38 | 5 | PP811673 |
| C. fargesii | 202,082 | 29.2 | 132 | 86 | 8 | 38 | 0 | NC_084418 |
| C. palangshanense | 207,381 | 29.2 | 130 | 77 | 8 | 38 | 7 | PP811671 |
| C. flavum | 190,225 | 30.4 | 132 | 75 | 8 | 38 | 11 | PP811665 |
| C. formosanum | 178,131 | 33.9 | 134 | 87 | 8 | 39 | 0 | NC_026772 |
| C. japonicum | 176,864 | 34.1 | 133 | 85 | 8 | 38 | 2 | PP811668 |
| C. calceolus | 175,122 | 34.4 | 133 | 87 | 8 | 38 | 0 | NC_045400 |
| C. calcicola | 200,587 | 30 | 133 | 87 | 8 | 38 | 0 | OR698926 |
| C. farreri | 181,739 | 33.1 | 133 | 86 | 8 | 38 | 1 | PP811664 |
| C. franchetii | 196,239 | 30.7 | 133 | 87 | 8 | 38 | 0 | OR698932 |
| C. henryi | 188,171 | 32 | 133 | 85 | 8 | 38 | 2 | PP811667 |
| C. himalaicum | 189,472 | 31.8 | 133 | 87 | 8 | 38 | 0 | OR698935 |
| C. ludlowii | 203,989 | 29.6 | 133 | 87 | 8 | 38 | 0 | OR698937 |
| C. macranthos | 197,070 | 30.6 | 133 | 86 | 8 | 38 | 1 | PP811669 |
| C. shanxiense | 191,697 | 31.5 | 133 | 87 | 8 | 38 | 0 | OR698940 |
| C. tibeticum | 197,815 | 30.5 | 134 | 88 | 8 | 38 | 0 | NC_053552 |
| C. × ventricosum | 175,385 | 34.5 | 131 | 85 | 8 | 38 | 0 | PP448181 |
| C. yunnanense | 197,618 | 30.6 | 133 | 87 | 8 | 38 | 1 | OR698943 |
| Variables | Total Length | LSC Length | SSC Length | IR Length | CDS Length | NCDS Length | SSRs | TRs | Intron Length |
|---|---|---|---|---|---|---|---|---|---|
| K | 0.24 ** | 0.27 ** | 0.33 ** | 0.44 ** | 0.34 ** | 0.24 ** | 0.14 * | 0.52 ** | 0.15 ** |
| lambda | 0.90 ** | 0.92 ** | 0.90 ** | 0.89 ** | 0.95 ** | 0.88 ** | 0.86 ** | 0.95 ** | 0.93 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Jacquemyn, H.; Wang, Y.; Hu, Y.; He, X.; Zhang, Y.; Zhang, Y.; Huang, Y.; Chen, W. Comparative Analysis of the Chloroplast Genomes of Cypripedium: Assessing the Roles of SSRs and TRs in the Non-Coding Regions of LSC in Shaping Chloroplast Genome Size. Int. J. Mol. Sci. 2025, 26, 3691. https://doi.org/10.3390/ijms26083691
Liu H, Jacquemyn H, Wang Y, Hu Y, He X, Zhang Y, Zhang Y, Huang Y, Chen W. Comparative Analysis of the Chloroplast Genomes of Cypripedium: Assessing the Roles of SSRs and TRs in the Non-Coding Regions of LSC in Shaping Chloroplast Genome Size. International Journal of Molecular Sciences. 2025; 26(8):3691. https://doi.org/10.3390/ijms26083691
Chicago/Turabian StyleLiu, Huanchu, Hans Jacquemyn, Yanlin Wang, Yuanman Hu, Xingyuan He, Ying Zhang, Yue Zhang, Yanqing Huang, and Wei Chen. 2025. "Comparative Analysis of the Chloroplast Genomes of Cypripedium: Assessing the Roles of SSRs and TRs in the Non-Coding Regions of LSC in Shaping Chloroplast Genome Size" International Journal of Molecular Sciences 26, no. 8: 3691. https://doi.org/10.3390/ijms26083691
APA StyleLiu, H., Jacquemyn, H., Wang, Y., Hu, Y., He, X., Zhang, Y., Zhang, Y., Huang, Y., & Chen, W. (2025). Comparative Analysis of the Chloroplast Genomes of Cypripedium: Assessing the Roles of SSRs and TRs in the Non-Coding Regions of LSC in Shaping Chloroplast Genome Size. International Journal of Molecular Sciences, 26(8), 3691. https://doi.org/10.3390/ijms26083691







