The Role of microRNAs in Lung Cancer: Mechanisms, Diagnostics and Therapeutic Potential
Abstract
1. Introduction
1.1. Overview of Lung Cancer
1.2. Introduction to microRNAs (miRNAs)
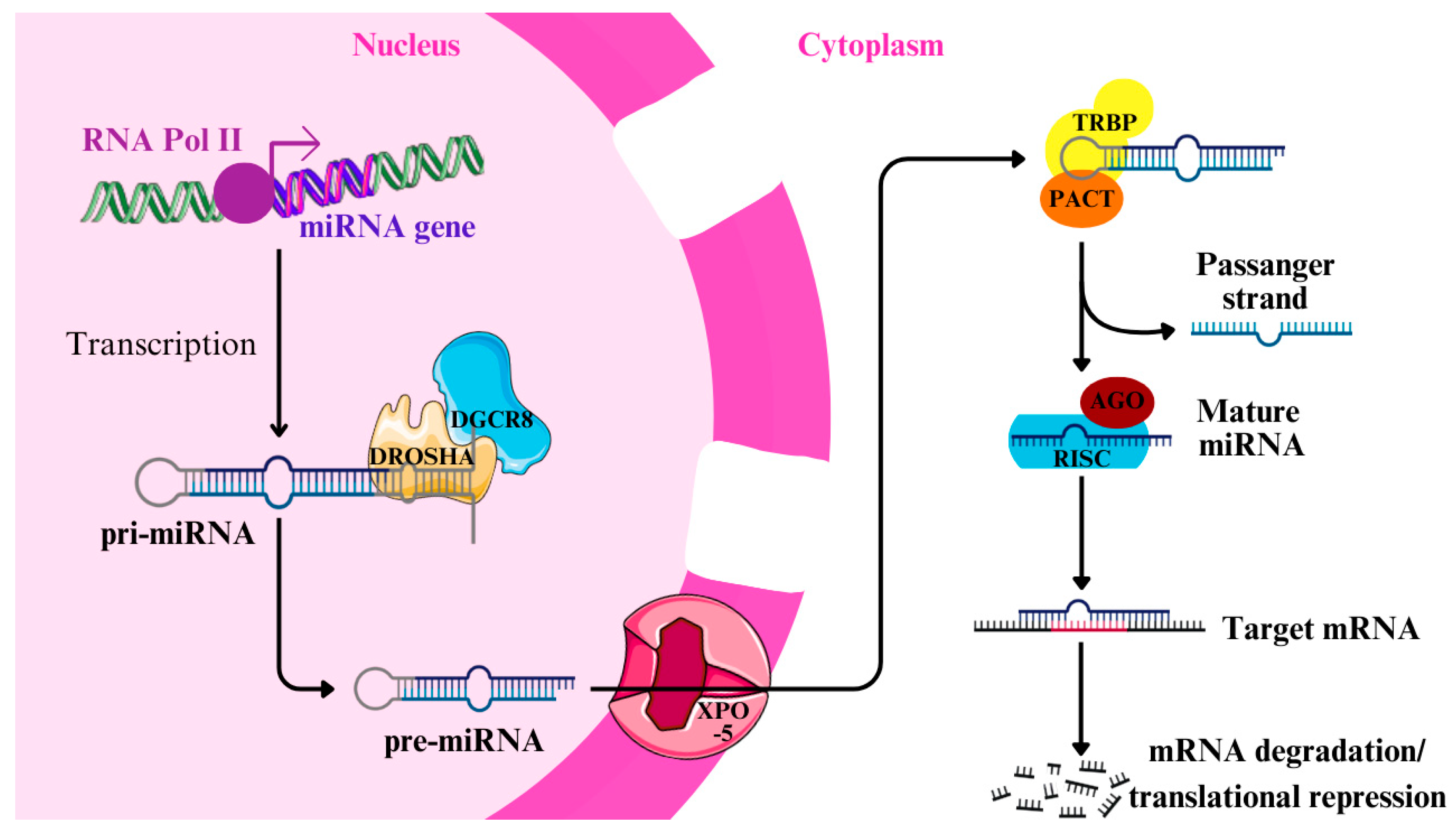
| Origins of miRNAs | —Introns and Exons: miRNAs can originate from introns and noncoding exons. —50% Noncoding: About 50% come from non-protein-coding transcripts, regulated by their promoters. —Intragenic miRNAs: Derived from intragenic regions, mostly introns. —Clusters: Some miRNAs are clustered and expressed as polycistronic transcripts. |
| Examples of miRNA Clusters | —miR-17/92 Cluster: Contains miR-17, miR-18, miR-19, miR-19b, miR-20a, and miR-92a, located on chromosome 13. —Paralogues: miR-106b/25 on chromosome 7 and miR-106a/363 on chromosome X. |
| Canonical vs. Noncanonical Pathway | —Canonical: Involves Drosha, Dicer, and XPO-5. —Noncanonical: Includes mirtrons (spliced introns) and m7G-capped pre-miRNAs, bypassing Drosha/DGCR8 or Dicer. |
| Biogenesis Pathway | 1. Transcription: RNA polymerase II transcribes pri-miRNA. 2. Processing in Nucleus: Microprocessor complex (Drosha + DGCR8) cleaves pri-miRNA into pre-miRNA. 3. Export: Exportin 5 (XPO-5) transports pre-miRNA to the cytoplasm. 4. Processing in Cytoplasm: Dicer processes pre-miRNA into mature miRNA. |
| Role of Proteins in Biogenesis | —Drosha: Cleaves pri-miRNA. —Dicer: Processes pre-miRNA to mature miRNA. —XPO-5: Transports pre-miRNA. —RISC: RNA-induced silencing complex involving Argonaute (AGO1-4) proteins. |
| Functions in RISC | —Guide Strand (-5p): Loaded into RISC, binds target mRNA. —Passenger Strand (-3p): Usually degraded, though some have functions. —Mechanism: Targets mRNAs for degradation or translational repression. |
| Determinants of Function | —Complementarity: Full complementarity leads to mRNA degradation; partial complementarity results in translational repression. —Cell State: Influences miRNA abundance and target mRNA availability. |
| Dual Roles in Regulation | —Repression: Inhibits gene expression in the cytoplasm. —Activation (RNAa): Some miRNAs activate gene expression in the nucleus by recruiting transcription factors (e.g., miR-24-1). |
| Biological Processes | —Development: Nervous system development, differentiation (e.g., let-7 and miR-290-295). —Cancer: p53 regulation (miR-34, miR-145, etc.). —Mitochondrial Function: MitomiRs regulate oxidative phosphorylation (e.g., miR-378). |
2. Mechanisms of miRNA Involvement in Lung Cancer
2.1. Tumor Suppressor miRNAs
2.2. Oncogenic miRNAs (oncomiRs)
| OncomiRs | Targets/Regulators | Function | Ref. |
|---|---|---|---|
| OncomiRs Related to Proliferation and Growth | |||
| miR-31 | LATS2, PPP2R2A | Through repressing tumor suppressors, LATS2 and PPP2R2A, miR-31 promotes cancer growth. | [52] |
| miR-411-5p, miR-411-3p | SPRY4, TXNIP | Targeting suppressor genes SPRY4 and TXNIP results in carcinogenesis promotion. | [46] |
| miR-1290, miR-1246 | P53, THBS2 | miR-1246 and miR-1290 inhibition decreases stemness markers and EMT markers, and thus, anti-miR-1246 and anti-miR-1290 suppress proliferation and invasion of NSCLC. | [53] |
| miR-211 | SRCIN1 | Downregulating SRCIN1 expression by miR-211 promotes NSCLC proliferation. | [54] |
| miR-196a | FoxO1, p27, HOXA9 | NSCLC proliferation and migration are stimulated through direct FoxO1, p27, and HOXA9 targeting. | [55] |
| miR-324-5p, miR-324-3p | miR-324-5p promotes both cell proliferation and invasion in lung cancer cells. miR-324-3p significantly increases cell proliferation but does not alter the invasive profile of cancer cells. | [56] | |
| miR-19 | CBX7 | miR-19 plays the role of a tumor accelerator, as it promotes lung cancer cell proliferation by inhibiting the expression of CBX7. | [57] |
| miR-1269a | SOX6 | Through downregulating SOX6 expression, miR-1269a promotes NSCLC growth. | [58] |
| miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, miR-92-1 | In cooperation with the c-Myc miR-17-29 cluster promotes tumor development and neovascularization. miR-20a may regulate genes associated with TGF-β and VEGF. | [50,59] | |
| OncomiRs involved in metastasis and invasion | |||
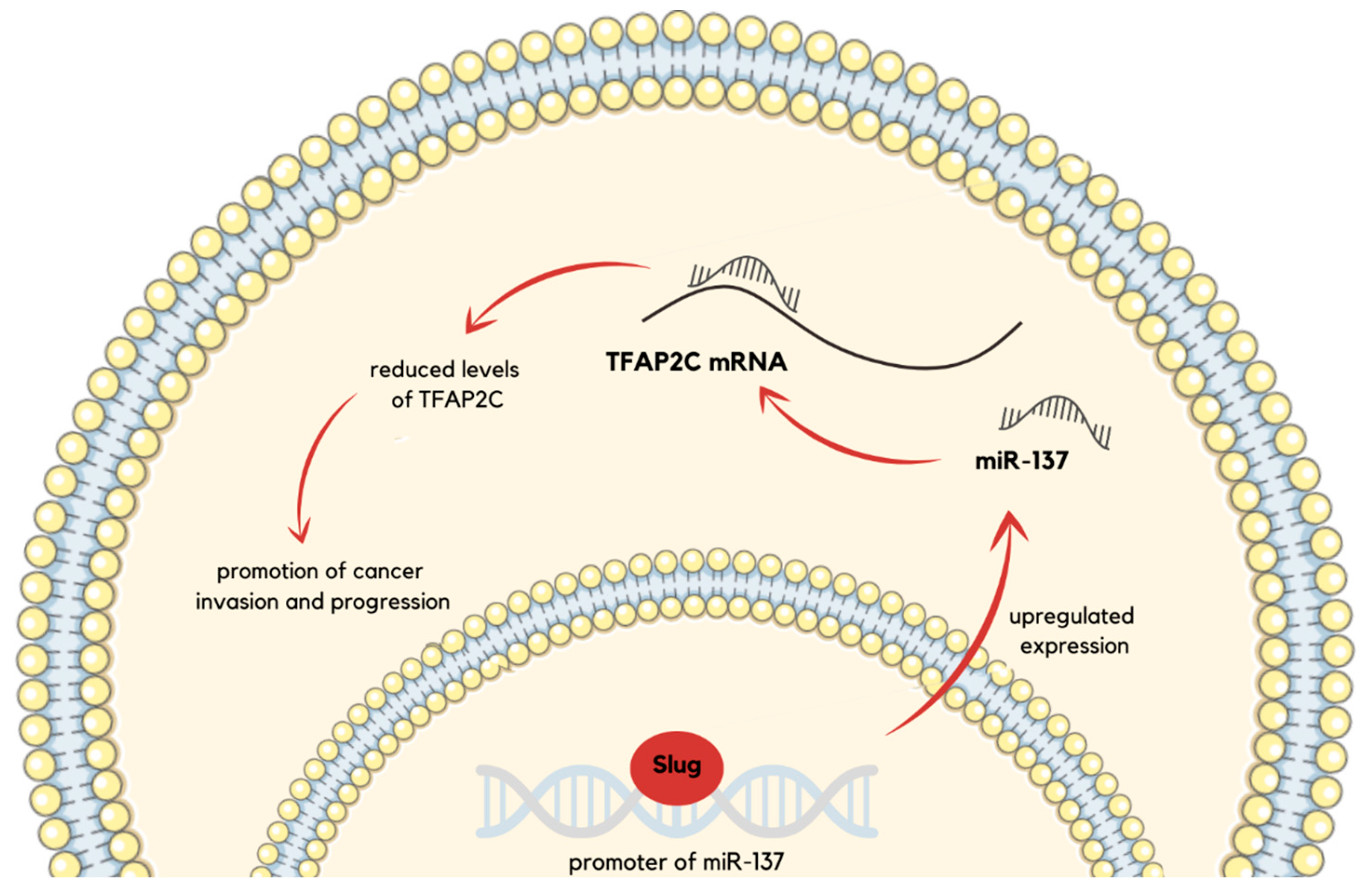
| miR-137 | TFAP2C | Cancer invasion and progression are promoted through TFAP2C suppression. | [47] |
| miR-490-3p | PCBP1 | Targeting PCBP1 regulates cancer metastasis. | [60] |
| miR-451a | Increased level of miR-451a is associated with lymph node metastasis and vascular invasion. | [61] | |
| miR-574-5p | PTPRU | miR-574-5p is overexpressed in patients with advanced metastatic NSCLC. It promotes both the migration and the invasion of cancer cells as well as enhances the tyrosine phosphorylation of β-catenin by repressing PTPRU expression in vitro. | [62] |
| miR-21, miR-155 | SOCS1, SOCS6, PTEN; DGKB | miR-21 and miR-155 promote the development of NSCLC by downregulating SOCS1, SOCS6, and PTEN. miR-21 also promotes metastasis to the brain through the ERK-STAT3 signaling pathway, which inhibits DGKB. | [51,63] |
| OncomiRs connected with immune modulation | |||
| miR-320a | STAT4 | mir-320a secreted by neutrophils of high-risk heavy smokers promotes the M2-like immunosuppressive phenotype of macrophages through STAT4 downregulation. | [64] |
| OncomiRs related to apoptosis and therapy resistance | |||
| miR-208a | p21 | miR-208a increases the proliferation of lung cancer cells and decreases cellular apoptosis. What is more, it enhances the radioresistance of the A549 lung cancer cells. | [65] |
| miR-146a | CHOP | Though downregulating CHOP expression, miR-146a induces chemotherapy resistance in lung cancer. | [66] |
| Prognostic markers | |||
| miR-23b-3p, miR-10b-5p, miR-21–5p | Elevated levels of miR-23b-3p, miR-10b-5p, and miR-21-5p are independently associated with poor overall survival in NSLCC patients. miR-21-5p might be involved in tumor progression; however, its exact pathological mechanism needs to be further examined. | [67] | |
3. miRNAs as Biomarkers in Lung Cancer Diagnostics
3.1. miRNA Detection Methods
Novel Technologies for microRNA Detection and Analysis
3.2. Early Detection—The Potential of Circulating miRNAs in Early Lung Cancer Diagnosis
| miRNA/miRNAs | Connection to Cancer/Cancer Type | Characteristic Features | Ref. |
|---|---|---|---|
| miRNA-223 | Early detection of lung cancer. | Potential biomarker detectable in body fluids. | [110] |
| miRNA-141 | Early detection of NSCLC. | Described as an auspicious biomarker for early NSCLC diagnosis. | [113] |
| miRNA-155 | Early detection of lung cancer; linked to metastasis. | Overexpressed in plasma; expression increases with lung cancer metastasis. | [109,110] |
| miRNA-1254 | Early detection of lung cancer. | Part of an early-diagnosis miRNA set detectable in body fluids. | [110] |
| miRNAs-125a-5p, 25, and 126 | Early detection of lung cancer. | The combined panel of miRNAs-125a-5p, -25, and -126 may be potentially used in early lung cancer diagnosis. | [76] |
| miRNA-145, 210, and 205-5p | Early detection of lung cancer. | Differentially expressed in lung cancer vs. cancer-free smokers. | [78,111] |
| miRNA-143, let-7g, and let-7a | Early detection of lung cancer. | Key serum biomarker. | [78] |
| miRNAs-7a-5p and 375 | Early-stage NSCLC. | Downregulated compared to healthy controls in NSCLC, and they are part of the same 5-miRNA panel. | [112] |
| miRNA-1-3p, 1291, and 214-3p | Early-stage NSCLC. | Upregulated compared to healthy controls in NSCLC, and they are part of the same 5-miRNA panel. | [112] |
| 34-miRNAs model | NSCLC, all stages. | Detects asymptomatic NSCLC and distinguishes benign from malignant lesions with better AUC for stage I than for stages II-IV. | [79] |
| miRNA-197 | Early detection of lung cancer; linked to metastasis. | Overexpressed in plasma; higher expression in metastatic disease. | [109] |
| miRNA-182 | Early detection of lung cancer. | Overexpressed in plasma. | [109] |
| miRNA-21 | NSCLC, correlation with stage also sputum-based detection. | Expression increases with the advancing stage; in sputum, shows higher sensitivity vs. cytology for lung cancer diagnosis. | [113,122] |
| miRNA-942 | NSCLC early diagnosis. | Significantly upregulated, outperforms traditional markers (CEA, CYFRA21-1, SCCA). | [114] |
| miRNA-601 | NSCLC early diagnosis. | Significantly upregulated, outperforms traditional markers, and combining with miR-942 improves early detection. | [114] |
| miRNA-21-5p | Male lung squamous cell carcinoma (LUCS); also part of a protein–miRNA panel. | Differential expression between early- and late-stage LUCS. In a separate study, combined with a 4MP panel to enhance sensitivity at 95% specificity. | [108,115] |
| miRNA-181-5p and 155-5p | Male LUCS. | Differential expression between early- and late-stage disease. | [115] |
| miRNA-3692-3p | NSCLC early diagnosis. | Upregulated in NSCLC but not correlated with treatment response or survival. | [116] |
| miRNAs-199a-3p, chr17_10932, 148a-3p, 210-3p, chr1_1402, 378d, and 138-5p | LUAD especially with ground-glass nodules (GGNs). | Overexpressed and part of a 7-miRNA early-detection panel. | [117,119] |
| miRNA-146a-5p | Early detection of lung cancer. | The addition to the mRNA panel improves AUC from 0.66 to 0.71 (diagnostic accuracy). | [118] |
| miRNA-301a-5p | Early-stage LUAD and LUCS. | Potential biomarker; upregulated in early disease. | [119] |
| miRNA-205 | Early-stage squamous cell lung cancer. | Overexpressed in sputum of squamous cell lung cancer patients; offers good early detection accuracy. | [123] |
| miRNA-324-3p | Early detection of stage I LUCS. | Significantly upregulated; AUC = 0.79 alone and improves to 0.89 when combined with miR-1285; independent prognostic indicator. | [124] |
| miRNA-1285 | Early detection of stage I LUCS. | Significantly downregulated; combining with miR-324-3p improves diagnostic accuracy. | [124] |
| miRNAs-320a-3p, 92a-3p, and 140-3p | Early detection of lung cancer. | Combined with a 4-protein marker panel (4MP) to improve sensitivity by 19% at 95% specificity. | [108] |
3.3. Prognostic Biomarkers—miRNA Signature Correlated with Patient Outcomes
| miRNA/miRNAs | Connection with Patient Outcomes | Characteristic Features | Ref. |
|---|---|---|---|
| miRNA-21 | Increased expression in lung cancer patients with LN metastasis. Low expression correlates with a longer median survival time. However, higher expression correlates with shorter DFS and OS. | Contradictorily impact on patient outcomes according to different references. | [82,124,143] |
| miRNA-31 | Increased expression in lung cancer patients with LN metastasis; lower expression correlates with longer median survival. | Associated with metastasis and shorter survival when overexpressed. | [124] |
| miRNA-let-7 | Increased expression in lung cancer patients with LN metastasis; lower expression correlates with longer median survival. | Higher miRNA-let-7 is linked to metastasis. | [124,135] |
| miRNA-25-3p | Linked to shorter OS in lung cancer patients. | May be grouped in panels for prognostic or diagnostic use. | [134] |
| miRNA-182-5p | Overexpression is strongly associated with LN metastasis in NSCLC/LUAD, but no significant survival difference on Kaplan–Meier curves. | miRNA-182-5p may be more useful as a metastatic marker than a clear prognostic factor for OS or DFS. | [125] |
| miRNAs-320a, 25-3p, and 148a-3p | Expression levels are significantly related to tumor stage in NSCLC. | miRNA-25-3p especially is related to shorter OS in lung cancer. | [133,134] |
| miRNAs-let-7f, 30e-3p, and 20b | Related to NSCLC stage and LN metastases. Elevated expression of miRNAs-let-7f and 30e-3p are correlated with unresectable tumors. Elevated miRNA-let-7f is linked to lower OS in advanced NSCLC; high miRNA-30e-3p expression is associated with shorter DFS. | miRNA-let-7f is considered a poor prognostic indicator for nonresectable NSCLC. | [139] |
| miRNA-424 | Elevated expression is associated with tumor stage as well as shorter survival. | Associated with aggressive metastasis and advanced clinical stage. | [141] |
| miRNA-942 and 601 | High serum levels are associated with worse outcomes in NSCLC. | Elevated expression is linked to a worse prognosis. | [114] |
| miRNA-223 | Significantly linked to time to disease progression (TTP); elevated miRNA-223 expression is associated with worse outcomes. | May predict treatment response and TTP. | [83] |
| miRNA-128 and 155 | High expression is associated with shorter OS in NSCLC. | Elevated miRNAs-21, 128, 155, and 181a are linked to worse outcomes in NSCLC patients. | [143] |
| miRNA-181a | Increased expression is associated with worse outcomes in squamous cell NSCLC. | - | [143] |
| miRNA-202 | Elevated expression is correlated with disease progression; predicts shorter PFS and OS in NSCLC; high miR-202 is associated with shorter OS in non-squamous patients. | Independent predictor of poor outcome. | [144] |
| miRNA-26a | High expression is correlated with worse OS in squamous NSCLC patients. | Histological-specific association with poor prognosis. | [144] |
| miRNA-181a-5p, and 630 | Reduced expression in NSCLC tissue is linked to longer PFS and OS. | Elevated levels are associated with worse outcomes; serve as independent prognostic markers in NSCLC. | [148] |
| miRNA-20a-5p | Higher expression is linked with lower DFS in LUCS patients. | Potentially prognostic biomarker in LUCS. | [147] |
| has-miRNA-197 | Upregulation correlates with a lower risk of metastases post-lung resection. | Expression in metastasis-developing patients remains near control levels, suggesting a protective role. | [130] |
| miRNA-221 | Lower expression in recurrent tumors; higher expression correlates with no recurrence. | May help identify patients at high risk of post-surgical relapse. | [81] |
| miRNA-3195 | Higher expression is associated with longer OS in NSCLC; identified as an independent prognostic factor. | May have a prognostic role. | [140] |
| miRNA-665 | Strongest predictive marker for tumor shrinkage in advanced non-squamous NSCLC patients treated with bevacizumab/erlotinib chemotherapy. | High expression might be linked with better tumor response. | [83] |
| miRNA-148a | Lower expression is related to LN metastasis and advanced clinical stage. | Low expression correlates with more aggressive disease. | [82] |
| miRNA-30c | Lower concentration is associated with shorter OS and RFS. | Potentially useful as a predictive biomarker for treatment response and survival. | [77] |
| miRNA-30a-5p | Decreased in LUAD; associated with TN stage, pathologic stage, residual tumor, primary therapy outcome, and OS. | Negatively correlates with BCL-2. Associated with favorable outcomes. | [128,129] |
| miRNA-125b-5p | Decreased expression is correlated with poor prognosis in LUAD. | Potentially tumor-suppressive role in LUAD. | [132] |
| miRNA-375 | Decreased expression is correlated with worse survival in NSCLC. | Often discussed as a tumor-suppressor miRNA in lung cancer. | [138] |
| miRNA-126-3p | Lower expression is associated with poor DFS in LUAD patients. | Often considered a tumor suppressor in various lung cancer contexts. | [147] |
| miRNA-152-3p and 199a-5p | Lower expression is associated with lower DFS in LUCS. | Decreased expression is associated with worse outcomes in squamous cell subtype patients. | [147] |
| miRNAs-9-2, 125, 193a | Methylation status correlates with OS in NSCLC. | Higher methylation is often associated with worse outcomes. | [136] |
| miRNA-127 | Patients with the highest level of methylated miR-127 have a longer median survival. | Increased methylation is associated with shorter survival. | [137] |
| miRNAs-19a-3p, 126-5p, 556-3p, 671-5p, 937-3p, 4664-3p, and 4746-5p | Expression levels are associated with OS in LUAD; high-risk groups show higher mortality. | Part of a multi-miRNA prognostic signature specific to LUAD. | [131] |
| 31 miRNAs (except miRNA-135b) | All are associated with LUAD patient survival regardless of tumor stage | Part of a broad prognostic signature in LUAD. | [135] |
| miRNAs-148b, 365, 32, 375, 21, 125b, and 155 | Display prognostic ability in NSCLC; high-risk signature is linked to shorter median OS. Decreased miR-375 is correlated with worse survival. | Combining miRNA signatures with tumor stage enhances prognostic accuracy. | [138] |
| miRNA-150 and 886-3p | miRNAs signature may predict OS and PFS in early-stage SCLC patients treated with surgery and adjuvant chemotherapy. | Downregulated in SCLC tissues. Potentially serve as independent prognostic biomarkers for patient stratification. | [145] |
| miRNAs-143, 100, 101-1, 101-2, 182, 183, 205, 21, 30a, and 30d | Identified via machine learning as significantly correlated with survival in LUSC; strong predictors of patient outcome | miRNA signature identified via machine learning is associated with LUSC prognosis. | [146] |
| miRNA-155-5p and 223-3p | Elevated expression is linked to poor DFS in LUAD patients. | DFS may be lower when this miRNA signature is overexpressed. | [147] |
| miRNA-6777-5p, 6780a-5p, and 877-5p | Expression in exhaled breath condensate may serve as potential prediction biomarkers in lung cancer. | Might be used as a noninvasive diagnostic in the future. | [142] |
3.4. Predictive Biomarkers—The Role of miRNAs in Predicting Treatment Response
| miRNA/miRNAs | Predictive Role | Characteristic Features | Ref. |
|---|---|---|---|
| miRNA-25, 145 and 210 | Predicts response to pemetrexed in advanced NSCLC. | Serum levels, together with miRNA-145 and miRNA-210, may help identify which patients respond to pemetrexed. However, further validation is needed due to a small sample size. | [150] |
| miRNA-1249-3p | Predicts chemotherapy response in NSCLC. | Higher expression in responders compared to non-responders, indicating potential use as a biomarker to differentiate treatment response in NSCLC. | [141] |
| miRNA-30a-5p | Predicts sensitivity to paclitaxel in NSCLC. | Increases NSCLC sensitivity to paclitaxel in vitro and in vivo by promoting apoptosis through BCL-2 suppression, suggesting it could be a useful biomarker for chemotherapy. | [130] |
| miRNA-10a-3p | Novel non-invasive biomarker for short-term NSCLC prognosis. | Identified in NSCLC cells cultured in 3D. miRNA-10a-3p circulating levels may help predict short-term survival outcomes. | [154] |
| miRNA-200b | Potential immunotherapy predictor due to correlation with PD-L1. | Negatively correlated with PD-L1 expression in NSCLC tissues thus may improve predictive value beyond PD-L1 tumor proportion scores (TPS) for selecting patients for immune checkpoint inhibitor therapy. | [153] |
| miRNA-let-7f and 30e | Distinguish resectable vs. non-resectable NSCLC. | Elevated plasma levels correlate with advanced disease and non-surgical cases, helping to determine surgical eligibility. | [140] |
| miRNA-455-5p | Predicts cisplatin resistance. | Inversely correlated with PD-L1 expression and regulates cisplatin resistance in NSCLC, suggesting it could be a predictive biomarker of chemotherapy outcomes. | [158] |
| miRNA-486-5p | Predicts progression after platinum-based chemotherapy in NSCLC. | Elevated serum levels are associated with a longer time to progression, as confirmed by Cox hazard regression analysis, suggesting a potentially more favorable response. | [159] |
| miRNA-34a | Predicts survival in NSCLC patients on platinum and gemcitabine chemotherapy. | High miRNA-34a expression is linked to shorter OS in patients treated with chemotherapy and radiotherapy. Nonetheless, immunotherapy responders exhibit elevated miRNA-34a expression, which correlated with improved outcomes. | [160,162] |
| miRNA-224 | Predicts OS in NSCLC patients receiving chemotherapy with radiotherapy. | High miRNA-224 expression correlates with shorter OS in combined chemo-radiotherapy. | [160] |
| miRNAs-93, 138-5p, 200, 27a, 424, 28, 106b, 193a-3p, and 181a | Predict immunotherapy response and survival; higher miRNA expression in responders is associated with better PFS and OS. | Identified in a 27-miRNA panel that distinguishes responders from nonresponders to checkpoint inhibitors. Patients with high expression experience longer PFS and improved OS. | [162] |
| miRNA-22, 24, and 34a | Correlate with pemetrexed response in NSCLC; miRNA-22 upregulation linked to progressive disease. | Upregulated in patients receiving pemetrexed. miRNA-22 is specifically associated with progressive disease, while higher miRNA-24 and miRNA-34a levels correlate with pemetrexed sensitivity. However, miRNA-34a is correlated with poor outcomes in chemotherapy patients. | [163] |
| miRNA-142-3p | Predicts early-stage LUAD recurrence and poor outcome with adjuvant therapy. | More predictive than miRNA-29b for LUAD recurrence and upregulated in patients whose adjuvant therapy outcome is poor, indicating its potential as a biomarker for relapse risk. | [152] |
| miRNA-29b | Associated with early-stage LUAD recurrence. | Elevated levels during LUAD recurrence, though less predictive than miRNA-142-3p, may help identify patients at higher risk of early relapse. | [152] |
| miRNA-105-5p and 767-5p | Predict immunotherapy survival outcomes in LUAD. | High levels are associated with poorer survival in LUAD patients treated with immune checkpoint inhibitors. It might be used to distinguish long responders from short responders. | [119] |
| miRNA-101-2, 139, 182, 183, 190, 326, and 944 | Predict outcomes in LUSC. | A seven-miRNA signature correlated with lower overall survival in high-score LUSC patients, suggesting its possible prognostic utility in this subtype. | [151] |
| miRNA-92a-2 | Predicts chemoresistance in SCLC. | Elevated levels are linked to chemoresistance and reduced survival, suggesting its use as a possible biomarker for de novo resistance in SCLC. | [157] |
| miRNA-147 and 574-5p | Linked to chemoresistance in lung cancer. | Elevated miRNA-147 and 574-5p tumor levels are associated with treatment resistance and reduced survival. | [157] |
| miRNA-375, 200c and 30c | Predicts response to concurrent chemoradiotherapy (cCRT) or PI3K-targeted therapy. | Lower expression in non-responders to cCRT, influences phosphatidylinositol-mediated signaling and autophagy and may also be relevant for immune regulation and treatment resistance. | [128] |
4. miRNAs as Therapeutic Targets
4.1. miRNA Replacement Therapy
4.2. Strategies to Restore Tumor-Suppressive miRNAs in Lung Cancer
4.3. Approaches to Inhibit Oncogenic miRNAs
4.3.1. AntagomiRs
4.3.2. miRNA Sponges
4.4. Challenges and Opportunities
4.4.1. Delivery Systems for miRNA-Based Therapies
Viral-Based Vectors
Non-Viral-Based Vectors
Extracellular Vesicles
4.5. Current Clinical Trials and Prospects
4.6. Hurdles in miRNA-Based Therapies in Lung Cancer
5. Future Directions and Challenges
5.1. Advances in miRNA Research in the Context of Personalized Medicine
5.2. Challenges in Translating miRNA-Based Diagnostics and Therapies into Clinical Practice
5.3. Emerging Trends in miRNA Research: Multi-Omics Approaches and Integration with Other Biomarkers
5.4. Conflicting miRNA Expression Patterns in Lung Cancer Subtypes
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, T.; Peng, X.; Geng, Y.; Song, C.; Zhou, Z.; Huang, Y. Frailty and Prognosis in Lung Cancer: Systematic Review and Meta-Analysis. BMJ Support. Palliat. Care 2024, 14, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Wadowska, K.; Bil-Lula, I.; Trembecki, Ł.; Śliwińska-Mossoń, M. Genetic Markers in Lung Cancer Diagnosis: A Review. Int. J. Mol. Sci. 2020, 21, 4569. [Google Scholar] [CrossRef] [PubMed]
- Deshpand, R.; Chandra, M.; Rauthan, A. Evolving Trends in Lung Cancer: Epidemiology, Diagnosis, and Management. Indian J. Cancer 2022, 59, S90–S105. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, B.; He, S. Advances and Challenges in the Treatment of Lung Cancer. Biomed. Pharmacother. 2023, 169, 115891. [Google Scholar] [CrossRef]
- Lung Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/lung-cancer (accessed on 6 November 2024).
- Lung Cancer Statistics | How Common Is Lung Cancer? | American Cancer Society. Available online: https://www.cancer.org/cancer/types/lung-cancer/about/key-statistics.html (accessed on 2 December 2024).
- Schabath, M.B.; Cote, M.L. Cancer Progress and Priorities: Lung Cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1563–1579. [Google Scholar] [CrossRef]
- Seijo, L.M.; Peled, N.; Ajona, D.; Boeri, M.; Field, J.K.; Sozzi, G.; Pio, R.; Zulueta, J.J.; Spira, A.; Massion, P.P.; et al. Biomarkers in Lung Cancer Screening: Achievements, Promises and Challenges. J. Thorac Oncol. 2018, 14, 343. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, Y. Small RNA Modifications: Regulatory Molecules and Potential Applications. J. Hematol. Oncol. 2023, 16, 64. [Google Scholar] [CrossRef]
- Occhipinti, C.; La Russa, R.; Iacoponi, N.; Lazzari, J.; Costantino, A.; Di Fazio, N.; Del Duca, F.; Maiese, A.; Fineschi, V. MiRNAs and Substances Abuse: Clinical and Forensic Pathological Implications: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 17122. [Google Scholar] [CrossRef]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Szyłło, K.; Hogendorf, P. MiRNAs in Cancer (Review of Literature). Int. J. Mol. Sci. 2022, 23, 2805. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, Z.; Lin, Q.; Luo, Q.; Cen, Y.; Li, J.; Fang, X.; Gong, C. MiRNAs and Cancer: Key Link in Diagnosis and Therapy. Genes 2021, 12, 1289. [Google Scholar] [CrossRef]
- Iacomino, G. MiRNAs: The Road from Bench to Bedside. Genes 2023, 14, 314. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. MiRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef] [PubMed]
- Conti, I.; Varano, G.; Simioni, C.; Laface, I.; Milani, D.; Rimondi, E.; Neri, L.M. MiRNAs as Influencers of Cell–Cell Communication in Tumor Microenvironment. Cells 2020, 9, 220. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Yang, C. The MicroRNA-200 Family: Small Molecules with Novel Roles in Cancer Development, Progression and Therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef]
- Otmani, K.; Lewalle, P. Tumor Suppressor MiRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Front. Oncol. 2021, 11, 708765. [Google Scholar] [CrossRef]
- Musumeci, M.; Coppola, V.; Addario, A.; Patrizii, M.; Maugeri-Saccá, M.; Memeo, L.; Colarossi, C.; Francescangeli, F.; Biffoni, M.; Collura, D.; et al. Control of Tumor and Microenvironment Cross-Talk by MiR-15a and MiR-16 in Prostate Cancer. Oncogene 2011, 30, 4231–4242. [Google Scholar] [CrossRef]
- Sadeghi, M.S.; Lotfi, M.; Soltani, N.; Farmani, E.; Fernandez, J.H.O.; Akhlaghitehrani, S.; Mohammed, S.H.; Yasamineh, S.; Kalajahi, H.G.; Gholizadeh, O. Recent Advances on High-Efficiency of MicroRNAs in Different Types of Lung Cancer: A Comprehensive Review. Cancer Cell Int. 2023, 23, 284. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 388354. [Google Scholar] [CrossRef]
- Wu, K.L.; Tsai, Y.M.; Lien, C.T.; Kuo, P.L.; Hung, J.Y. The Roles of MicroRNA in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 1611. [Google Scholar] [CrossRef]
- Kang, Y.; Jia, Y.; Wang, Q.; Zhao, Q.; Song, M.; Ni, R.; Wang, J. Long Noncoding RNA KCNQ1OT1 Promotes the Progression of Non-Small Cell Lung Cancer via Regulating MiR-204-5p/ATG3 Axis. Onco. Targets Ther. 2019, 12, 10787–10797. [Google Scholar] [CrossRef]
- Sun, L.L.; Li, W.D.; Lei, F.R.; Li, X.Q. The Regulatory Role of MicroRNAs in Angiogenesis-Related Diseases. J. Cell. Mol. Med. 2018, 22, 4568–4587. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Wang, H.; Liang, Y.; Zhao, L.; Li, Y.; Yan, Y.; Zhao, H.; Zhang, X.; Zou, F. MiR-15a-5p Enhances the Malignant Phenotypes of Colorectal Cancer Cells through the STAT3/TWIST1 and PTEN/AKT Signaling Pathways by Targeting SIRT4. Cell Signal. 2023, 101, 110517. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Yang, Y.; Ran, J.; Zhang, L.; Yao, M.; Liu, Z.; Zhang, L. MiR-15a-5p Inhibits Metastasis and Lipid Metabolism by Suppressing Histone Acetylation in Lung Cancer. Free Radic. Biol. Med. 2020, 161, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Naidu, S.; Garofalo, M. MicroRNAs: An Emerging Paradigm in Lung Cancer Chemoresistance. Front. Med. 2015, 2, 166854. [Google Scholar] [CrossRef]
- Wu, S.G.; Chang, T.H.; Liu, Y.N.; Shih, J.Y. MicroRNA in Lung Cancer Metastasis. Cancers 2019, 11, 265. [Google Scholar] [CrossRef]
- Frydrychowicz, M.; Kuszel, Ł.; Dworacki, G.; Budna-Tukan, J. MicroRNA in Lung Cancer—A Novel Potential Way for Early Diagnosis and Therapy. J. Appl. Genet. 2023, 64, 459–477. [Google Scholar] [CrossRef]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of Non-Small Cell Lung Tumor Development by the Let-7 MicroRNA Family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef]
- Feng, H.; Ge, F.; Du, L.; Zhang, Z.; Liu, D. MiR-34b-3p Represses Cell Proliferation, Cell Cycle Progression and Cell Apoptosis in Non-Small-Cell Lung Cancer (NSCLC) by Targeting CDK4. J. Cell. Mol. Med. 2019, 23, 5282–5291. [Google Scholar] [CrossRef]
- Xiong, S.; Zheng, Y.; Jiang, P.; Liu, R.; Liu, X.; Chu, Y. MicroRNA-7 Inhibits the Growth of Human Non-Small Cell Lung Cancer A549 Cells through Targeting BCL-2. Int. J. Biol. Sci. 2011, 7, 805–814. [Google Scholar] [CrossRef]
- Chu, Y. Abstract 4183: MicroRNA-7 Targeting PSME3 Inhibits the Growth of Non-Small Cell Lung Cancer. Cancer Res. 2013, 73, 4183. [Google Scholar] [CrossRef]
- CHENG, T.; HU, C.; YANG, H.; CAO, L.; AN, J. Transforming Growth Factor-β-Induced MiR-143 Expression in Regulation of Non-Small Cell Lung Cancer Cell Viability and Invasion Capacity in Vitro and in Vivo. Int. J. Oncol. 2014, 45, 1977–1988. [Google Scholar] [CrossRef] [PubMed]
- Arechaga-Ocampo, E.; Lopez-Camarillo, C.; Villegas-Sepulveda, N.; Gonzalez-De la Rosa, C.H.; Perez-Añorve, I.X.; Roldan-Perez, R.; Flores-Perez, A.; Peña-Curiel, O.; Angeles-Zaragoza, O.; Rangel Corona, R.; et al. Tumor Suppressor MiR-29c Regulates Radioresistance in Lung Cancer Cells. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, Y.; Jin, M.; Wang, J.; Li, S.; Chen, Z.; Yu, W. MicroRNA-134 Reverses Multidrug Resistance in Human Lung Adenocarcinoma Cells by Targeting FOXM1. Oncol. Lett. 2017, 13, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.-C.; Li, S.-J.; Li, D.-J.; Sun, C.-C.; Li, S.-J.; Li, D.-J. Hsa-MiR-134 Suppresses Non-Small Cell Lung Cancer (NSCLC) Development through down-Regulation of CCND1. Oncotarget 2016, 7, 35960–35978. [Google Scholar] [CrossRef]
- Kwon, J.J.; Factora, T.D.; Dey, S.; Kota, J. A Systematic Review of MiR-29 in Cancer. Mol. Ther. Oncolytics 2019, 12, 173–194. [Google Scholar] [CrossRef]
- Chi, Y.; Jin, Q.; Liu, X.; Xu, L.; He, X.; Shen, Y.; Zhou, Q.; Zhang, J.; Jin, M. MiR-203 Inhibits Cell Proliferation, Invasion, and Migration of Non-Small-Cell Lung Cancer by Downregulating RGS17. Cancer Sci. 2017, 108, 2366–2372. [Google Scholar] [CrossRef]
- Liu, B.; Qu, J.; Xu, F.; Guo, Y.; Wang, Y.; Yu, H.; Qian, B.; Liu, B.; Qu, J.; Xu, F.; et al. MiR-195 Suppresses Non-Small Cell Lung Cancer by Targeting CHEK1. Oncotarget 2015, 6, 9445–9456. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, L.; Hu, Q.; Xia, J.; Sun, J.; Wang, X.; Xiong, H.; Gurbani, D.; Li, L.; Liu, Y.; et al. MicroRNA-218 Functions as a Tumor Suppressor in Lung Cancer by Targeting IL-6/STAT3 and Negatively Correlates with Poor Prognosis. Mol. Cancer 2017, 16, 141. [Google Scholar] [CrossRef]
- Meng, W.; Li, Y.; Chai, B.; Liu, X.; Ma, Z. MiR-199a: A Tumor Suppressor with Noncoding RNA Network and Therapeutic Candidate in Lung Cancer. Int. J. Mol. Sci. 2022, 23, 8518. [Google Scholar] [CrossRef]
- Yeh, M.; Oh, C.S.; Yoo, J.Y.; Kaur, B.; Lee, T.J. Pivotal Role of MicroRNA-138 in Human Cancers. Am. J. Cancer Res. 2019, 9, 1118. [Google Scholar]
- Chen, T.M.; Xiao, Q.; Wang, X.J.; Wang, Z.Q.; Hu, J.W.; Zhang, Z.; Gong, Z.N.; Chen, S.L. MiR-16 Regulates Proliferation and Invasion of Lung Cancer Cells via the ERK/MAPK Signaling Pathway by Targeted Inhibition of MAPK Kinase 1 (MEK1). J. Int. Med. Res. 2019, 47, 5194–5204. [Google Scholar] [CrossRef]
- Liang, L.; Xu, W.y.; Shen, A.; Cen, H.y.; Chen, Z.j.; Tan, L.; Zhang, L.m.; Zhang, Y.; Fu, J.j.; Qin, A.p.; et al. Promoter Methylation-Regulated MiR-148a-3p Inhibits Lung Adenocarcinoma (LUAD) Progression by Targeting MAP3K9. Acta Pharmacol. Sin. 2022, 43, 2946–2955. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Min, L.; Ren, C.; Xu, X.; Yang, J.; Sun, X.; Wang, T.; Wang, F.; Sun, C.; Zhang, X. MiRNA-148a Serves as a Prognostic Factor and Suppresses Migration and Invasion through Wnt1 in Non-Small Cell Lung Cancer. PLoS ONE 2017, 12, e0171751. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, H.; Liu, X.; Hu, Y.; Ding, L.; Zhang, X.; Sun, Q.; Li, Y. Oncogenic MicroRNA-411 Promotes Lung Carcinogenesis by Directly Targeting Suppressor Genes SPRY4 and TXNIP. Oncogene 2018, 38, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.H.; Tsai, M.F.; Gow, C.H.; Wu, S.G.; Liu, Y.N.; Chang, Y.L.; Yu, S.L.; Tsai, H.C.; Lin, S.W.; Chen, Y.W.; et al. Upregulation of MicroRNA-137 Expression by Slug Promotes Tumor Invasion and Metastasis of Non-Small Cell Lung Cancer Cells through Suppression of TFAP2C. Cancer Lett. 2017, 402, 190–202. [Google Scholar] [CrossRef]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A MicroRNA Polycistron as a Potential Human Oncogene. Nature 2005, 435, 828. [Google Scholar] [CrossRef]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A Polycistronic MicroRNA Cluster, MiR-17-92, Is Overexpressed in Human Lung Cancers and Enhances Cell Proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef]
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of Tumor Angiogenesis by a Myc-Activated MicroRNA Cluster. Nat. Genet. 2006, 38, 1060. [Google Scholar] [CrossRef]
- Tiong, T.Y.; Chan, M.l.; Wang, C.H.; Yadav, V.K.; Pikatan, N.W.; Fong, I.H.; Yeh, C.T.; Kuo, K.T.; Huang, W.C. Exosomal MiR-21 Determines Lung-to-Brain Metastasis Specificity through the DGKB/ERK Axis within the Tumor Microenvironment. Life Sci. 2023, 329, 121945. [Google Scholar] [CrossRef]
- Liu, X.; Sempere, L.F.; Ouyang, H.; Memoli, V.A.; Andrew, A.S.; Luo, Y.; Demidenko, E.; Korc, M.; Shi, W.; Preis, M.; et al. MicroRNA-31 Functions as an Oncogenic MicroRNA in Mouse and Human Lung Cancer Cells by Repressing Specific Tumor Suppressors. J. Clin. Investig. 2010, 120, 1298. [Google Scholar] [CrossRef]
- Kim, G.; An, H.J.; Lee, M.J.; Song, J.Y.; Jeong, J.Y.; Lee, J.H.; Jeong, H.C. Hsa-MiR-1246 and Hsa-MiR-1290 Are Associated with Stemness and Invasiveness of Non-Small Cell Lung Cancer. Lung Cancer 2016, 91, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Wang, H.; Liu, B. MiR-211 Promotes Non-Small Cell Lung Cancer Proliferation by Targeting SRCIN1. Tumor Biol. 2016, 37, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, I.; D’Angelo, D.; Pallante, P.; Santos, M.; Scrima, M.; Malanga, D.; De Marco, C.; Ravo, M.; Weisz, A.; Laudanna, C.; et al. Analysis of MiRNA Profiles Identified MiR-196a as a Crucial Mediator of Aberrant PI3K/AKT Signaling in Lung Cancer Cells. Oncotarget 2016, 8, 19172–19191. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Chen, Y.Z.; Lee, M.Y.; Weng, K.P.; Chang, H.T.; Yu, S.Y.; Dong, B.J.; Kuo, F.R.; Hung, L.T.; Liu, L.F.; et al. Comprehensive Identification of MicroRNA Arm Selection Preference in Lung Cancer: MiR-324-5p and -3p Serve Oncogenic Functions in Lung Cancer. Oncol. Lett. 2018, 15, 9818–9826. [Google Scholar] [CrossRef]
- Peng, X.; Guan, L.; Gao, B. MiRNA-19 Promotes Non-Small-Cell Lung Cancer Cell Proliferation via Inhibiting CBX7 Expression. Onco. Targets Ther. 2018, 11, 8865–8874. [Google Scholar] [CrossRef]
- Jin, R.H.; Yu, D.J.; Zhong, M. MiR-1269a Acts as an Onco-MiRNA in Non-Small Cell Lung Cancer via down-Regulating SOX6. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4888–4897. [Google Scholar] [CrossRef]
- Chaniad, P.; Trakunran, K.; Geater, S.L.; Keeratichananont, W.; Thongsuksai, P.; Raungrut, P. Serum MiRNAs Associated with Tumor-Promoting Cytokines in Non-Small Cell Lung Cancer. PLoS ONE 2020, 15, e0241593. [Google Scholar] [CrossRef]
- Li, J.; Feng, Q.; Wei, X.; Yu, Y. MicroRNA-490 Regulates Lung Cancer Metastasis by Targeting Poly r(C)-Binding Protein 1. Tumor Biol. 2016, 37, 15221–15228. [Google Scholar] [CrossRef]
- Kanaoka, R.; Iinuma, H.; Dejima, H.; Sakai, T.; Uehara, H.; Matsutani, N.; Kawamura, M. Usefulness of Plasma Exosomal MicroRNA-451a as a Noninvasive Biomarker for Early Prediction of Recurrence and Prognosis of Non-Small Cell Lung Cancer. Oncology 2018, 94, 311–323. [Google Scholar] [CrossRef]
- Zhou, R.; Zhou, X.; Yin, Z.; Guo, J.; Hu, T.; Jiang, S.; Liu, L.; Dong, X.; Zhang, S.; Wu, G. MicroRNA-574-5p Promotes Metastasis of Non-Small Cell Lung Cancer by Targeting PTPRU. Sci. Rep. 2016, 6, 35714. [Google Scholar] [CrossRef]
- Xue, X.; Liu, Y.; Wang, Y.; Meng, M.; Wang, K.; Zang, X.; Zhao, S.; Sun, X.; Cui, L.; Pan, L.; et al. MiR-21 and MiR-155 Promote Non-Small Cell Lung Cancer Progression by Downregulating SOCS1, SOCS6, and PTEN. Oncotarget 2016, 7, 84508–84519. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, O.; Borzi, C.; Milione, M.; Centonze, G.; Conte, D.; Boeri, M.; Verri, C.; Moro, M.; Facchinetti, F.; Andriani, F.; et al. Circulating Mir-320a Promotes Immunosuppressive Macrophages M2 Phenotype Associated with Lung Cancer Risk. Int. J. Cancer 2019, 144, 2746–2761. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Cui, Y.; Li, Z.; Jiao, Z.; Zhang, Y.; He, Y.; Cheng, G.; Zhou, Q.; Wang, W.; Zhou, X.; et al. Radiation-Induced MiR-208a Increases the Proliferation and Radioresistance by Targeting P21 in Human Lung Cancer Cells. J. Exp. Clin. Cancer Res. 2016, 35, 7. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Liao, Y.; Qiu, Y.; Liu, H.; Tan, D.; Wu, T.; Tang, M.; Zhang, S.; Wang, H. MiRNA 146a Promotes Chemotherapy Resistance in Lung Cancer Cells by Targeting DNA Damage Inducible Transcript 3 (CHOP). Cancer Lett. 2018, 428, 55–68. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, Z.; Yuan, S.; Xie, W.; Li, C.; Hu, Z.; Xiang, Y.; Wu, N.; Wu, L.; Bai, L.; et al. Circulating Exosomal MicroRNAs as Prognostic Biomarkers for Non-Small-Cell Lung Cancer. Oncotarget 2016, 8, 13048. [Google Scholar] [CrossRef]
- Jin, X.; Guan, Y.; Zhang, Z.; Wang, H. Microarray Data Analysis on Gene and MiRNA Expression to Identify Biomarkers in Non-Small Cell Lung Cancer. BMC Cancer 2020, 20, 329. [Google Scholar] [CrossRef]
- Geng, X.; Tsou, J.H.; Stass, S.A.; Jiang, F. Utilizing MiSeq Sequencing to Detect Circulating MicroRNAs in Plasma for Improved Lung Cancer Diagnosis. Int. J. Mol. Sci. 2023, 24, 10277. [Google Scholar] [CrossRef]
- Usó, M.; Jantus-Lewintre, E.; Sirera, R.; Bremnes, R.M.; Camps, C. MiRNA Detection Methods and Clinical Implications in Lung Cancer. Future Oncol. 2014, 10, 2279–2292. [Google Scholar] [CrossRef]
- Shaterabadi, D.; Zamani Sani, M.; Rahdan, F.; Taghizadeh, M.; Rafiee, M.; Dorosti, N.; Dianatinasab, A.; Taheri-Anganeh, M.; Asadi, P.; Khatami, S.H.; et al. MicroRNA Biosensors in Lung Cancer. Clin. Chim. Acta 2024, 552, 117676. [Google Scholar] [CrossRef]
- Lampignano, R.; Kloten, V.; Krahn, T.; Schlange, T. Integrating Circulating MiRNA Analysis in the Clinical Management of Lung Cancer: Present or Future? Mol. Aspects Med. 2020, 72, 100844. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical Relevance of Circulating Cell-Free MicroRNAs in Cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood Cell Origin of Circulating MicroRNAs: A Cautionary Note for Cancer Biomarker Studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Peng, Q.; Zhu, J.; Shen, Y.; Lin, K.; Shen, Y.; Zhu, Y. Identification of MiR-210 and Combination Biomarkers as Useful Agents in Early Screening Non-Small Cell Lung Cancer. Gene 2020, 729, 144225. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yang, D.; Zhang, H.; Wei, X.; Ma, T.; Cheng, Z.; Hong, Q.; Hu, J.; Zhuo, H.; Song, Y.; et al. Early Detection of Lung Cancer in Serum by a Panel of MicroRNA Biomarkers. Clin. Lung Cancer 2015, 16, 313–319.e1. [Google Scholar] [CrossRef]
- Gao, L.; Yan, S.B.; Yang, J.; Kong, J.L.; Shi, K.; Ma, F.C.; Huang, L.Z.; Luo, J.; Yin, S.Y.; He, R.Q.; et al. MiR-182-5p and Its Target HOXA9 in Non-Small Cell Lung Cancer: A Clinical and in-Silico Exploration with the Combination of RT-QPCR, MiRNA-Seq and MiRNA-Chip. BMC Med. Genomics 2020, 13, 3. [Google Scholar] [CrossRef]
- Tulinský, L.; Dzian, A.; Mataková, T.; Ihnát, P. Overexpression of the MiR-143/145 and Reduced Expression of the Let-7 and MiR-126 for Early Lung Cancer Diagnosis. J. Appl. Biomed. 2022, 20, 1–6. [Google Scholar] [CrossRef]
- Bianchi, F.; Nicassio, F.; Marzi, M.; Belloni, E.; Dall’Olio, V.; Bernard, L.; Pelosi, G.; Maisonneuve, P.; Veronesi, G.; Di Fiore, P.P. A Serum Circulating MiRNA Diagnostic Test to Identify Asymptomatic High-Risk Individuals with Early Stage Lung Cancer. EMBO Mol. Med. 2011, 3, 495–503. [Google Scholar] [CrossRef]
- Abd-El-Fattah, A.A.; Sadik, N.A.H.; Shaker, O.G.; Aboulftouh, M.L. Differential MicroRNAs Expression in Serum of Patients with Lung Cancer, Pulmonary Tuberculosis, and Pneumonia. Cell Biochem. Biophys. 2013, 67, 875–884. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Z.; Fu, J.; Wang, Z.; Fan, Z.; Lei, T. Endogenous MicroRNA-424 Predicts Clinical Outcome and Its Inhibition Acts as Cancer Suppressor in Human Non-Small Cell Lung Cancer. Biomed. Pharmacother. 2017, 89, 208–214. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Z.; Wang, Q.; Cui, Y.; Luo, S. Clinical Significance of the Expression of MiRNA-21, MiRNA-31 and MiRNA-Let7 in Patients with Lung Cancer. Saudi J. Biol. Sci. 2018, 26, 777. [Google Scholar] [CrossRef]
- Pérez-Sánchez, C.; Barbarroja, N.; Pantaleão, L.C.; López-Sánchez, L.M.; Ozanne, S.E.; Jurado-Gámez, B.; Aranda, E.; Lopez-Pedrera, C.; Rodríguez-Ariza, A. Clinical Utility of MicroRNAs in Exhaled Breath Condensate as Biomarkers for Lung Cancer. J. Pers. Med. 2021, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.Q.; Zhao, R.C.; Morris, K.V. Profiling MicroRNA Expression with Microarrays. Trends Biotechnol. 2008, 26, 70–76. [Google Scholar] [CrossRef]
- Wanunu, M.; Bhattacharya, S.; Xie, Y.; Tor, Y.; Aksimentiev, A.; Drndic, M. Nanopore Analysis of Individual RNA/Antibiotic Complexes. ACS Nano 2011, 5, 9345–9353. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Newbigging, A.M.; Reid, M.S.; Uppal, J.S.; Xu, J.; Zhang, H.; Le, X.C. Signal Amplification in Living Cells: A Review of Microrna Detection and Imaging. Anal. Chem. 2020, 92, 292–308. [Google Scholar] [CrossRef]
- Válóczi, A.; Hornyik, C.; Varga, N.; Burgyán, J.; Kauppinen, S.; Havelda, Z. Sensitive and Specific Detection of MicroRNAs by Northern Blot Analysis Using LNA-Modified Oligonucleotide Probes. Nucleic Acids Res. 2004, 32, e175. [Google Scholar] [CrossRef]
- Nelson, P.T.; Baldwin, D.A.; Scearce, L.M.; Oberholtzer, J.C.; Tobias, J.W.; Mourelatos, Z. Microarray-Based, High-Throughput Gene Expression Profiling of MicroRNAs. Nat. Methods 2004, 1, 155–161. [Google Scholar] [CrossRef]
- Ma, F.; Zhang, Q.; Zhang, C.Y. Catalytic Self-Assembly of Quantum-Dot-Based MicroRNA Nanosensor Directed by Toehold-Mediated Strand Displacement Cascade. Nano Lett. 2019, 19, 6370–6376. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-Time Quantification of MicroRNAs by Stem–Loop RT–PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Hackenberg, M.; Sturm, M.; Langenberger, D.; Falcón-Pérez, J.M.; Aransay, A.M. MiRanalyzer: A MicroRNA Detection and Analysis Tool for next-Generation Sequencing Experiments. Nucleic Acids Res. 2009, 37, W68–W76. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, D.; Tan, Q.; Wang, M.X.; Gu, L.Q. Nanopore-Based Detection of Circulating MicroRNAs in Lung Cancer Patients. Nat. Nanotechnol. 2011, 6, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.; He, Z.; Wang, Y.; Chen, S.J.; Gu, L.Q. Designing a Polycationic Probe for Simultaneous Enrichment and Detection of MicroRNAs in a Nanopore. ACS Nano 2013, 7, 3962–3969. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA Profiling: Approaches and Considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Wu, Y.; Liu, G.; Gooding, J.J. CRISPR Mediated Biosensing Toward Understanding Cellular Biology and Point-of-Care Diagnosis. Angew. Chem. Int. Ed. 2020, 59, 20754–20766. [Google Scholar] [CrossRef]
- Rouhi, S.; Ghasemi, H.; Alizadeh, M.; Movahedpour, A.; Vahedi, F.; Fattahi, M.; Aiiashi, S.; Khatami, S.H. MiRNA-Based Electrochemical Biosensors for Ovarian Cancer. Clinica Chimica Acta 2025, 564, 119946. [Google Scholar] [CrossRef]
- Moazampour, M.; Zare, H.R.; Shekari, Z. Femtomolar Determination of an Ovarian Cancer Biomarker (MiR-200a) in Blood Plasma Using a Label Free Electrochemical Biosensor Based on l -Cysteine Functionalized ZnS Quantum Dots. Anal. Methods 2021, 13, 2021–2029. [Google Scholar] [CrossRef]
- Meng, X.; Pang, X.; Yang, J.; Zhang, X.; Dong, H. Recent Advances in Electrochemiluminescence Biosensors for MicroRNA Detection. Small 2024, 20, 2307701. [Google Scholar] [CrossRef]
- Hesari, M.; Ding, Z. Review—Electrogenerated Chemiluminescence: Light Years Ahead. J. Electrochem. Soc. 2016, 163, H3116–H3131. [Google Scholar] [CrossRef]
- Yang, Y.T.; Liu, J.L.; Sun, M.F.; Yuan, R.; Chai, Y.Q. Highly Efficient Electrochemiluminescence of MnS:CdS@ZnS Core− Shell Quantum Dots for Ultrasensitive Detection of MicroRNA. Anal. Chem. 2022, 94, 6874–6881. [Google Scholar] [CrossRef]
- Cao, W.; Liu, L.; Yuan, R.; Wang, H. High Efficiency Electrochemiluminescence of 3D Porous G-C3N4 with Dissolved O2 as Co-Reactant and Its Sensing Application for Ultrasensitive Detection of MicroRNA in Tumor Cells. Biosens. Bioelectron. 2022, 214, 114506. [Google Scholar] [CrossRef]
- Kerr, E.; Farr, R.; Doeven, E.H.; Nai, Y.H.; Alexander, R.; Guijt, R.M.; Prieto-Simon, B.; Francis, P.S.; Dearnley, M.; Hayne, D.J.; et al. Amplification-Free Electrochemiluminescence Molecular Beacon-Based MicroRNA Sensing Using a Mobile Phone for Detection. Sens. Actuators B Chem. 2021, 330, 129261. [Google Scholar] [CrossRef]
- Qi, H.; Zhang, C. Electrogenerated Chemiluminescence Biosensing. Anal. Chem. 2020, 92, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, J.; Sharma, R.; Dutta, P.; Bhunia, B. Artificial Intelligence in Healthcare: A Mastery. Biotechnol. Genet. Eng. Rev. 2023, 2023, 37013913. [Google Scholar] [CrossRef] [PubMed]
- Christou, C.D.; Mitsas, A.C.; Vlachavas, I.; Tsoulfas, G. The Use of Machine Learning in MicroRNA Diagnostics: Current Perspectives. Microrna 2022, 11, 175–184. [Google Scholar] [CrossRef]
- Muthamilselvan, S.; Ramasami Sundhar Baabu, P.; Palaniappan, A. Microfluidics for Profiling MiRNA Biomarker Panels in AI-Assisted Cancer Diagnosis and Prognosis. Technol. Cancer Res. Treat. 2023, 22, 37365928. [Google Scholar] [CrossRef]
- Vykoukal, J.; Fahrmann, J.F.; Patel, N.; Shimizu, M.; Ostrin, E.J.; Dennison, J.B.; Ivan, C.; Goodman, G.E.; Thornquist, M.D.; Barnett, M.J.; et al. Contributions of Circulating MicroRNAs for Early Detection of Lung Cancer. Cancers 2022, 14, 4221. [Google Scholar] [CrossRef]
- Zheng, D.; Haddadin, S.; Wang, Y.; Gu, L.Q.; Perry, M.C.; Freter, C.E.; Wang, M.X. Plasma MicroRNAs as Novel Biomarkers for Early Detection of Lung Cancer. Int. J. Clin. Exp. Pathol. 2011, 4, 575. [Google Scholar]
- Eggert, J.A.; Palavanzadeh, M.; Blanton, A. Screening and Early Detection of Lung Cancer. Semin. Oncol. Nurs. 2017, 33, 129–140. [Google Scholar] [CrossRef]
- Leng, Q.; Lin, Y.; Jiang, F.; Lee, C.J.; Zhan, M.; Fang, H.B.; Wang, Y.; Jiang, F. A Plasma MiRNA Signature for Lung Cancer Early Detection. Oncotarget 2017, 8, 111902. [Google Scholar] [CrossRef]
- Ying, L.; Du, L.; Zou, R.; Shi, L.; Zhang, N.; Jin, J.; Xu, C.; Zhang, F.; Zhu, C.; Wu, J.; et al. Development of a Serum MiRNA Panel for Detection of Early Stage Non-Small Cell Lung Cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 25036–25042. [Google Scholar] [CrossRef]
- Arab, A.; Karimipoor, M.; Irani, S.; Kiani, A.; Zeinali, S.; Tafsiri, E.; Sheikhy, K. Potential Circulating MiRNA Signature for Early Detection of NSCLC. Cancer Genet. 2017, 216–217, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Chen, Z.; Zhao, L.; Zhao, W.; Zhu, Y.; Liu, J.; Zhao, X. A Novel Circulating MiRNA-based Signature for the Early Diagnosis and Prognosis Prediction of Non–Small-cell Lung Cancer. J. Clin. Lab. Anal. 2020, 34, e23505. [Google Scholar] [CrossRef] [PubMed]
- Bica, C.; Jurj, A.; Harangus, A.; Ciocan, C.; Moldovan, A.; Zanoaga, O.; Burz, C.; Ferracin, M.; Raduly, L.; Berindan-Neagoe, I. MiRNA Patterns in Male LUSC Patients— The 3-Way Mirror: Tissue, Plasma and Exosomes. Transl. Oncol. 2024, 44, 101951. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Gaur, V.; Mir, I.A.; Saikia, J.; Kumar, S. MicroRNA-3692-3p Is Overexpressed in Lung Tumors but May Not Serve as a Prognostic Biomarker in Lung Cancer Patients. Mol. Biol. Rep. 2023, 50, 1147–1156. [Google Scholar] [CrossRef]
- He, Y.; Yang, Y.; Kuang, P.; Ren, S.; Rozeboom, L.; Rivard, C.J.; Li, X.; Zhou, C.; Rhirsch, F. Seven-MicroRNA Panel for Lung Adenocarcinoma Early Diagnosis in Patients Presenting with Ground-Glass Nodules. Onco Targets Ther. 2017, 10, 5915. [Google Scholar] [CrossRef]
- Pavel, A.B.; Campbell, J.D.; Liu, G.; Elashoff, D.; Dubinett, S.; Smith, K.; Whitney, D.; Lenburg, M.E.; Spira, A. Alterations in Bronchial Airway MiRNA Expression for Lung Cancer Detection. Cancer Prev. Res. 2017, 10, 651–659. [Google Scholar] [CrossRef]
- Abdipourbozorgbaghi, M.; Vancura, A.; Radpour, R.; Haefliger, S. Circulating MiRNA Panels as a Novel Non-Invasive Diagnostic, Prognostic, and Potential Predictive Biomarkers in Non-Small Cell Lung Cancer (NSCLC). Br. J. Cancer 2024, 131, 1350–1362. [Google Scholar] [CrossRef]
- Pastorino, U.; Boeri, M.; Sestini, S.; Sabia, F.; Milanese, G.; Silva, M.; Suatoni, P.; Verri, C.; Cantarutti, A.; Sverzellati, N.; et al. Baseline Computed Tomography Screening and Blood MicroRNA Predict Lung Cancer Risk and Define Adequate Intervals in the BioMILD Trial. Ann. Oncol. 2022, 33, 395–405. [Google Scholar] [CrossRef]
- Roa, W.H.; Kim, J.O.; Razzak, R.; Du, H.; Guo, L.; Singh, R.; Gazala, S.; Ghosh, S.; Wong, E.; Joy, A.A.; et al. Sputum MicroRNA Profiling: A Novel Approach for the Early Detection of Non-Small Cell Lung Cancer. Clin. Investig. Med. 2012, 35, E271. [Google Scholar] [CrossRef]
- Xie, Y.; Todd, N.W.; Liu, Z.; Zhan, M.; Fang, H.B.; Peng, H.; Alattar, M.; Deepak, J.; Stass, S.A.; Jiang, F. Altered MiRNA Expression in Sputum for Diagnosis of Non-Small Cell Lung Cancer. Lung Cancer 2010, 67, 170–176. [Google Scholar] [CrossRef]
- Xing, L.; Todd, N.W.; Yu, L.; Fang, H.; Jiang, F. Early Detection of Squamous Cell Lung Cancer in Sputum by a Panel of MicroRNA Markers. Mod. Pathol. 2010, 23, 1157–1164. [Google Scholar] [CrossRef]
- Gao, X.; Wang, Y.; Zhao, H.; Wei, F.; Zhang, X.; Su, Y.; Wang, C.; Li, H.; Ren, X. Plasma MiR-324-3p and MiR-1285 as Diagnostic and Prognostic Biomarkers for Early Stage Lung Squamous Cell Carcinoma. Oncotarget 2016, 7, 59664–59675. [Google Scholar] [CrossRef] [PubMed]
- CHEN, Y.; MIN, L.; ZHANG, X.; HU, S.; WANG, B.; LIU, W.; WANG, R.; GU, X.; SHEN, W.; LV, H.; et al. Decreased MiRNA-148a Is Associated with Lymph Node Metastasis and Poor Clinical Outcomes and Functions as a Suppressor of Tumor Metastasis in Non-Small Cell Lung Cancer. Oncol. Rep. 2013, 30, 1832–1840. [Google Scholar] [CrossRef] [PubMed]
- Xiao-chun, W.; Wei, W.; Zhu-Bo, Z.; Jing, Z.; Xiao-Gang, T.; Jian-Chao, L. Overexpression of MiRNA-21 Promotes Radiation-Resistance of Non-Small Cell Lung Cancer. Radiat. Oncol. 2013, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Cui, E.H.; Li, H.J.; Hua, F.; Wang, B.; Mao, W.; Feng, X.R.; Li, J.Y.; Wang, X. Serum MicroRNA 125b as a Diagnostic or Prognostic Biomarker for Advanced NSCLC Patients Receiving Cisplatin-Based Chemotherapy. Acta Pharmacol. Sin. 2012, 34, 309–313. [Google Scholar] [CrossRef]
- de Miguel-Perez, D.; Ortega, F.G.; Tejada, R.G.; Martínez-Única, A.; Peterson, C.B.; Russo, A.; Gunasekaran, M.; Cardona, A.F.; Amezcua, V.; Lorente, J.A.; et al. Baseline Extracellular Vesicle MiRNA-30c and Autophagic CTCs Predict Chemoradiotherapy Resistance and Outcomes in Patients with Lung Cancer. Biomark Res. 2023, 11, 1–5. [Google Scholar] [CrossRef]
- Jiang, X.; Yuan, Y.; Tang, L.; Wang, J.; Zhang, D.; Cho, W.C.; Duan, L. Identification and Validation Prognostic Impact of MiRNA-30a-5p in Lung Adenocarcinoma. Front. Oncol. 2022, 12, 831997. [Google Scholar] [CrossRef]
- Xu, X.; Jin, S.; Ma, Y.; Fan, Z.; Yan, Z.; Li, W.; Song, Q.; You, W.; Lyu, Z.; Song, Y.; et al. MiR-30a-5p Enhances Paclitaxel Sensitivity in Non-Small Cell Lung Cancer through Targeting BCL-2 Expression. J. Mol. Med. 2017, 95, 861–871. [Google Scholar] [CrossRef]
- Leidinger, P.; Galata, V.; Backes, C.; Stähler, C.; Rheinheimer, S.; Huwer, H.; Meese, E.; Keller, A. Longitudinal Study on Circulating MiRNAs in Patients after Lung Cancer Resection. Oncotarget 2015, 6, 16674. [Google Scholar] [CrossRef]
- Li, T.; Liu, H.; Dong, C.; Lyu, J. Application of MiRNA Biomarkers in Predicting Overall Survival Outcomes for Lung Adenocarcinoma. Biomed. Res. Int. 2022, 2022, 5249576. [Google Scholar] [CrossRef]
- Tang, L.; Yuan, Y.; Zhai, H.; Wang, J.; Zhang, D.; Liang, H.; Shi, Y.; Duan, L.; Jiang, X. MicroRNA-125b-5p Correlates With Prognosis and Lung Adenocarcinoma Progression. Front. Mol. Biosci. 2022, 8, 788690. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharawat, S.K.; Ali, A.; Gaur, V.; Malik, P.S.; Kumar, S.; Mohan, A.; Guleria, R. Identification of Differentially Expressed Circulating Serum MicroRNA for the Diagnosis and Prognosis of Indian Non–Small Cell Lung Cancer Patients. Curr. Probl. Cancer 2020, 44, 100540. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.P.; Cinegaglia, N.C.; Felix, T.F.; Evangelista, A.F.; Oliveira, R.A.; Hasimoto, E.N.; Cataneo, D.C.; Cataneo, A.J.M.; Neto, C.S.; Viana, C.R.; et al. Deregulated MicroRNAs Are Associated with Patient Survival and Predicted to Target Genes That Modulate Lung Cancer Signaling Pathways. Cancers 2020, 12, 2711. [Google Scholar] [CrossRef] [PubMed]
- Vucic, E.A.; Thu, K.L.; Pikor, L.A.; Enfield, K.S.S.; Yee, J.; English, J.C.; MacAulay, C.E.; Lam, S.; Jurisica, I.; Lam, W.L. Smoking Status Impacts MicroRNA Mediated Prognosis and Lung Adenocarcinoma Biology. BMC Cancer 2014, 14, 1–14. [Google Scholar] [CrossRef]
- Tan, W.; Gu, J.; Huang, M.; Wu, X.; Hildebrandt, M.A.T. Epigenetic Analysis of MicroRNA Genes in Tumors from Surgically Resected Lung Cancer Patients and Association with Survival. Mol. Carcinog. 2015, 54, E45–E51. [Google Scholar] [CrossRef]
- Wang, N.; Guo, H.; Dong, Z.; Chen, Q.; Zhang, X.; Shen, W.; Bao, Y.; Wang, X. Cancer Management and Research Dovepress Establishment and Validation of a 7-MicroRNA Prognostic Signature for Non-Small Cell Lung Cancer. Cancer Manag. Res. 2018, 10, 3463. [Google Scholar] [CrossRef]
- Yu, H.; Jiang, L.; Sun, C.; Guo, L.; Lin, M.; Huang, J.; Zhu, L. Decreased Circulating MiR-375: A Potential Biomarker for Patients with Non-Small-Cell Lung Cancer. Gene 2014, 534, 60–65. [Google Scholar] [CrossRef]
- Silva, J.; García, V.; Zaballos, Á.; Provencio, M.; Lombardía, L.; Almonacid, L.; García, J.M.; Domínguez, G.; Peña, C.; Diaz, R.; et al. Vesicle-Related MicroRNAs in Plasma of Nonsmall Cell Lung Cancer Patients and Correlation with Survival. Eur. Respir. J. 2011, 37, 617–623. [Google Scholar] [CrossRef]
- Kumar, S.; Sharawat, S.K.; Ali, A.; Gaur, V.; Malik, P.S.; Pandey, M.; Kumar, S.; Mohan, A.; Guleria, R. Differential Expression of Circulating Serum MiR-1249-3p, MiR-3195, and MiR-3692-3p in Non-Small Cell Lung Cancer. Hum. Cell 2020, 33, 839–849. [Google Scholar] [CrossRef]
- Duncavage, E.; Goodgame, B.; Sezhiyan, A.; Govindan, R.; Pfeifer, J. Use of MicroRNA Expression Levels to Predict Outcomes in Resected Stage I Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2010, 5, 1755–1763. [Google Scholar] [CrossRef]
- Joerger, M.; Baty, F.; Früh, M.; Droege, C.; Stahel, R.A.; Betticher, D.C.; von Moos, R.; Ochsenbein, A.; Pless, M.; Gautschi, O.; et al. Circulating MicroRNA Profiling in Patients with Advanced Non-Squamous NSCLC Receiving Bevacizumab/Erlotinib Followed by Platinum-Based Chemotherapy at Progression (SAKK 19/05). Lung Cancer 2014, 85, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, C.; Monastirioti, A.; Rounis, K.; Makrakis, D.; Kalbakis, K.; Nikolaou, C.; Mavroudis, D.; Agelaki, S. Circulating MicroRNAs Regulating DNA Damage Response and Responsiveness to Cisplatin in the Prognosis of Patients with Non-Small Cell Lung Cancer Treated with First-Line Platinum Chemotherapy. Cancers 2020, 12, 1282. [Google Scholar] [CrossRef] [PubMed]
- Monastirioti, A.; Papadaki, C.; Rounis, K.; Kalapanida, D.; Mavroudis, D.; Agelaki, S. A Prognostic Role for Circulating Micrornas Involved in Macrophage Polarization in Advanced Non-Small Cell Lung Cancer. Cells 2021, 10, 1988. [Google Scholar] [CrossRef] [PubMed]
- Bi, N.; Cao, J.; Song, Y.; Shen, J.; Liu, W.; Fan, J.; He, J.; Shi, Y.; Zhang, X.; Lu, N.; et al. A MicroRNA Signature Predicts Survival in Early Stage Small-Cell Lung Cancer Treated with Surgery and Adjuvant Chemotherapy. PLoS ONE 2014, 9, e91388. [Google Scholar] [CrossRef]
- Ye, Z.; Sun, B.; Xiao, Z. Machine Learning Identifies 10 Feature MiRNAs for Lung Squamous Cell Carcinoma. Gene 2020, 749, 144669. [Google Scholar] [CrossRef]
- Sanfiorenzo, C.; Ilie, M.I.; Belaid, A.; Barlési, F.; Mouroux, J.; Marquette, C.H.; Brest, P.; Hofman, P. Two Panels of Plasma MicroRNAs as Non-Invasive Biomarkers for Prediction of Recurrence in Resectable NSCLC. PLoS ONE 2013, 8, e54596. [Google Scholar] [CrossRef]
- Simiene, J.; Dabkeviciene, D.; Stanciute, D.; Prokarenkaite, R.; Jablonskiene, V.; Askinis, R.; Normantaite, K.; Cicenas, S.; Suziedelis, K. Potential of MiR-181a-5p and MiR-630 as Clinical Biomarkers in NSCLC. BMC Cancer 2023, 23, 857. [Google Scholar] [CrossRef]
- Shi, S.B.; Wang, M.; Tian, J.; Li, R.; Chang, C.X.; Qi, J.L. MicroRNA 25, MicroRNA 145, and MicroRNA 210 as Biomarkers for Predicting the Efficacy of Maintenance Treatment with Pemetrexed in Lung Adenocarcinoma Patients Who Are Negative for Epidermal Growth Factor Receptor Mutations or Anaplastic Lymphoma Kinase Translocations. Transl. Res. 2016, 170, 1–7. [Google Scholar] [CrossRef]
- Gao, X.; Wu, Y.; Yu, W.; Li, H. Identification of a Seven-MiRNA Signature as Prognostic Biomarker for Lung Squamous Cell Carcinoma. Oncotarget 2016, 7, 81670. [Google Scholar] [CrossRef]
- Kaduthanam, S.; Gade, S.; Meister, M.; Brase, J.C.; Johannes, M.; Dienemann, H.; Warth, A.; Schnabel, P.A.; Herth, F.J.F.; Sültmann, H.; et al. Serum MiR-142-3p Is Associated with Early Relapse in Operable Lung Adenocarcinoma Patients. Lung Cancer 2013, 80, 223–227. [Google Scholar] [CrossRef]
- Katakura, S.; Kobayashi, N.; Hashimoto, H.; Kamimaki, C.; Tanaka, K.; Kubo, S.; Nakashima, K.; Teranishi, S.; Manabe, S.; Watanabe, K.; et al. MicroRNA-200b Is a Potential Biomarker of the Expression of PD-L1 in Patients with Lung Cancer. Thorac. Cancer 2020, 11, 2975. [Google Scholar] [CrossRef] [PubMed]
- Simiene, J.; Kunigenas, L.; Prokarenkaite, R.; Dabkeviciene, D.; Strainiene, E.; Stankevicius, V.; Cicenas, S.; Suziedelis, K. Prognostic Value of MiR-10a-3p in Non-Small Cell Lung Cancer Patients. Onco Targets Ther. 2024, 17, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xu, P.; Mi, Y.; Wang, W.; Pan, X.; Wu, X.; He, Q.; Liu, H.; Tang, W.; An, H. Plasma MiRNA Alterations between NSCLC Patients Harboring Del19 and L858R EGFR Mutations. Oncotarget 2016, 7, 54965. [Google Scholar] [CrossRef] [PubMed]
- Reguart, N.; Remon, J. Common EGFR-Mutated Subgroups (Del19/L858R) in Advanced Non-Small-Cell Lung Cancer: Chasing Better Outcomes with Tyrosine Kinase Inhibitors. Future Oncol. 2015, 11, 1245–1257. [Google Scholar] [CrossRef]
- Ranade, A.R.; Cherba, D.; Sridhar, S.; Richardson, P.; Webb, C.; Paripati, A.; Bowles, B.; Weiss, G.J. MicroRNA 92a-2*: A Biomarker Predictive for Chemoresistance and Prognostic for Survival in Patients with Small Cell Lung Cancer. J. Thorac. Oncol. 2010, 5, 1273–1278. [Google Scholar] [CrossRef]
- Cuttano, R.; Colangelo, T.; Guarize, J.; Dama, E.; Cocomazzi, M.P.; Mazzarelli, F.; Melocchi, V.; Palumbo, O.; Marino, E.; Belloni, E.; et al. MiRNome Profiling of Lung Cancer Metastases Revealed a Key Role for MiRNA-PD-L1 Axis in the Modulation of Chemotherapy Response. J. Hematol. Oncol. 2022, 15, 178. [Google Scholar] [CrossRef]
- Petriella, D.; De Summa, S.; Lacalamita, R.; Galetta, D.; Catino, A.; Logroscino, A.F.; Palumbo, O.; Carella, M.; Zito, F.A.; Simone, G.; et al. MiRNA Profiling in Serum and Tissue Samples to Assess Noninvasive Biomarkers for NSCLC Clinical Outcome. Tumor Biol. 2016, 37, 5503–5513. [Google Scholar] [CrossRef]
- Kulda, V.; Svaton, M.; Mukensnabl, P.; Hrda, K.; Dvorak, P.; Houdek, Z.; Houfkova, K.; Vrzakova, R.; Babuska, V.; Pesek, M.; et al. Predictive Relevance of MiR-34a, MiR-224 and MiR-342 in Patients with Advanced Squamous Cell Carcinoma of the Lung Undergoing Palliative Chemotherapy. Oncol. Lett. 2017, 15, 592. [Google Scholar] [CrossRef]
- Kasinski, A.L.; Slack, F.J. MiRNA-34 Prevents Cancer Initiation and Progression in a Therapeutically Resistant K-Ras and P53-Induced Mouse Model of Lung Adenocarcinoma. Cancer Res. 2012, 72, 5576–5587. [Google Scholar] [CrossRef]
- Fan, J.; Yin, Z.; Xu, J.; Wu, F.; Huang, Q.; Yang, L.; Jin, Y.; Yang, G. Circulating MicroRNAs Predict the Response to Anti-PD-1 Therapy in Non-Small Cell Lung Cancer. Genomics 2020, 112, 2063–2071. [Google Scholar] [CrossRef]
- Franchina, T.; Amodeo, V.; Bronte, G.; Savio, G.; Ricciardi, G.R.R.; Picciotto, M.; Russo, A.; Giordano, A.; Adamo, V. Circulating MiR-22, MiR-24 and MiR-34a as Novel Predictive Biomarkers to Pemetrexed-Based Chemotherapy in Advanced Non-Small Cell Lung Cancer. J. Cell Physiol. 2014, 229, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Petrek, H.; Yu, A.M. MicroRNAs in Non-Small Cell Lung Cancer: Gene Regulation, Impact on Cancer Cellular Processes, and Therapeutic Potential. Pharmacol. Res. Perspect. 2019, 7, e00528. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, Y.; Chen, L.; Zhao, J.; Guo, M.; Zhao, X.; Wen, Z.; He, Z.; Chen, C.; Xu, L. MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges? Biomolecules 2023, 13, 877. [Google Scholar] [CrossRef] [PubMed]
- Gambari, R.; Brognara, E.; Spandidos, D.A.; Fabbri, E. Targeting OncomiRNAs and Mimicking Tumor Suppressor MiRNAs: Ew Trends in the Development of MiRNA Therapeutic Strategies in Oncology (Review). Int. J. Oncol. 2016, 49, 5–32. [Google Scholar] [CrossRef]
- Ye, Q.; Raese, R.; Luo, D.; Cao, S.; Wan, Y.W.; Qian, Y.; Guo, N.L. MicroRNA, MRNA, and Proteomics Biomarkers and Therapeutic Targets for Improving Lung Cancer Treatment Outcomes. Cancers 2023, 15, 2294. [Google Scholar] [CrossRef]
- Hussen, B.M.; Saleem, S.J.; Abdullah, S.R.; Mohamadtahr, S.; Hidayat, H.J.; Rasul, M.F.; Taheri, M.; Kiani, A. Current Landscape of MiRNAs and TGF-β Signaling in Lung Cancer Progression and Therapeutic Targets. Mol. Cell. Probes 2023, 72, 101929. [Google Scholar] [CrossRef]
- Tang, S.; Li, S.; Liu, T.; He, Y.; Hu, H.; Zhu, Y.; Tang, S.; Zhou, H. MicroRNAs: Emerging Oncogenic and Tumor-Suppressive Regulators, Biomarkers and Therapeutic Targets in Lung Cancer. Cancer Lett. 2021, 502, 71–83. [Google Scholar] [CrossRef]
- Qian, H.; Maghsoudloo, M.; Kaboli, P.J.; Babaeizad, A.; Cui, Y.; Fu, J.; Wang, Q.; Imani, S. Decoding the Promise and Challenges of MiRNA-Based Cancer Therapies: An Essential Update on MiR-21, MiR-34, and MiR-155. Int. J. Med. Sci. 2024, 21, 2781–2798. [Google Scholar] [CrossRef]
- Mizuno, K.; Seki, N.; Mataki, H.; Matsushita, R.; Kamikawaji, K.; Kumamoto, T.; Takagi, K.; Goto, Y.; Nishikawa, R.; Kato, M.; et al. Tumor-Suppressive MicroRNA-29 Family Inhibits Cancer Cell Migration and Invasion Directly Targeting LOXL2 in Lung Squamous Cell Carcinoma. Int. J. Oncol. 2016, 48, 450–460. [Google Scholar] [CrossRef]
- Asghariazar, V.; Sakhinia, E.; Mansoori, B.; Mohammadi, A.; Baradaran, B. Tumor Suppressor MicroRNAs in Lung Cancer: An Insight to Signaling Pathways and Drug Resistance. J. Cell. Biochem. 2019, 120, 19274–19289. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, J.; Huang, Z. Recent Progress in MicroRNA-Based Delivery Systems for the Treatment of Human Disease. ExRNA 2019, 1, 24. [Google Scholar] [CrossRef]
- Nguyen, D.-D.; Chang, S. Molecular Sciences Development of Novel Therapeutic Agents by Inhibition of Oncogenic MicroRNAs. Int. J. Mol. Sci. 2018, 19, 65. [Google Scholar] [CrossRef]
- Tay, F.C.; Lim, J.K.; Zhu, H.; Hin, L.C.; Wang, S. Using Artificial MicroRNA Sponges to Achieve MicroRNA Loss-of-Function in Cancer Cells. Adv. Drug Deliv. Rev. 2015, 81, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, I.; Chatterjee, A. Recent Advances in MiRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Brillante, S.; Volpe, M.; Indrieri, A. Advances in MicroRNA Therapeutics: From Preclinical to Clinical Studies. Hum. Gene Ther. 2024, 35, 17–18. [Google Scholar] [CrossRef]
- Tani, H. Recent Advances and Prospects in RNA Drug Development. Int. J. Mol. Sci. 2024, 25, 12284. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 Study of MRX34, a Liposomal MiR-34a Mimic, in Patients with Advanced Solid Tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and Activity of MicroRNA-Loaded Minicells in Patients with Recurrent Malignant Pleural Mesothelioma: A First-in-Man, Phase 1, Open-Label, Dose-Escalation Study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Zhang, N.; Hu, G.; Myers, T.G.; Williamson, P.R. Protocols for the Analysis of MicroRNA Expression, Biogenesis and Function in Immune Cells. Curr. Protoc. Immunol. 2019, 126, e78. [Google Scholar] [CrossRef]
- Detassis, S.; Grasso, M.; Del Vescovo, V.; Denti, M.A. MicroRNAs Make the Call in Cancer Personalized Medicine. Front. Cell Dev. Biol. 2017, 5, 291696. [Google Scholar] [CrossRef]
- Angele, P.; Zellner, J.; Pattappa, G.; Ho, P.T.B.; Clark, I.M.; Le, L.T.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Sarkar, B.K.; Lee, S.S. The Novel Strategies for Next-Generation Cancer Treatment: MiRNA Combined with Chemotherapeutic Agents for the Treatment of Cancer. Oncotarget 2018, 9, 10164. [Google Scholar] [CrossRef] [PubMed]
- Latini, A.; Borgiani, P.; Novelli, G.; Ciccacci, C. MiRNAs in Drug Response Variability: Potential Utility as Biomarkers for Personalized Medicine. Pharmacogenomics 2019, 20, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- HOTCHI, M.; SHIMADA, M.; KURITA, N.; IWATA, T.; SATO, H.; MORIMOTO, S.; YOSHIKAWA, K.; HIGASHIJIMA, J.; MIYATANI, T. MicroRNA Expression Is Able to Predict Response to Chemoradiotherapy in Rectal Cancer. Mol. Clin. Oncol. 2012, 1, 137. [Google Scholar] [CrossRef]
- Lv, J.F.; Hu, L.; Zhuo, W.; Zhang, C.M.; Zhou, H.H.; Fan, L. Epigenetic Alternations and Cancer Chemotherapy Response. Cancer Chemother. Pharmacol. 2015, 77, 673–684. [Google Scholar] [CrossRef]
- Sayed, S.R.E.; Cristante, J.; Guyon, L.; Denis, J.; Chabre, O.; Cherradi, N. MicroRNA Therapeutics in Cancer: Current Advances and Challenges. Cancers 2021, 13, 2680. [Google Scholar] [CrossRef]
- Zhang, S.; Cheng, Z.; Wang, Y.; Han, T. The Risks of MiRNA Therapeutics: In a Drug Target Perspective. Drug Des. Devel. Ther. 2021, 15, 721. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic Advances of MiRNAs: A Preclinical and Clinical Update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef]
- Seyhan, A.A. Trials and Tribulations of MicroRNA Therapeutics. Int. J. Mol. Sci. 2024, 25, 1469. [Google Scholar] [CrossRef]
- Segal, M.; Slack, F.J. Challenges Identifying Efficacious MiRNA Therapeutics for Cancer. Expert Opin. Drug. Discov. 2020, 15, 987–992. [Google Scholar] [CrossRef]
- Cortez, M.A.; Anfossi, S.; Ramapriyan, R.; Menon, H.; Atalar, S.C.; Aliru, M.; Welsh, J.; Calin, G.A. Role of MiRNAs in Immune Responses and Immunotherapy in Cancer. Genes Chromosomes Cancer 2019, 58, 244. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. (2024, November 15). Nonclinical Safety Assessment of Oligonucleotide-Based Therapeutics; Draft Guidance for Industry; Availability. Federal Register, 89(221), 90296–90298. Available online: https://www.govinfo.gov/content/pkg/FR-2024-11-15/html/2024-26682.htm (accessed on 10 April 2025).
- Piergentili, R.; Basile, G.; Nocella, C.; Carnevale, R.; Marinelli, E.; Patrone, R.; Zaami, S. Using NcRNAs as Tools in Cancer Diagnosis and Treatment—The Way towards Personalized Medicine to Improve Patients’ Health. Int. J. Mol. Sci. 2022, 23, 9353. [Google Scholar] [CrossRef]
- Beale, D.J.; Karpe, A.V.; Ahmed, W. Beyond Metabolomics: A Review of Multi-Omics-Based Approaches. Microbial Metabolomics: Applications in Clinical, Environmental, and Industrial Microbiology; Springer: Berlin/Heidelberg, Germany, 2016; pp. 289–312. [Google Scholar] [CrossRef]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using Machine Learning Approaches for Multi-Omics Data Analysis: A Review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef] [PubMed]
- Aswathy, R.; Chalos, V.A.; Suganya, K.; Sumathi, S. Advancing MiRNA Cancer Research through Artificial Intelligence: From Biomarker Discovery to Therapeutic Targeting. Med. Oncol. 2025, 42, 30. [Google Scholar] [CrossRef] [PubMed]
- Klimentová, E.; Hejret, V.; Krčmář, J.; Grešová, K.; Giassa, I.C.; Alexiou, P. MiRBind: A Deep Learning Method for MiRNA Binding Classification. Genes 2022, 13, 2323. [Google Scholar] [CrossRef]
- He, Q.; Qiao, W.; Fang, H.; Bao, Y. Improving the Identification of MiRNA-Disease Associations with Multi-Task Learning on Gene-Disease Networks. Brief. Bioinform. 2023, 24, bbad203. [Google Scholar] [CrossRef]
- Su, Y.; Fang, H.B.; Jiang, F. Integrating DNA Methylation and MicroRNA Biomarkers in Sputum for Lung Cancer Detection. Clin. Epigenetics 2016, 8, 109. [Google Scholar] [CrossRef]
- Saiyed, A.N.; Vasavada, A.R.; Johar, S.R.K. Recent Trends in MiRNA Therapeutics and the Application of Plant MiRNA for Prevention and Treatment of Human Diseases. Future J. Pharm. Sci. 2022, 8, 24. [Google Scholar] [CrossRef]
- Markou, A.; Tsaroucha, E.G.; Kaklamanis, L.; Fotinou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic Value of Mature MicroRNA-21 and MicroRNA-205 Overexpression in Non–Small Cell Lung Cancer by Quantitative Real-Time RT-PCR. Clin. Chem. 2008, 54, 1696–1704. [Google Scholar] [CrossRef]
- Pereira, D.M.; Rodrigues, P.M.; Borralho, P.M.; Rodrigues, C.M.P. Delivering the Promise of MiRNA Cancer Therapeutics. Drug Discov. Today 2013, 18, 282–289. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Liu, C.; Zhang, X.; Wu, Y.; Diao, M.; Tan, S.; Huang, S.; Cheng, Y.; You, T. MicroRNA-21 as a Diagnostic and Prognostic Biomarker of Lung Cancer: A Systematic Review and Meta-Analysis. Biosci. Rep. 2022, 42, BSR20211653. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zeng, X.; Ma, R.; Wang, L. MicroRNA-21 Promotes the Proliferation, Migration and Invasion of Non-Small Cell Lung Cancer A549 Cells by Regulating Autophagy Activity via AMPK/ULK1 Signaling Pathway. Exp. Ther. Med. 2018, 16, 2038–2045. [Google Scholar] [CrossRef] [PubMed]
- Kunz, M.; Göttlich, C.; Walles, T.; Nietzer, S.; Dandekar, G.; Dandekar, T. MicroRNA-21 versus MicroRNA-34: Lung Cancer Promoting and Inhibitory MicroRNAs Analysed in Silico and in Vitro and Their Clinical Impact. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Jenike, A.E.; Halushka, M.K. MiR-21: A Non-specific Biomarker of All Maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS Is Regulated by the Let-7 MicroRNA Family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Goto, Y.; Tanai, C.; Yoh, K.; Hosomi, Y.; Sakai, H.; Kato, T.; Kaburagi, T.; Nishio, M.; Kim, Y.H.; Inoue, A.; et al. Continuing EGFR-TKI beyond Radiological Progression in Patients with Advanced or Recurrent, EGFR Mutation-Positive Non-Small-Cell Lung Cancer: An Observational Study. ESMO Open 2017, 2, e000214. [Google Scholar] [CrossRef]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gullà, A.; Tagliaferri, P.; Tassone, P.; et al. Mir-34: A New Weapon against Cancer? Mol. Ther. Nucleic Acids 2014, 3, e195. [Google Scholar] [CrossRef]
- Raponi, M.; Dossey, L.; Jatkoe, T.; Wu, X.; Chen, G.; Fan, H.; Beer, D.G. MicroRNA Classifiers for Predicting Prognosis of Squamous Cell Lung Cancer. Cancer Res. 2009, 69, 5776–5783. [Google Scholar] [CrossRef]
| Tumor Suppressor miRNA | Targets/Regulators | Mechanism | Ref. |
|---|---|---|---|
| Apoptosis | |||
| miR-7 | PSME3, BCL-2, Pax6, PTK2, CCND1, FAK | Mir-7 downregulates PSME3, leading to reduced cell proliferation; it targets BCL-2, promoting apoptosis by reducing anti-apoptotic signals; it inhibits Pax6, which downregulates the ERK/MAPK signaling pathway, and PTK2 (FAK), which regulates cell migration and survival; it also downregulates CCND1 to arrest the cell cycle and reduce proliferation. | [31,32] |
| miR-34b-3p | CDK4 | Acts as a tumor suppressor in NSCLC by targeting the 3′-UTR of CDK4 mRNA, leading to decreased CDK4 expression, which inhibits cell proliferation and induces cell cycle arrest and apoptosis. | [30] |
| Apoptosis, cell cycle arrest | |||
| mir-143 | PSME3, BCL-2, Pax6, PTK2 (FAK), CCND1 | miR-143 downregulates PSME3 to reduce cell proliferation; targets BCL-2 to promote apoptosis by reducing anti-apoptotic signals; inhibits Pax6, suppressing the ERK/MAPK pathway; downregulates PTK2 (FAK) to limit migration and survival; targets CCND1 to arrest the cell cycle and reduce proliferation. | [33] |
| Apoptosis, metastasis | |||
| miR-29c | DNA methyltransferases (DNMT3A, DNMT3B) MMPs, VEGF, pro-apoptotic genes (e.g., Bax, Bak), Bcl-2, Mcl-1, Cyclins, CDKs | Regulates DNA methylation by targeting DNMT3A and DNMT3B, helping maintain proper epigenetic regulation, and preventing tumor suppressor gene silencing. Inhibits metastasis by targeting genes involved in extracellular matrix remodeling, such as MMPs, reducing tumor cell migration and invasion. Promotes apoptosis by regulating pro-apoptotic proteins like Bax and Bak, Downregulates anti-apoptotic proteins (Bcl-2, Mcl-1), inducing apoptosis and sensitizing cells to radiation-induced cell death. | [34] |
| miR-134 | FOXM1, CCND1, EGFR, ITGB1, Cyclin D1, Cyclin D2, CDK4, MMP-7, MMP-9, Caspase-3, Bcl-2 | Suppresses EMT, migration, and invasion by targeting FOXM1 and CCND1; reduces EGFR expression, inhibiting cell growth and proliferation; promotes apoptosis via Caspase-3 and Bcl-2 regulation; reduces MMP-7 and MMP-9 for anti-migration effects. | [35,36] |
| Angiogenesis, metastasis | |||
| miR-29 | LOXL2, Wnt-1, DNMT3A, DNMT1 | Suppresses cancer progression by targeting LOXL2 (oncogene) and by demethylating Wnt-1. It also targets DNMTs to reduce hypermethylation of tumor suppressor genes. | [37] |
| Antiproliferation, metastasis | |||
| miR-203 | RGS17 | miR-203 targets the 3′-UTR of RGS17 mRNA, leading to post-transcriptional downregulation of RGS17 expression. The downregulation of RGS17 inhibits cell proliferation, migration, and invasion in NSCLC cells. RGS17 contains a palmitoylation site that regulates its subcellular localization and G-protein receptor selectivity, promoting tumor progression. Overexpression of RGS17 reverses the effects of miR-203. | [38] |
| Cell cycle arrest | |||
| miR-195 | CHECK1 | Downregulation of CHEK1 expression and delays cell cycle progression. | [39] |
| Drug resistance | |||
| miR-218 | SLIT2/3, IL-6R, JAK3, STAT3 (phosphorylated) | It regulates the expression of its host gene SLIT2/3 and targets IL-6R, JAK3, and phosphorylated STAT3. Overexpression of miR-218 inhibits cell survival, migration, and invasion. miR-218 expression also influences the ALDH1A1-positive lung cancer cell population, affecting their tumorigenic potential. | [40] |
| miR-199a-3p, miR-199a-5p | Rheb, MAP3K11 | Regulates mTOR signaling pathway, inhibits cell proliferation, and migration, and promotes apoptosis; regulatory axis with MAP3K11 in NSCLC. | [41] |
| EMT, metastasis | |||
| miR-138 | SIRT1, ZEB2, EGFR, EZH2, GPR124, YAP1 | miR-138 is downregulated by TGFβ1 exposure, leading to increased stemness via EMT. It targets SIRT1 to reduce AMPK signaling, ZEB2 to suppress EMT, and EGFR/EZH2 to inhibit proliferation. MiR-138 also targets GPR124 to restore EGFR TKI sensitivity in NSCLC. Additionally, lncRNAs like PFAR and SNHG12 regulate miR-138 expression, impacting lung fibrosis and cancer progression. | [42] |
| Metastasis | |||
| miR-15a | ACSS2, fatty acid synthesis | Acts as a tumor suppressor by inhibiting the proliferation, migration, and invasion of lung cancer cells. It targets the 3′-UTR region of ACSS2, reducing acetate uptake and acetyl-CoA activity, leading to decreased histone H4 acetylation. This disrupts fatty acid synthesis and lipid metabolism, impairing metastasis. Under hypoxic conditions, miR-15a-5p is transported into the nucleus, further inhibiting ACSS2 and reducing acetyl-CoA activity and histone acetylation, thus suppressing cancer progression. | [25] |
| miR-16 | MEK1 (MAPK kinase 1) | miR-16 downregulates MEK1 expression, inhibiting the ERK/MAPK signaling pathway and suppressing lung cancer cell proliferation, migration, and invasion. | [43] |
| miR-148 | Wnt1, ROCK1, CEA | Suppresses cancer progression by targeting Wnt1 and ROCK1, reducing cell invasion and migration, and reversing EMT in NSCLC. It also inhibits CEA in cancer cells. Overexpression of miR-148a-3p or MAP3K9 silencing inhibits tumor growth and metastasis in vitro and in vivo. | [44,45] |
| miRNA Panel | AUC | Sensitivity | Specificity | Ref. |
|---|---|---|---|---|
| miRNAs-25, 223, 141, 155, 1254 | - | - | - | - |
| miRNAs-125a-5p, 25, 126 | 0.936 | 87.5% | 87.5% | [76] |
| miRNAs-126, 145, 210, 205-5p | - | 91.5% | 96.2% | [77] |
| miR-143, let-7g, miR-126, let-7a, miR-145 | 0.90–0.93 | 75–85% | 75–85% | [78] |
| miRNAs-7a-5p, 375, 1-3p, 1291, 214-3p | - | - | - | - |
| 34-miRNA model | 0.89 (Stage I); 0.88 (Stage II–IV) | 59% (Stage I); 92% (Stage II–IV) | 90% (Stage I); 90% (Stage II–IV) | [79] |
| miRNAs-155, 197, 182 | 0.92, 0.84 0.98 | miRNA-182 100% | miRNA-182 86.5% | [80] |
| miRNAs-21-5p, 181-5p, 155-5p | - | - | - | - |
| miRNAs-199a−3p, chr17_10932, 148a−3p, 210−3p, chr1_1402, 378d, 138−5p | - | - | - | - |
| miRNAs-210-3p, 301a-5p | - | - | - | - |
| miRNAs-324-3p, 1285 | 0.89 | 85.4% | 81.8% | [81] |
| miRNAs-320a-3p, 210-3p, 92a-3p, 21-5p, 140-3p + 4MP | - | 19.1% | 95% | [82] |
| miRNAs-6777-5p, 6780a-5p, 877-5p | 0.98 | 88% | 100% | [83] |
| miRNA-150, miRNA-886-3p | - | - | - | - |
| miRNAs-143, 100, 101-1, 101-2, 182, 183, 205, 21, 30a, 30d | - | - | - | - |
| miRNAs-155-5p, 223-3p, 126-3p | - | - | - | - |
| miRNAs-20a-5p, 152-3p, 199a-5p | - | - | - | - |
| miRNAs-25, 145, 210 | - | - | - | - |
| miRNAs-375, 200c, 30c | - | - | - | - |
| miRNAs-101-2, 139, 182, 183, 190, 326, 944 | - | - | - | - |
| miRNAs-142-3p, 29b | - | - | - | - |
| miRNAs-92a-2, 147, 574-5p | - | - | - | - |
| miRNAs-105-5p, 767-5p | - | - | - | - |
| miRNAs-22, 24, 34a | - | - | - | - |
| Delivery System | Advantages | Disadvantages |
|---|---|---|
| Viral vectors | ||
| Adenoviruses | High delivery efficiency; well researched. | Significant immunogenicity; immune response limits therapy effectiveness and repeated use. |
| Adeno-associated viruses | Lower immunogenic potential. | Still triggers an immune response in some cases. |
| Retroviruses and lentiviruses | Stable gene expression through integration into the host genome; long-term therapy potential. | Risk of insertional mutagenesis and carcinogenesis. |
| Non-viral vectors | ||
| Lipid-based vectors | Low immunogenicity; high availability and variety. | Low delivery efficiency; low tissue specificity; off-target effects; susceptible to phagocytosis. |
| Polymeric vectors | Low immunogenicity; flexibility for modification and synthesis. | Cytotoxicity of common polymers. |
| Extracellular vesicles | ||
| Exosomes | Low immunogenicity and cytotoxicity; high delivery efficiency. | High production costs. |
| Microvesicles and apoptotic bodies | Low immunogenicity and cytotoxicity. | Limited research. |
| Study Type | miRNA | Current Status of the Trial | Key Information and Conclusions | Ref. |
|---|---|---|---|---|
| Interventional (MRX34) | miR-34 | Terminated | This trial ended early due to immune-related serious adverse events, including the death of four patients. Further studies to understand systemic immune activation and discover new delivery systems are crucial. | [179] |
| Interventional (MesomiR-1) | miR-16 | Completed Phase I | The study showed promising results in patients with mesothelioma, although more data and research are needed to determine the effectiveness of NSCLC. | [180] |
| Observational | miR-21, miR-155 | Completed | Current research indicates potential and the need for further studies on the application of these molecules. | [170] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartoszewska, E.; Misiąg, P.; Czapla, M.; Rakoczy, K.; Tomecka, P.; Filipski, M.; Wawrzyniak-Dzierżek, E.; Choromańska, A. The Role of microRNAs in Lung Cancer: Mechanisms, Diagnostics and Therapeutic Potential. Int. J. Mol. Sci. 2025, 26, 3736. https://doi.org/10.3390/ijms26083736
Bartoszewska E, Misiąg P, Czapla M, Rakoczy K, Tomecka P, Filipski M, Wawrzyniak-Dzierżek E, Choromańska A. The Role of microRNAs in Lung Cancer: Mechanisms, Diagnostics and Therapeutic Potential. International Journal of Molecular Sciences. 2025; 26(8):3736. https://doi.org/10.3390/ijms26083736
Chicago/Turabian StyleBartoszewska, Elżbieta, Piotr Misiąg, Melania Czapla, Katarzyna Rakoczy, Paulina Tomecka, Michał Filipski, Elżbieta Wawrzyniak-Dzierżek, and Anna Choromańska. 2025. "The Role of microRNAs in Lung Cancer: Mechanisms, Diagnostics and Therapeutic Potential" International Journal of Molecular Sciences 26, no. 8: 3736. https://doi.org/10.3390/ijms26083736
APA StyleBartoszewska, E., Misiąg, P., Czapla, M., Rakoczy, K., Tomecka, P., Filipski, M., Wawrzyniak-Dzierżek, E., & Choromańska, A. (2025). The Role of microRNAs in Lung Cancer: Mechanisms, Diagnostics and Therapeutic Potential. International Journal of Molecular Sciences, 26(8), 3736. https://doi.org/10.3390/ijms26083736






