NSP6 of SARS-CoV-2 Dually Regulates Autophagic–Lysosomal Degradation
Abstract
1. Introduction
2. Results
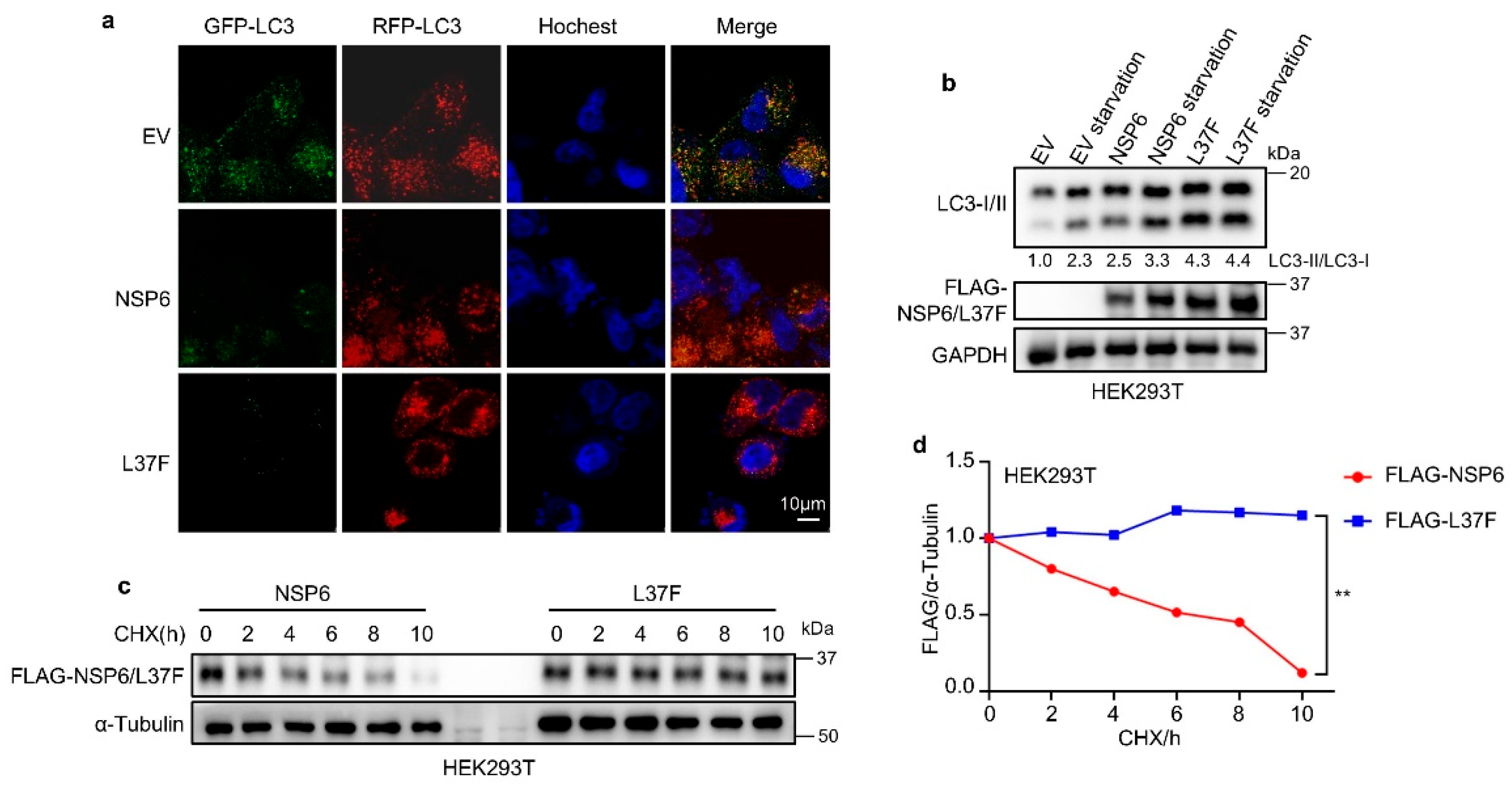
2.1. NSP6 and Its SNP L37F Promote the Initiation of Autophagy
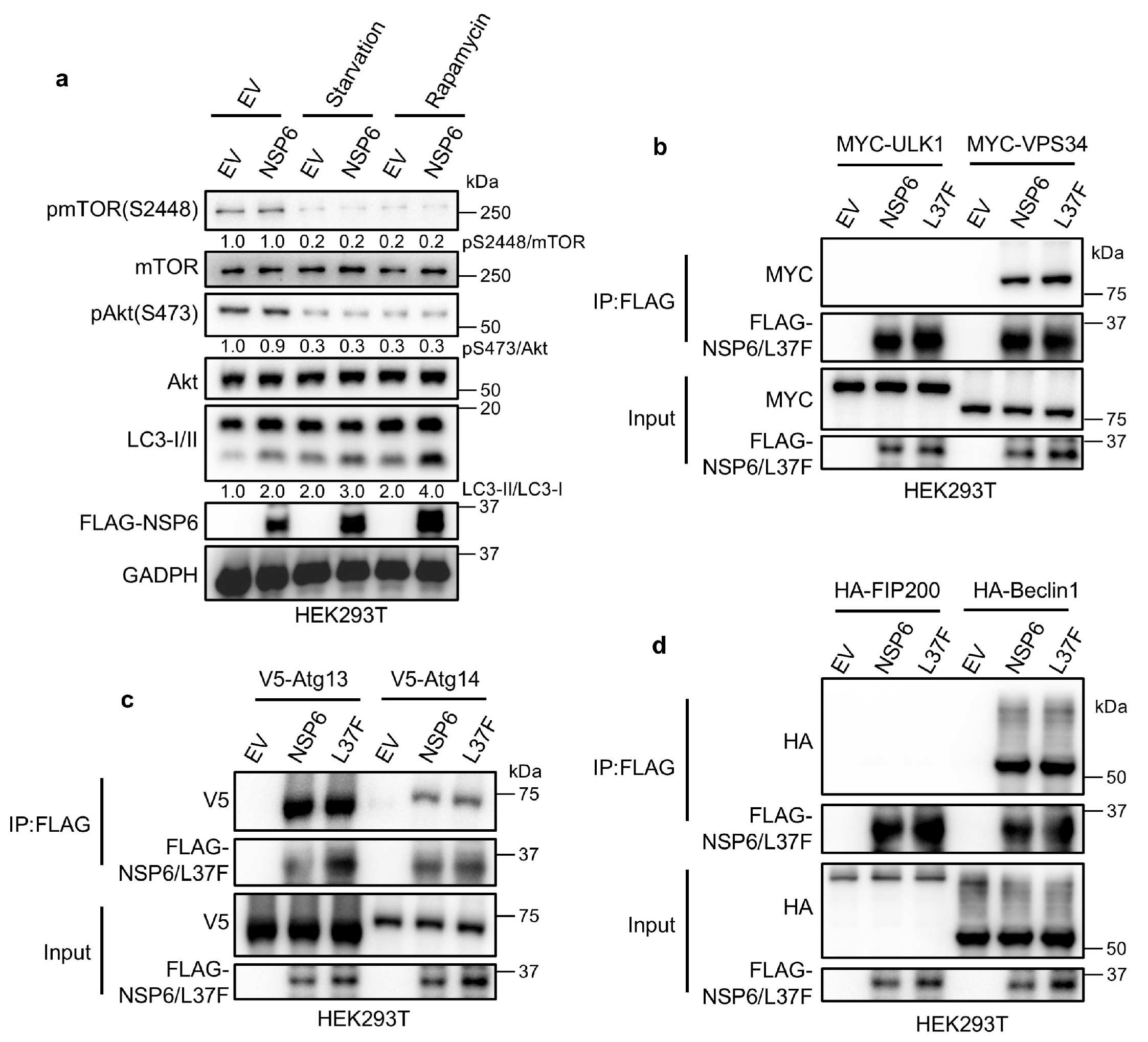
2.2. NSP6 and L37F Can Interact with the ATG14/Beclin1/VPS34 Complex
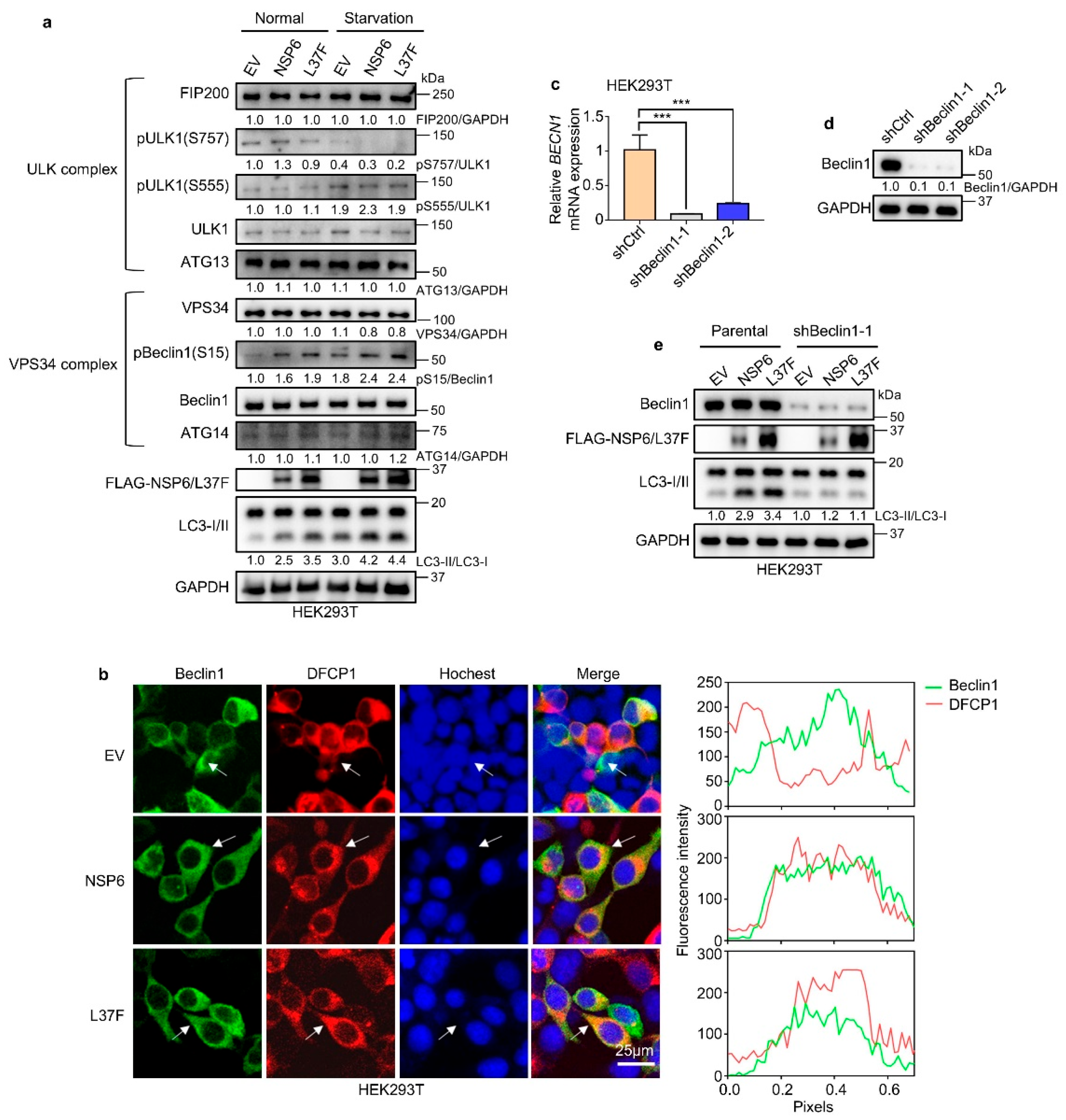
2.3. NSP6 and L37F Enhance the Initiation of Autophagy by Activating Beclin1
2.4. NSP6 Inhibits Autophagic–Lysosomal Degradation Through Inhibition of MLN1
3. Discussion
4. Materials and Methods
4.1. Key Materials
4.2. Cell Lines
4.3. Clones and Constructs
4.4. Antibodies and Immunoblotting
4.5. Cycloheximide-Blocking Assay
4.6. Cell Transfection
4.7. Lentivirus Production and Infection
4.8. Immunofluorescence Co-Localization Assay
4.9. Reverse Transcription and Quantitative Real-Time PCR (RT-qPCR)
4.10. FBS Starvation and Rapamycin Treatment Assay
4.11. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Viswanathan, T.; Arya, S.; Chan, S.H.; Qi, S.; Dai, N.; Misra, A.; Park, J.G.; Oladunni, F.; Kovalskyy, D.; Hromas, R.A.; et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020, 11, 3718. [Google Scholar] [CrossRef] [PubMed]
- Madhugiri, R.; Fricke, M.; Marz, M.; Ziebuhr, J. Coronavirus—Acting RNA Elements. Adv. Virus Res. 2016, 96, 127–163. [Google Scholar] [PubMed]
- Hasan, S.S.; Capstick, T.; Ahmed, R.; Kow, C.S.; Mazhar, F.; Merchant, H.A.; Zaidi, S.T.R. Mortality in COVID-19 patients with acute respiratory distress syndrome and corticosteroids use: A systematic review and meta-analysis. Expert Rev. Respir. Med. 2020, 14, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.J.; Yu, H.D.; Gu, W.H.; Luo, X.L.; Li, R.; Zhang, J.; Xu, Y.F.; Yang, L.J.; Shen, N.; Feng, L.; et al. Autophagy Negatively Regulates Transmissible Gastroenteritis Virus Replication. Sci. Rep. 2016, 6, 23864. [Google Scholar] [CrossRef]
- Richetta, C.; Faure, M. Autophagy in antiviral innate immunity. Cell. Microbiol. 2013, 15, 368–376. [Google Scholar] [CrossRef]
- Jain, K.; Paranandi, K.S.; Sridharan, S.; Basu, A. Autophagy in breast cancer and its implications for therapy. Am. J. Cancer Res. 2013, 3, 251–265. [Google Scholar]
- Morselli, E.; Galluzzi, L.; Kepp, O.; Vicencio, J.M.; Criollo, A.; Maiuri, M.C.; Kroemer, G. Anti- and pro-tumor functions of autophagy. BBA Mol. Cell Res. 2009, 1793, 1524–1532. [Google Scholar] [CrossRef]
- Ding, M.; Zhou, F.; Li, Y.; Liu, C.; Gu, Y.; Wu, J.; Fan, G.; Li, Y.; Li, X. Cassiae Semen improves non-alcoholic fatty liver disease through autophagy-related pathway. J. Chin. Herb. Med. 2023, 15, 421–429. [Google Scholar] [CrossRef]
- Wu, W.K.K.; Yue, J.B. Autophagy in host-microbe interactions. Semin. Cell Dev. Biol. 2020, 101, 1–2. [Google Scholar] [CrossRef]
- Hu, B.L.; Zhang, Y.N.; Jia, L.; Wu, H.S.; Fan, C.F.; Sun, Y.T.; Ye, C.J.; Liao, M.; Zhou, J.Y. Binding of the pathogen receptor HSP90AA1 to avibirnavirus VP2 induces autophagy by inactivating the AKT-MTOR pathway. Autophagy 2015, 11, 503–515. [Google Scholar] [CrossRef]
- Alonso, C.; Galindo, I.; Cuesta-Geijo, M.A.; Cabezas, M.; Hernaez, B.; Muñoz-Moreno, R. African swine fever virus-cell interactions: From virus entry to cell survival. Virus Res. 2013, 173, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Kang, R.Y.; Wang, J.; Luo, G.X.; Yang, W.; Zhao, Z.D. Hepatitis C virus inhibits AKT-tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy 2013, 9, 175–195. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, T.; Mariienko, Y.; Mehterov, N.; Kazakova, M.; Sbirkov, Y.; Todorova, K.; Hayrabedyan, S.; Sarafian, V. Autophagy and SARS-CoV-2-Old Players in New Games. Int. J. Mol. Sci. 2023, 24, 7734. [Google Scholar] [CrossRef] [PubMed]
- Miao, G.Y.; Zhao, H.Y.; Li, Y.; Ji, M.M.; Chen, Y.; Shi, Y.; Bi, Y.H.; Wang, P.H.; Zhang, H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell 2021, 56, 427–442.e5. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.H.; Hozumi, Y.T.; Yin, C.C.; Wei, G.W. Decoding Asymptomatic COVID-19 Infection and Transmission. J. Phys. Chem. Lett. 2020, 11, 10007–10015. [Google Scholar] [CrossRef]
- Benvenuto, D.; Angeletti, S.; Giovanetti, M.; Bianchi, M.; Pascarella, S.; Cauda, R.; Ciccozzi, M.; Cassone, A. Evolutionary analysis of SARS-CoV-2: How mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020, 81, E24–E27. [Google Scholar] [CrossRef]
- Jia, K.; Li, Y.; Liu, T.; Gu, X.; Li, X. New insights for infection mechanism and potential targets of COVID-19: Three Chinese patent medicines and three Chinese medicine formulas as promising therapeutic approaches. J. Chin. Herb. Med. 2023, 15, 157–168. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef]
- Ricciardi, S.; Guarino, A.M.; Giaquinto, L.; Polishchuk, E.V.; Santoro, M.; Di Tullio, G.; Wilson, C.; Panariello, F.; Soares, V.C.; Dias, S.S.G.; et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature 2022, 606, 761–768. [Google Scholar] [CrossRef]
- Cottam, E.M.; Whelband, M.C.; Wileman, T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy 2014, 10, 1426–1441. [Google Scholar] [CrossRef]
- Dai, A.B.; Yu, L.; Wang, H.W. WHAMM initiates autolysosome tubulation by promoting actin polymerization on autolysosomes. Nat. Commun. 2019, 10, 3699. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Lu, Y.; Ren, Y.L.; Yuan, J.P.; Zhang, N.S.; Kimball, H.; Zhou, L.Q.; Yang, M. Starvation-induced suppression of DAZAP1 by miR-10b integrates splicing control into TSC2-regulated oncogenic autophagy in esophageal squamous cell carcinoma. Theranostics 2020, 10, 4983–4996. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Nakada, D.; Yilmaz, O.H.; Tothova, Z.; Joseph, N.M.; Lim, M.S.; Gilliland, D.G.; Morrison, S.J. mTOR Activation Induces Tumor Suppressors that Inhibit Leukemogenesis and Deplete Hematopoietic Stem Cells after Deletion. Cell Stem Cell 2010, 7, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Cullion, K.; Draheim, K.M.; Hermance, N.; Tammam, J.; Sharma, V.M.; Ware, C.; Nikov, G.; Krishnamoorthy, V.; Majumder, P.K.; Kelliher, M.A. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood 2009, 113, 6172–6181. [Google Scholar] [CrossRef]
- Deitersen, J.; Berning, L.; Stuhldreier, F.; Ceccacci, S.; Schlütermann, D.; Friedrich, A.; Wu, W.X.; Sun, Y.D.; Böhler, P.; Berleth, N.; et al. High-throughput screening for natural compound-based autophagy modulators reveals novel chemotherapeutic mode of action for arzanol. Cell Death Dis. 2021, 12, 560. [Google Scholar] [CrossRef]
- Moltedo, O.; Remondelli, P.; Amodio, G. The Mitochondria-Endoplasmic Reticulum Contacts and Their Critical Role in Aging and Age-Associated Diseases. Front. Cell Dev. Biol. 2019, 7, 172. [Google Scholar] [CrossRef]
- Mochizuki, Y.; Ohashi, R.; Kawamura, T.; Iwanari, H.; Kodama, T.; Naito, M.; Hamakubo, T. Phosphatidylinositol 3-Phosphatase Myotubularin-related Protein 6 (MTMR6) Is Regulated by Small GTPase Rab1B in the Early Secretory and Autophagic Pathways. J. Biol. Chem. 2013, 288, 1009–1021. [Google Scholar] [CrossRef]
- Zhang, H.; Dong, X.Q.; Zhao, R.; Zhang, R.J.; Xu, C.; Wang, X.X.; Liu, C.X.; Hu, X.Y.; Huang, S.L.; Chen, L. Cadmium results in accumulation of autophagosomes-dependent apoptosis through activating Akt-impaired autophagic flux in neuronal cells. Cell Signal. 2019, 55, 26–39. [Google Scholar] [CrossRef]
- Vergarajauregui, S.; Connelly, P.S.; Daniels, M.P.; Puertollano, R. Autophagic dysfunction in mucolipidosis type IV patients. Hum. Mol. Genet. 2008, 17, 2723–2737. [Google Scholar] [CrossRef]
- Sun, T.; Wang, X.W.; Lu, Q.; Ren, H.Y.; Zhang, H. CUP-5, the C. ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy 2011, 7, 1308–1315. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, P.; Saumya, K.U.; Giri, R. Investigating the conformational dynamics of SARS-CoV-2 NSP6 protein with emphasis on non-transmembrane 91–112 & 231–290 regions. Microb. Pathog. 2021, 161, 105236. [Google Scholar] [PubMed]
- Suleman, M.; Ishaq, I.; Khan, H.; Khan, S.U.; Masood, R.; Albekairi, N.A.; Alshammari, A.; Crovella, S. Elucidating the binding mechanism of SARS-CoV-2 NSP6-TBK1 and structure-based designing of phytocompounds inhibitors for instigating the host immune response. Front. Chem. 2024, 11, 1346796. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Xu, W.H.; Ma, J.; Wang, H.J.; Cui, Z.; Jiang, T.L.; Li, C.H. Inhibitory Mechanisms of DHA/CQ on pH and Iron Homeostasis of Erythrocytic Stage Growth of Plasmodium falciparum. Molecules 2019, 24, 1941. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, Y.Z.; Huang, Z.H.; Xu, W.Y.; Hu, W.; Yi, L.N.; Liu, Z.; Chan, H.; Zeng, J.D.; Liu, X.D.; et al. SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1. Cell Death Differ. 2022, 29, 1240–1254. [Google Scholar] [CrossRef]
- Nishitsuji, H.; Iwahori, S.; Ohmori, M.; Shimotohno, K.; Murata, T. Ubiquitination of SARS-CoV-2 NSP6 and ORF7a Facilitates NF-κB Activation. mBio 2022, 13, e0097122. [Google Scholar] [CrossRef]
- Jiao, P.T.; Fan, W.H.; Ma, X.Y.; Lin, R.S.; Zhao, Y.A.; Li, Y.B.; Zhang, H.; Jia, X.J.; Bi, Y.H.; Feng, X.L.; et al. SARS-CoV-2 nonstructural protein 6 triggers endoplasmic reticulum stress-induced autophagy to degrade STING1. Autophagy 2023, 19, 3113–3131. [Google Scholar] [CrossRef]
- Xia, H.J.; Cao, Z.G.; Xie, X.P.; Zhang, X.W.; Chen, J.Y.C.; Wang, H.L.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Rashid, F.; Xie, Z.X.; Suleman, M.; Shah, A.B.L.; Khan, S.; Luo, S.S. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol. 2022, 13, 940756. [Google Scholar] [CrossRef]
- Plante, J.A.; Mitchell, B.M.; Plante, K.S.; Debbink, K.; Weaver, S.C.; Menachery, V.D. The variant gambit: COVID-19’s next move. Cell Host Microbe 2021, 29, 508–515. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Chang, J.; Sheng, R. NSP6 of SARS-CoV-2 Dually Regulates Autophagic–Lysosomal Degradation. Int. J. Mol. Sci. 2025, 26, 3699. https://doi.org/10.3390/ijms26083699
Zhang H, Chang J, Sheng R. NSP6 of SARS-CoV-2 Dually Regulates Autophagic–Lysosomal Degradation. International Journal of Molecular Sciences. 2025; 26(8):3699. https://doi.org/10.3390/ijms26083699
Chicago/Turabian StyleZhang, Haijiao, Jianying Chang, and Ren Sheng. 2025. "NSP6 of SARS-CoV-2 Dually Regulates Autophagic–Lysosomal Degradation" International Journal of Molecular Sciences 26, no. 8: 3699. https://doi.org/10.3390/ijms26083699
APA StyleZhang, H., Chang, J., & Sheng, R. (2025). NSP6 of SARS-CoV-2 Dually Regulates Autophagic–Lysosomal Degradation. International Journal of Molecular Sciences, 26(8), 3699. https://doi.org/10.3390/ijms26083699




