Genome-Wide Identification of the ARF Gene Family in Safflower (Carthamus tinctorius L.) and Their Response Patterns to Exogenous Hormone Treatments
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Property Analysis of ARF Transcription Factor Family Members in Safflower
2.2. Chromosomal Distribution of ARF Family Members in Safflower
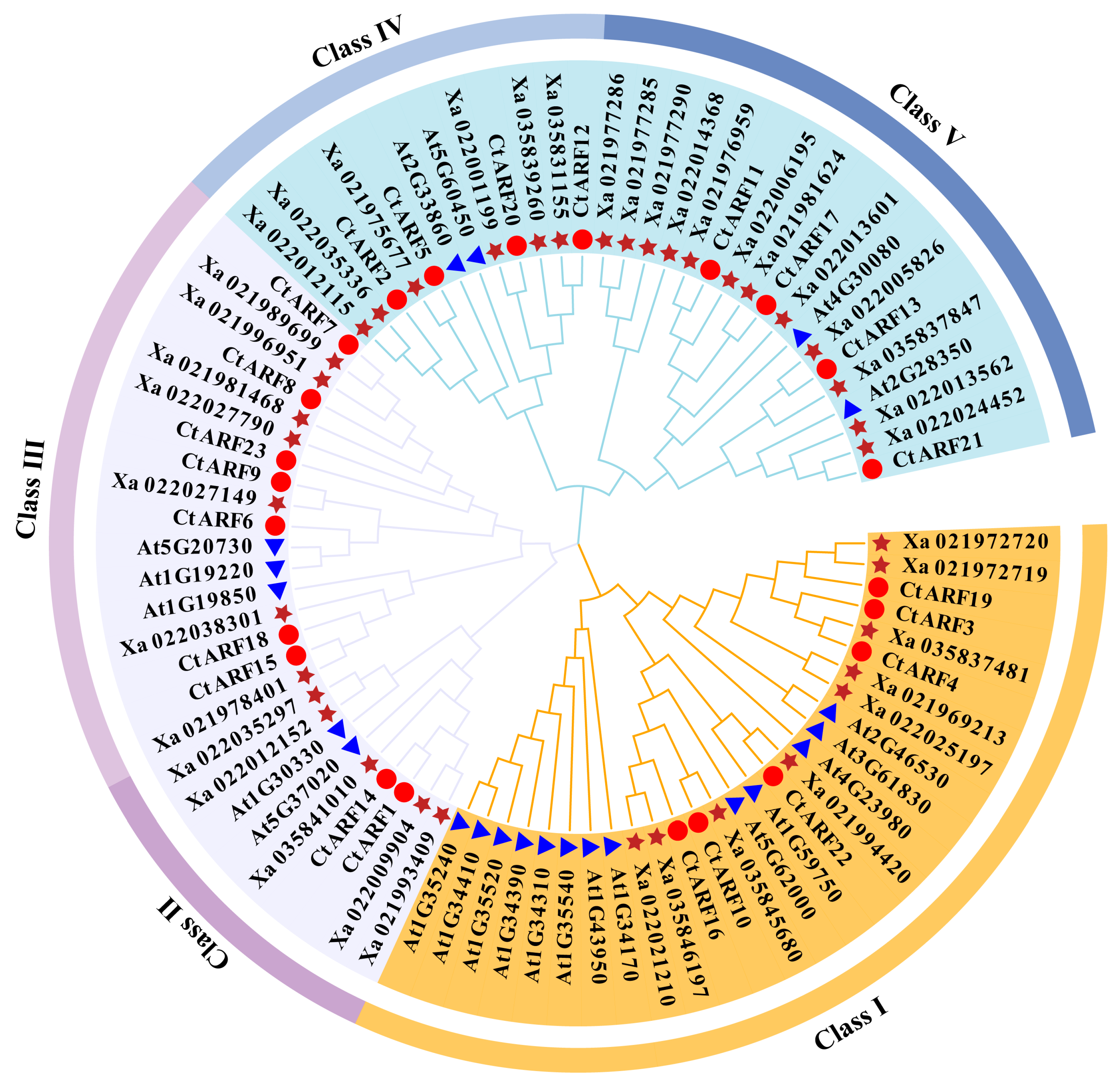
2.3. Phylogenetic Analysis of ARF Proteins in Safflower
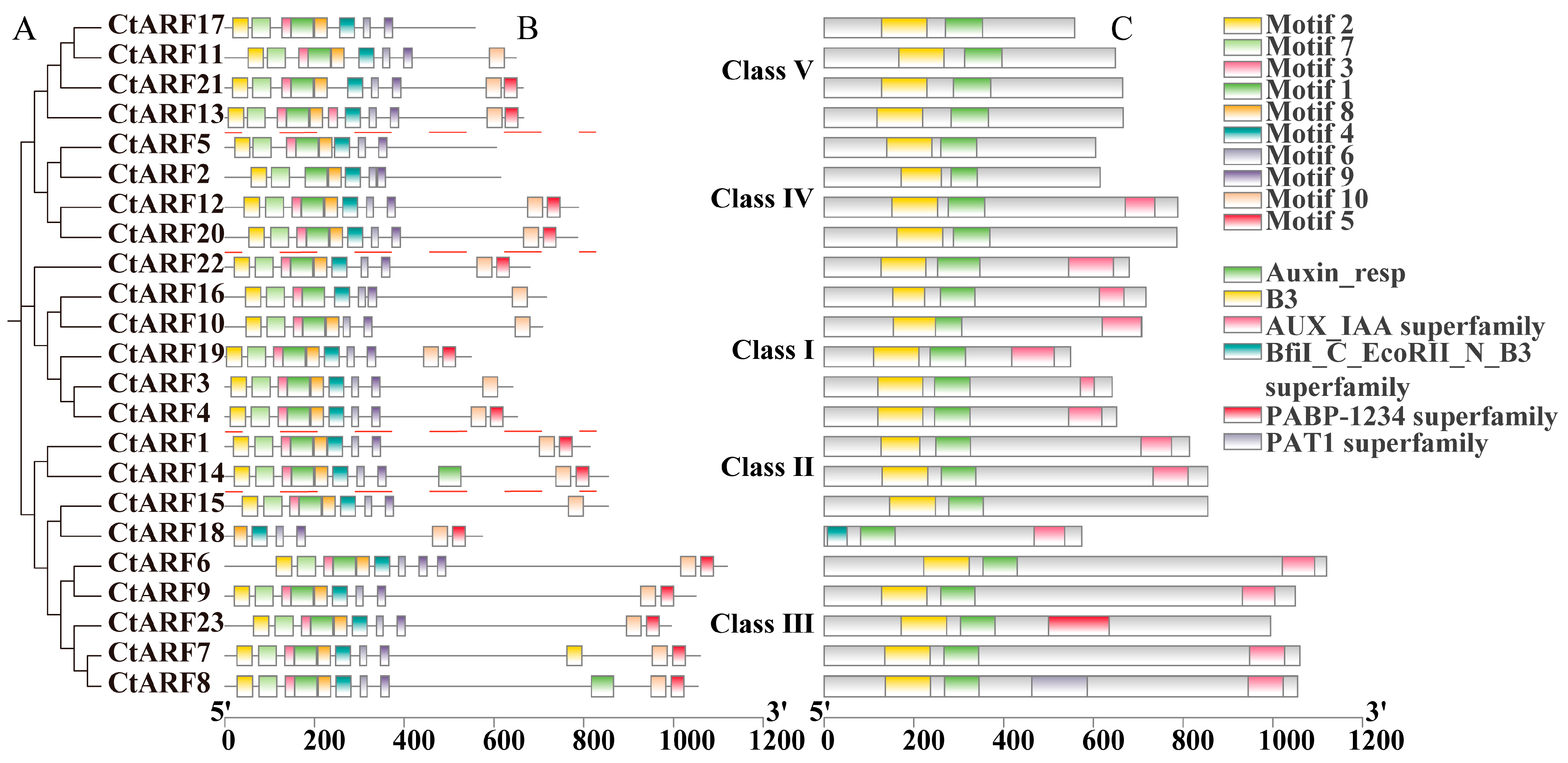
2.4. Gene Structure Analysis of ARF Proteins in Safflower
2.5. Analysis of Conserved Domains and Motifs in Safflower ARF Proteins
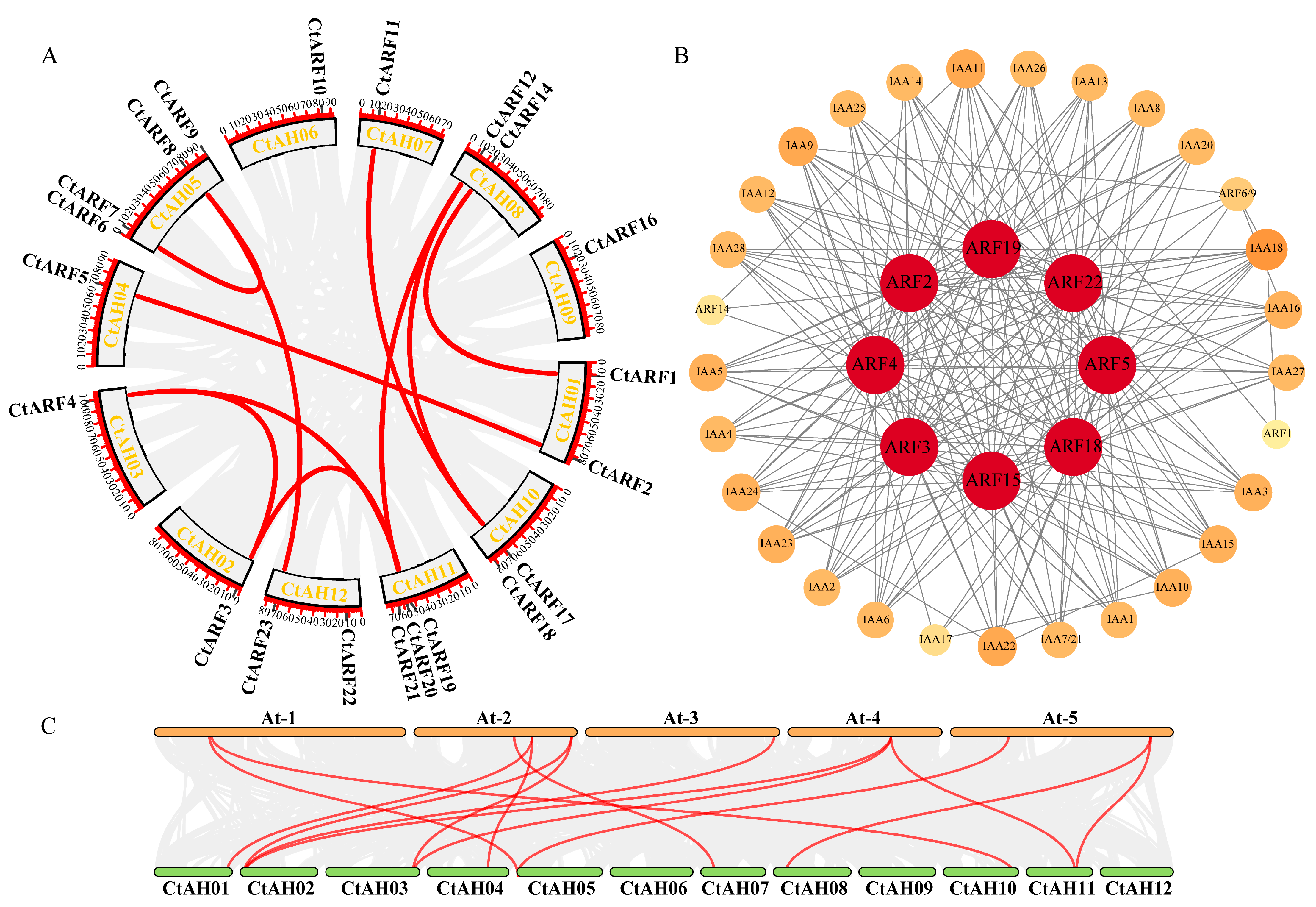
2.6. Analysis of Synteny and Interactions of Safflower ARF Proteins
2.7. Analysis of Cis-Acting Elements in the Promoter Regions of Safflower ARF Genes
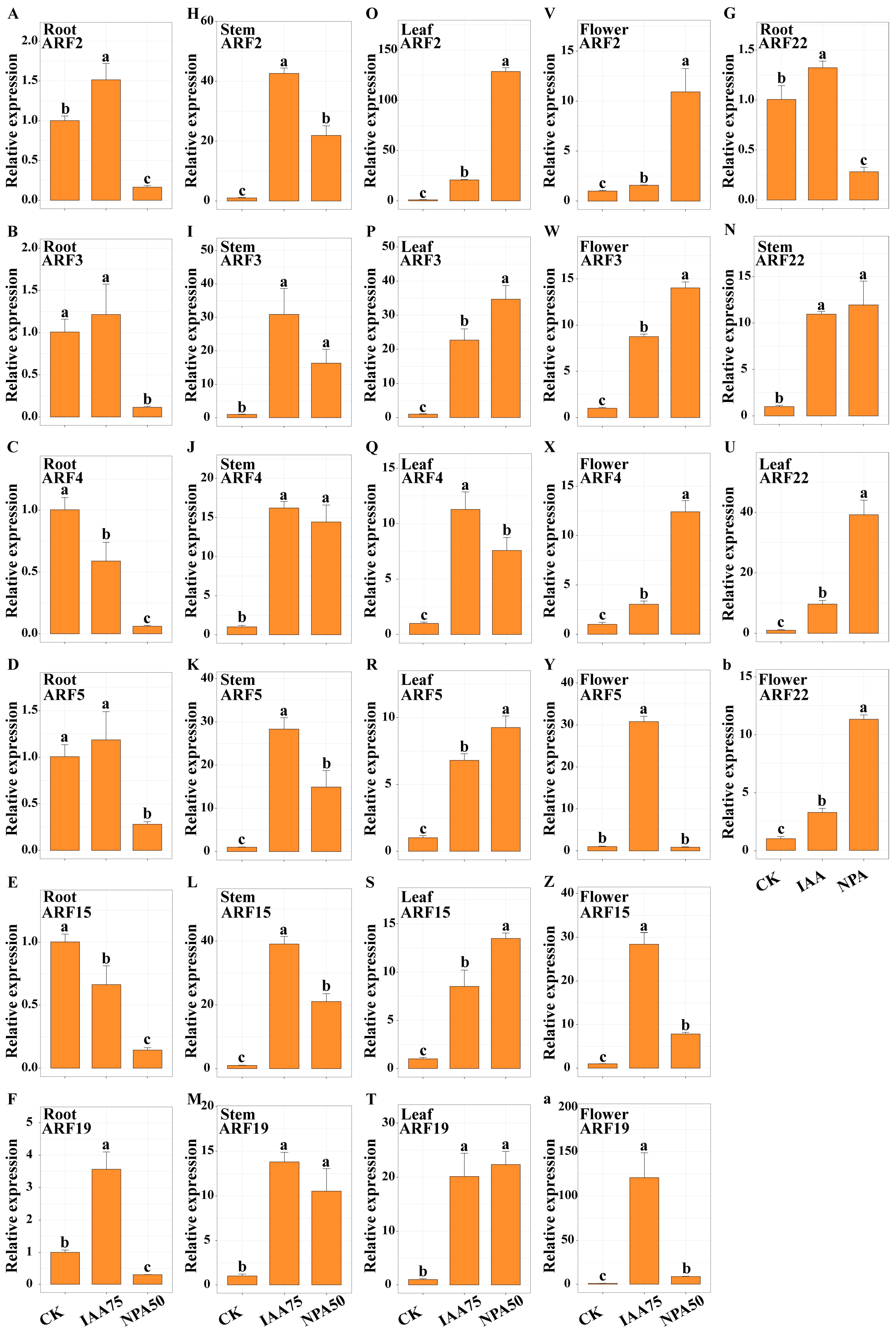
2.8. Expression Analysis of Safflower ARF Genes in Different Flower Colors
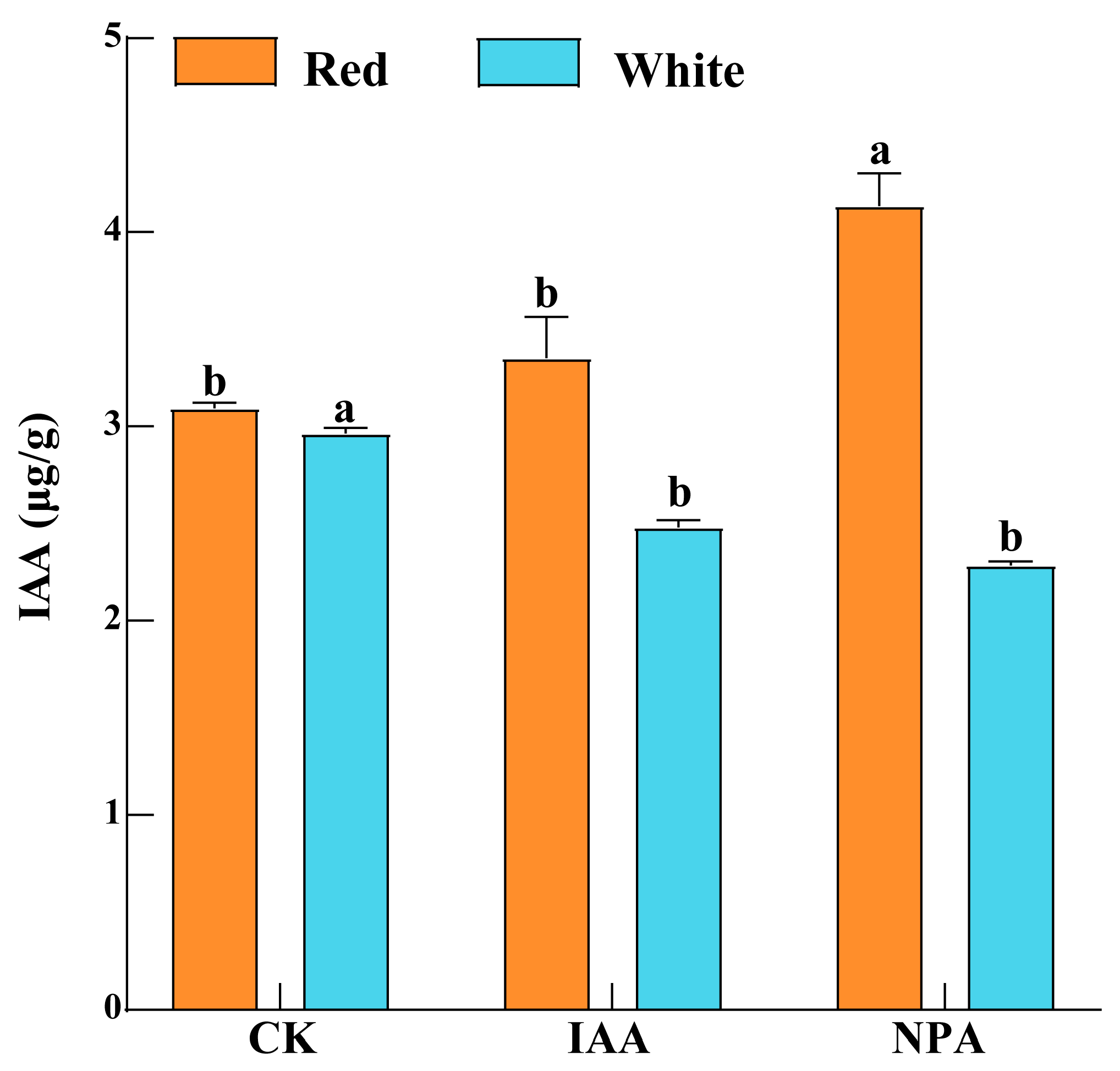
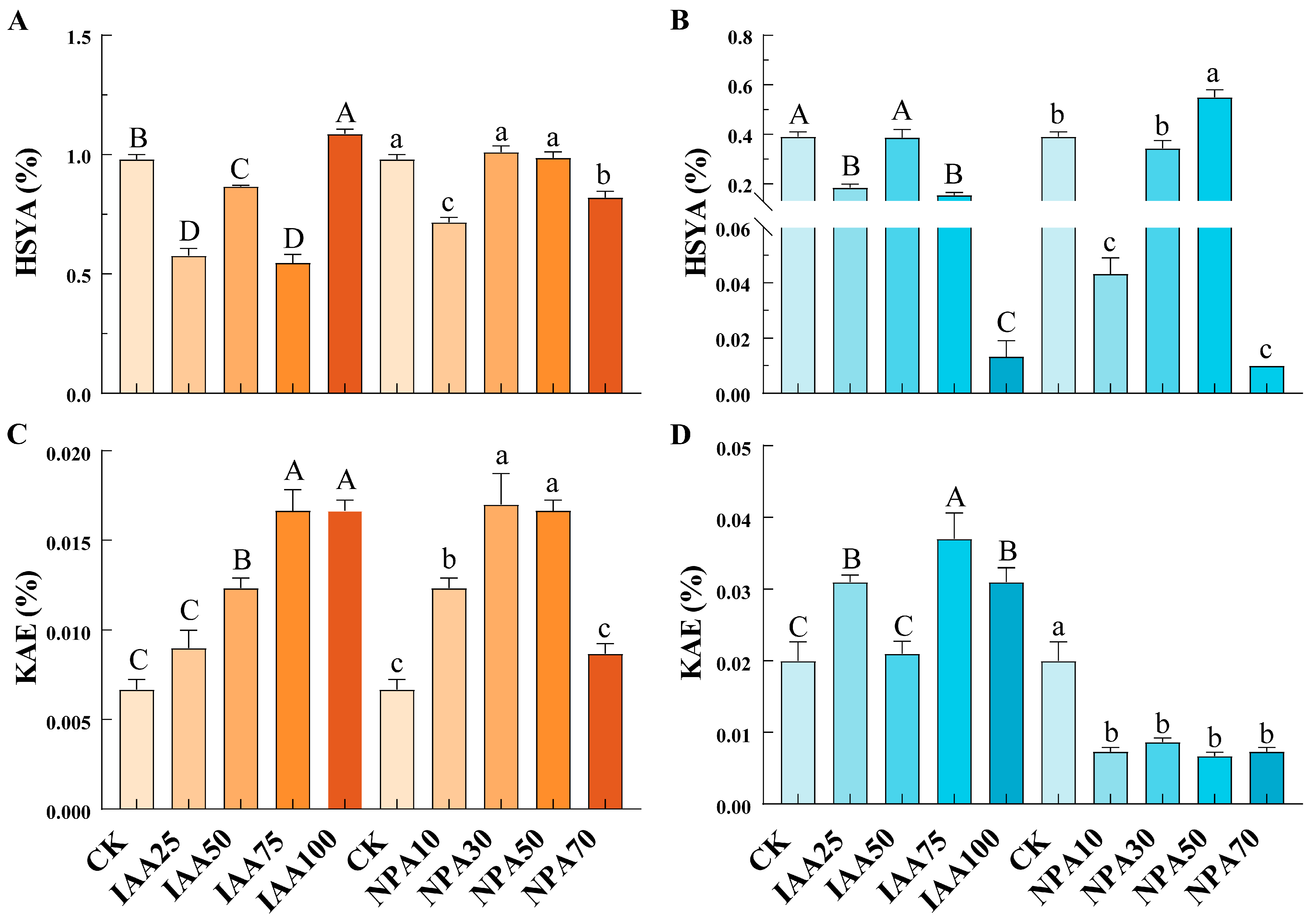
2.9. Content of Endogenous IAA and Active Components in Safflower with Different Flower Colors
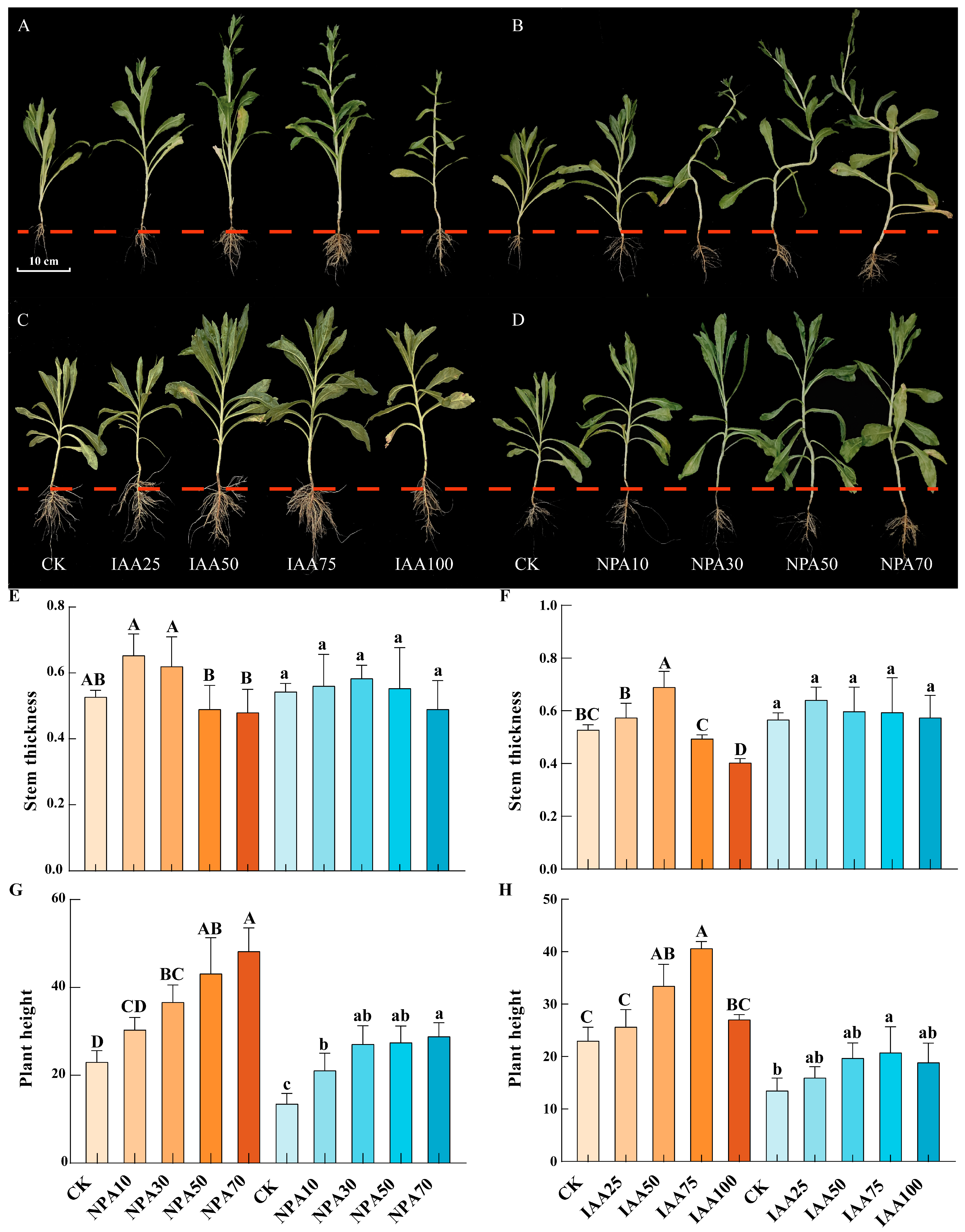
2.10. Effects of Different Exogenous Plant Hormone Treatments on Agronomic Traits of Safflower
2.11. Expression Analysis of CtARF Genes and Determination of Endogenous IAA and Active Component Content After IAA and NPA Treatment
3. Discussion
3.1. Identification and Structural Analysis of the CtARF
3.2. Structural Analysis of CtARF Protein
3.3. Cis-Acting Element and Interaction Network Analysis of CtARFs
3.4. The Response of Endogenous IAA and Active Ingredient Content in Safflower with Different Flower Colors to Exogenous Hormone Treatment
4. Materials and Methods
4.1. Materials
4.2. Identification and Physicochemical Properties and Structural Analysis of Safflower ARF Transcription Factor Family Members
4.3. Multiple Sequence Alignment and Phylogenetic Analysis
4.4. Chromosomal Distribution, Gene Structure, and Collinearity Analysis
4.5. Conserved Domain and Conserved Motif Analysis
4.6. Cis-Acting Element Analysis and Protein–Protein Interaction Prediction
4.7. Expression Pattern Analysis of ARF Genes in Different Flower Colors
4.8. Determination of Endogenous IAA and Active Component Content
4.9. RNA Extraction and Quantitative Real-Time Reverse Transcription PCR (qRT-PCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Orgah, J.O.; He, S.; Wang, Y.; Jiang, M.; Wang, Y.; Orgah, E.A.; Duan, Y.; Zhao, B.; Zhang, B.; Han, J. Pharmacological potential of the combination of Salvia miltiorrhiza (Danshen) and Carthamus tinctorius (Honghua) for diabetes mellitus and its cardiovascular complications. Pharmacol. Res. 2020, 153, 104654. [Google Scholar] [CrossRef] [PubMed]
- Baerle, A.; Savcenco, A.; Tatarov, P.; Fetea, F.; Ivanova, R.; Radu, O. Stability limits of a red Carthamin–cellulose complex as a potential food colourant. Food Funct. 2021, 12, 8037–8043. [Google Scholar] [CrossRef]
- Zanetti, F.; Angelini, L.G.; Berzuini, S.; Foschi, L.; Clemente, C.; Ferioli, F.; Vecchi, A.; Rossi, A.; Monti, A.; Tavarini, S. Safflower (Carthamus tinctorius L.) a winter multipurpose oilseed crop for the Mediterranean region: Lesson learnt from on-farm trials. Ind. Crops Prod. 2022, 184, 115042. [Google Scholar] [CrossRef]
- Khalid, N.; Khan, R.S.; Hussain, M.I.; Farooq, M.; Ahmad, A.; Ahmed, I. A comprehensive characterisation of safflower oil for its potential applications as a bioactive food ingredient—A review. Trends Food Sci. Technol. 2017, 66, 176–186. [Google Scholar] [CrossRef]
- Chen, G.; Li, C.; Zhang, L.; Yang, J.; Meng, H.; Wan, H.; He, Y. Hydroxysafflor yellow A and anhydrosafflor yellow B alleviate ferroptosis and parthanatos in PC12 cells injured by OGD/R. Free Radic. Biol. Med. 2022, 179, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Du, M.; Li, S.; Zhang, Y.; Ding, J.; Wang, J.; Wang, Y.; Liu, P. Hydroxysafflor yellow A regulates lymphangiogenesis and inflammation via the inhibition of PI3K on regulating AKT/mTOR and NF-κB pathway in macrophages to reduce atherosclerosis in ApoE-/-mice. Phytomedicine 2023, 112, 154684. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, M.; Fu, X.; Qi, M.; Zhu, F.; Fan, F.; Wang, Y.; Zhang, K.; Chu, S. Hydroxysafflor yellow A ameliorates alcohol-induced liver injury through PI3K/Akt and STAT3/NF-κB signaling pathways. Phytomedicine 2024, 132, 155814. [Google Scholar] [CrossRef]
- Yu, H.-C.; Bai, Q.-R.; Guo, J.-J.; Chen, M.-Y.; Wang, L.; Tong, F.-C.; Zhang, S.-L.; Wu, J. Elucidating hydroxysafflor yellow A’s multi-target mechanisms against alcoholic liver disease through integrative pharmacology. Phytomedicine 2024, 134, 155956. [Google Scholar] [CrossRef]
- Bao, D.; Chang, S.; Li, X.; Qi, Y. Advances in the study of auxin early response genes: Aux/IAA, GH3, and SAUR. Crop J. 2024, 12, 964–978. [Google Scholar] [CrossRef]
- Wang, J.; Hu, D.; Shi, X.; Luo, J.; Ren, G.; Dai, Z.; Qi, S.; Du, D. Different Responses of Invasive Weed Alternanthera philoxeroide s and Oryza sativa to Plant Growth Regulators. Life 2022, 12, 1069. [Google Scholar] [CrossRef]
- Kakei, Y.; Yamazaki, C.; Suzuki, M.; Nakamura, A.; Sato, A.; Ishida, Y.; Kikuchi, R.; Higashi, S.; Kokudo, Y.; Ishii, T. Small-molecule auxin inhibitors that target YUCCA are powerful tools for studying auxin function. Plant J. 2015, 84, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Huang, Y.; Qi, P.; Lian, G.; Hu, X.; Han, N.; Wang, J.; Zhu, M.; Qian, Q.; Bian, H. Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance. New Phytol. 2021, 229, 2676–2692. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Prigge, M.J.; Tao, S.; Shain, S.; Kuo, A.; Kirchsteiger, K.; Estelle, M. Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins. Elife 2016, 5, e13325. [Google Scholar] [CrossRef]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Getting a grasp on domain III/IV responsible for Auxin Response Factor–IAA protein interactions. Plant Sci. Amst. Neth. 2012, 190, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Berne-Dedieu, A.; Scutt, C.P.; Marlétaz, F. Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 2013, 30, 45–56. [Google Scholar] [CrossRef]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin response factors: Output control in auxin biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef]
- Li, Y.; Han, S.; Qi, Y. Advances in structure and function of auxin response factor in plants. J. Integr. Plant Biol. 2023, 65, 617–632. [Google Scholar] [CrossRef]
- Hu, W.; Zuo, J.; Hou, X.; Yan, Y.; Wei, Y.; Liu, J.; Li, M.; Xu, B.; Jin, Z. The auxin response factor gene family in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant Sci. 2015, 6, 742. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsögön, A.; Smouni, A. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dong, L.; Deng, X.; Liu, D.; Liu, Y.; Li, M.; Hu, Y.; Yan, Y. Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol. 2018, 18, 336. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. Auxin Response Factor1 and Auxin Response Factor2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef]
- Wang, B.; Xue, J.-S.; Yu, Y.-H.; Liu, S.-Q.; Zhang, J.-X.; Yao, X.-Z.; Liu, Z.-X.; Xu, X.-F.; Yang, Z.-N. Fine regulation of ARF17 for anther development and pollen formation. BMC Plant Biol. 2017, 17, 243. [Google Scholar] [CrossRef]
- Wang, C.-K.; Han, P.-L.; Zhao, Y.-W.; Yu, J.-Q.; You, C.-X.; Hu, D.-G.; Hao, Y.-J. Genome-wide analysis of auxin response factor (ARF) genes and functional identification of MdARF2 reveals the involvement in the regulation of anthocyanin accumulation in apple. N. Z. J. Crop Hortic. Sci. 2021, 49, 78–91. [Google Scholar] [CrossRef]
- Wan, W.; Jia, F.; Liu, Z.; Sun, W.; Zhang, X.; Su, J.; Guan, Z.; Chen, F.; Zhang, F.; Fang, W. Quantitative evaluation and genome-wide association studies of chrysanthemum flower color. Sci. Hortic. 2024, 338, 113561. [Google Scholar] [CrossRef]
- Pahlavani, M.; Mirlohi, A.; Saeidi, G. Inheritance of flower color and spininess in safflower (Carthamus tinctorius L.). J. Hered. 2004, 95, 265–267. [Google Scholar] [CrossRef]
- Durbin, M.L.; Lundy, K.E.; Morrell, P.L.; Torres-Martinez, C.L.; Clegg, M.T. Genes that determine flower color: The role of regulatory changes in the evolution of phenotypic adaptations. Mol. Phylogenet. Evol. 2003, 29, 507–518. [Google Scholar] [CrossRef]
- Wang, R.; Ren, C.; Dong, S.; Chen, C.; Xian, B.; Wu, Q.; Wang, J.; Pei, J.; Chen, J. Integrated metabolomics and transcriptome analysis of flavonoid biosynthesis in safflower (Carthamus tinctorius L.) with different colors. Front. Plant Sci. 2021, 12, 712038. [Google Scholar] [CrossRef]
- Feng, L.; Li, G.; He, Z.; Han, W.; Sun, J.; Huang, F.; Di, J.; Chen, Y. The ARF, GH3, and Aux/IAA gene families in castor bean (Ricinus communis L.): Genome-wide identification and expression profiles in high-stalk and dwarf strains. Ind. Crops Prod. 2019, 141, 111804. [Google Scholar] [CrossRef]
- Wilmoth, J.C.; Wang, S.; Tiwari, S.B.; Joshi, A.D.; Hagen, G.; Guilfoyle, T.J.; Alonso, J.M.; Ecker, J.R.; Reed, J.W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005, 43, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, M.-L.; Ma, T.-L.; Wang, Y. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 2016, 28, 3005–3019. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Dimerization and DNA binding of auxin response factors. Plant J. 1999, 19, 309–319. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, W.; Shen, H.; Yang, L. Genome-wide identification and expression analysis of ARF gene family in embryonic development of Korean pine (Pinus koraiensis). BMC Plant Biol. 2024, 24, 267. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, R.; Yu, J.; Huang, S.; Zhang, Y.; Wei, H.; Wei, Z. Genome-wide identification and characterization of auxin response factor (ARF) gene family involved in wood formation and response to exogenous hormone treatment in Populus trichocarpa. Int. J. Mol. Sci. 2023, 24, 740. [Google Scholar] [CrossRef]
- Hu, Z.; Zong, D.; Zhang, Q.; Zhang, X.; Lu, Y.; He, C. PyuARF16/33 are involved in the regulation of lignin synthesis and rapid growth in Populus yunnanensis. Genes 2023, 14, 278. [Google Scholar] [CrossRef]
- Jiao, Y. Double the genome, double the fun: Genome duplications in angiosperms. Mol. Plant 2018, 11, 357–358. [Google Scholar] [CrossRef]
- Boer, D.R.; Freire-Rios, A.; Van Den Berg, W.A.; Saaki, T.; Manfield, I.W.; Kepinski, S.; López-Vidrieo, I.; Franco-Zorrilla, J.M.; de Vries, S.C.; Solano, R. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 2014, 156, 577–589. [Google Scholar] [CrossRef]
- Yu, H.; Soler, M.; Mila, I.; San Clemente, H.; Savelli, B.; Dunand, C.; Paiva, J.A.; Myburg, A.A.; Bouzayen, M.; Grima-Pettenati, J. Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS ONE 2014, 9, e108906. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Teale, W.; Lang, D.; Paponov, M.; Reski, R.; Rensing, S.A.; Palme, K. The evolution of nuclear auxin signalling. BMC Evol. Biol. 2009, 9, 126. [Google Scholar] [CrossRef]
- Tian, C.e.; Muto, H.; Higuchi, K.; Matamura, T.; Tatematsu, K.; Koshiba, T.; Yamamoto, K.T. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004, 40, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef]
- Feng, S.; Li, N.; Chen, H.; Liu, Z.; Li, C.; Zhou, R.; Zhang, Y.; Cao, R.; Ma, X.; Song, X. Large-scale analysis of the ARF and Aux/IAA gene families in 406 horticultural and other plants. Mol. Hortic. 2024, 4, 13. [Google Scholar] [CrossRef]
- Hakman, I.; Hallberg, H.; Palovaara, J. The polar auxin transport inhibitor NPA impairs embryo morphology and increases the expression of an auxin efflux facilitator protein PIN during Picea abies somatic embryo development. Tree Physiol. 2009, 29, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Guseman, J.M.; Hellmuth, A.; Lanctot, A.; Feldman, T.P.; Moss, B.L.; Klavins, E.; Calderón Villalobos, L.I.A.; Nemhauser, J.L. Auxin-induced degradation dynamics set the pace for lateral root development. Development 2015, 142, 905–909. [Google Scholar]
- Li, P.; Chen, X.; Sun, F.; Dong, H. Tobacco TTG 2 and ARF 8 function concomitantly to control flower colouring by regulating anthocyanin synthesis genes. Plant Biol. 2017, 19, 525–532. [Google Scholar] [CrossRef]
- Solanki, M.; Shukla, L.I. Recent advances in auxin biosynthesis and homeostasis. 3 Biotech 2023, 13, 290. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, X.; Luo, K.; Tembrock, L.R.; Li, S.; Wu, Z. The Identification of Auxin Response Factors and Expression Analyses of Different Floral Development Stages in Roses. Genes 2025, 16, 41. [Google Scholar] [CrossRef]
- Ren, C.; Chen, C.; Dong, S.; Wang, R.; Xian, B.; Liu, T.; Xi, Z.; Pei, J.; Chen, J. Integrated metabolomics and transcriptome analysis on flavonoid biosynthesis in flowers of safflower (Carthamus tinctorius L.) during colour-transition. PeerJ 2022, 10, e13591. [Google Scholar] [PubMed]
- Tian, Y.; Chen, Z.; Jiang, Z.; Huang, X.; Zhang, L.; Zhang, Z.; Sun, P. Effects of plant growth regulators on flower abscission and growth of tea plant Camellia sinensis (L.) O. Kuntze. J. Plant Growth Regul. 2022, 41, 1161–1173. [Google Scholar]
- Hou, M.; Zhang, Y.; Xu, X.; Ai, H. Advances in auxin synthesis, transport, and signaling in rice: Implications for stress resilience and crop improvement. Front. Plant Sci. 2025, 15, 1516884. [Google Scholar]
- Wang, J.; Jin, D.; Deng, Z.; Zheng, L.; Guo, P.; Ji, Y.; Song, Z.; Zeng, H.Y.; Kinoshita, T.; Liao, Z. The apoplastic pH is a key determinant in the hypocotyl growth response to auxin dosage and light. Nat. Plants 2025, 11, 279–294. [Google Scholar] [PubMed]
- Kong, M.; Liu, X.; Sun, L.; Tan, S. Molecular mechanisms of N-1-naphthylphthalamic acid, a chemical tool in plant biology and agriculture. Mol. Hortic. 2022, 2, 22. [Google Scholar]
- Xian, B.; Wang, R.; Jiang, H.; Zhou, Y.; Yan, J.; Huang, X.; Chen, J.; Wu, Q.; Chen, C.; Xi, Z. Comprehensive review of two groups of flavonoids in Carthamus tinctorius L. Biomed. Pharmacother. 2022, 153, 113462. [Google Scholar]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005, 33, D501–D504. [Google Scholar]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar]
- Letunic, I.; Goodstadt, L.; Dickens, N.J.; Doerks, T.; Schultz, J.; Mott, R.; Ciccarelli, F.; Copley, R.R.; Ponting, C.P.; Bork, P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002, 30, 242–244. [Google Scholar]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar]
- Marchler-Bauer, A.; Anderson, J.B.; DeWeese-Scott, C.; Fedorova, N.D.; Geer, L.Y.; He, S.; Hurwitz, D.I.; Jackson, J.D.; Jacobs, A.R.; Lanczycki, C.J. CDD: A curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003, 31, 383–387. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2010, 39, D561–D568. [Google Scholar] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [PubMed]
- Mao, W.; Bao, C.; Cheng, Q.; Liang, N.; Wang, L.; Yang, H. All-year high IAA and ABA contents in rhizome buds may contribute to natural four-season shooting in woody bamboo cephalostachyum pingbianense. Plants 2024, 13, 410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, L.; Jin, W.; Li, C.; Wang, Y.; Wan, H.; Yang, J. Simultaneous optimization of the ultrasonic extraction method and determination of the antioxidant activities of hydroxysafflor yellow A and anhydrosafflor yellow B from safflower using a response surface methodology. Molecules 2020, 25, 1226. [Google Scholar] [CrossRef]
- Tan, Z.W.; Lu, D.D.; Yu, Y.L.; Li, L.; Xu, L.J.; Dong, W.; Li, C.M.; Yang, Q.; Liang, H.Z. Genome-Wide Analysis of the APETALA2/Ethylene-Responsive Factor Gene Family in Carthamus tinctorius L. Plant Direct 2025, 9, e70032. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar]

| Gene Name | Locus ID | Protein Length (aa) | Molecular Weight (KDa) | PI | Instability Indices | Aliphatic Indices | Hydrophobicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| CtARF1 | CtAH01T0069600 | 813 | 90.53341 | 5.85 | 60.62 | 73.14 | −0.468 | nucleus |
| CtARF2 | CtAH01T0211500 | 614 | 67.8474 | 8.38 | 53.06 | 71.04 | −0.436 | nucleus |
| CtARF3 | CtAH02T0026700 | 641 | 71.89815 | 6.41 | 56.42 | 70.83 | −0.582 | nucleus |
| CtARF4 | CtAH03T0268400 | 651 | 73.22105 | 5.51 | 55.27 | 73.01 | −0.437 | nucleus |
| CtARF5 | CtAH04T0143600 | 604 | 67.71435 | 6.66 | 53.22 | 68.08 | −0.483 | nucleus |
| CtARF6 | CtAH05T0001600 | 1119 | 125.48255 | 6.92 | 73.1 | 69.63 | −0.638 | nucleus |
| CtARF7 | CtAH05T0047600 | 1059 | 117.82049 | 6.16 | 59.13 | 72.83 | −0.551 | nucleus |
| CtARF8 | CtAH05T0213800 | 1054 | 116.50864 | 5.86 | 60.98 | 68.55 | −0.575 | nucleus |
| CtARF9 | CtAH05T0279800 | 1049 | 117.9647 | 6.36 | 75.38 | 65.73 | −0.765 | nucleus |
| CtARF10 | CtAH06T0272100 | 707 | 79.08847 | 6.08 | 68.44 | 63.41 | −0.688 | nucleus |
| CtARF11 | CtAH07T0101600 | 648 | 72.28166 | 6.82 | 52.99 | 69.95 | −0.44 | nucleus |
| CtARF12 | CtAH08T0107100 | 787 | 86.37022 | 5.94 | 56.73 | 70.32 | −0.433 | nucleus |
| CtARF13 | CtAH08T0109300 | 665 | 73.11407 | 5.9 | 47.25 | 74.6 | −0.362 | nucleus |
| CtARF14 | CtAH08T0176800 | 854 | 95.41481 | 5.81 | 63.57 | 70.2 | −0.557 | nucleus |
| CtARF15 | CtAH08T0303600 | 855 | 94.24119 | 5.71 | 55.24 | 68.87 | −0.521 | nucleus |
| CtARF16 | CtAH09T0132300 | 716 | 79.16385 | 6.84 | 58.87 | 65.21 | −0.6 | nucleus |
| CtARF17 | CtAH10T0191300 | 557 | 62.06441 | 8.21 | 51.29 | 71.92 | −0.447 | nucleus |
| CtARF18 | CtAH10T0242900 | 573 | 63.1186 | 5.13 | 57.16 | 71.61 | −0.43 | nucleus |
| CtARF19 | CtAH11T0158500 | 548 | 61.86642 | 6.11 | 45.67 | 75.71 | −0.451 | nucleus |
| CtARF20 | CtAH11T0163400 | 785 | 85.89191 | 6.06 | 54.01 | 69.87 | −0.414 | nucleus |
| CtARF21 | CtAH11T0194100 | 664 | 72.94625 | 7.64 | 46.99 | 66.49 | −0.462 | nucleus |
| CtARF22 | CtAH12T0040800 | 679 | 76.07008 | 5.75 | 57.36 | 76.91 | −0.462 | nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, S.; Wen, X.; Ma, M.; Wang, J.; Zhang, J.; Huang, M.; Sun, K.; Zhao, Y.; Zhao, M.; Ismayil, A.; et al. Genome-Wide Identification of the ARF Gene Family in Safflower (Carthamus tinctorius L.) and Their Response Patterns to Exogenous Hormone Treatments. Int. J. Mol. Sci. 2025, 26, 3773. https://doi.org/10.3390/ijms26083773
Qin S, Wen X, Ma M, Wang J, Zhang J, Huang M, Sun K, Zhao Y, Zhao M, Ismayil A, et al. Genome-Wide Identification of the ARF Gene Family in Safflower (Carthamus tinctorius L.) and Their Response Patterns to Exogenous Hormone Treatments. International Journal of Molecular Sciences. 2025; 26(8):3773. https://doi.org/10.3390/ijms26083773
Chicago/Turabian StyleQin, Shuwei, Xinrong Wen, Mengyuan Ma, Jiaxing Wang, Jianhang Zhang, Meihui Huang, Kexin Sun, Ya Zhao, Meng Zhao, Asigul Ismayil, and et al. 2025. "Genome-Wide Identification of the ARF Gene Family in Safflower (Carthamus tinctorius L.) and Their Response Patterns to Exogenous Hormone Treatments" International Journal of Molecular Sciences 26, no. 8: 3773. https://doi.org/10.3390/ijms26083773
APA StyleQin, S., Wen, X., Ma, M., Wang, J., Zhang, J., Huang, M., Sun, K., Zhao, Y., Zhao, M., Ismayil, A., Liu, M., & Cao, A. (2025). Genome-Wide Identification of the ARF Gene Family in Safflower (Carthamus tinctorius L.) and Their Response Patterns to Exogenous Hormone Treatments. International Journal of Molecular Sciences, 26(8), 3773. https://doi.org/10.3390/ijms26083773


_Kim.png)




