Integrating Machine Learning and Follow-Up Variables to Improve Early Detection of Hepatocellular Carcinoma in Tyrosinemia Type 1: A Multicenter Study
Abstract
1. Introduction
2. Results
2.1. Patient Cohort Characterization
2.2. AFP Association with Follow-Up Variables
2.3. Relevance of Biochemical Variables as Features Related to AFP Levels
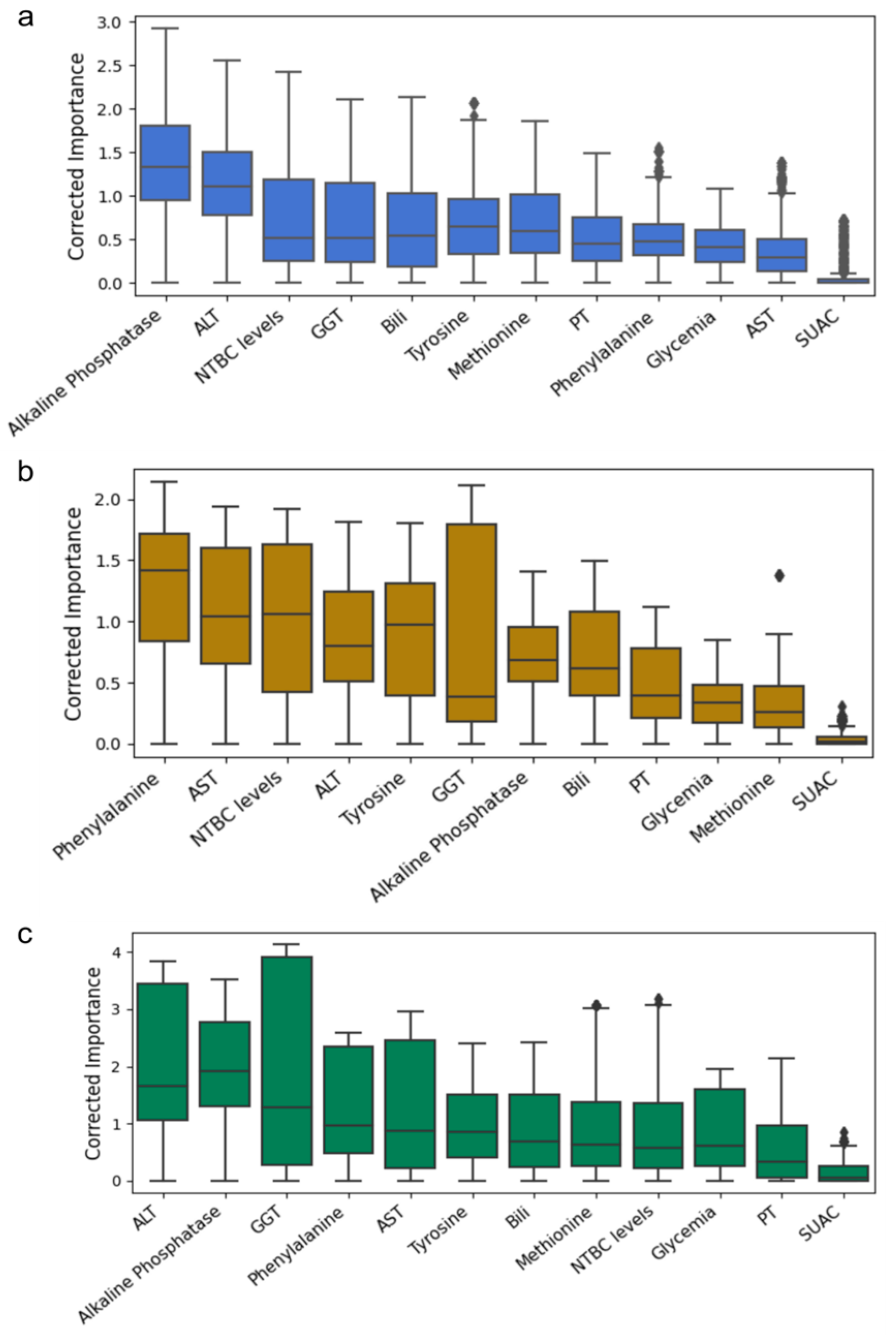
2.4. Feature Importance Based on Biochemical and Age-Related Variables
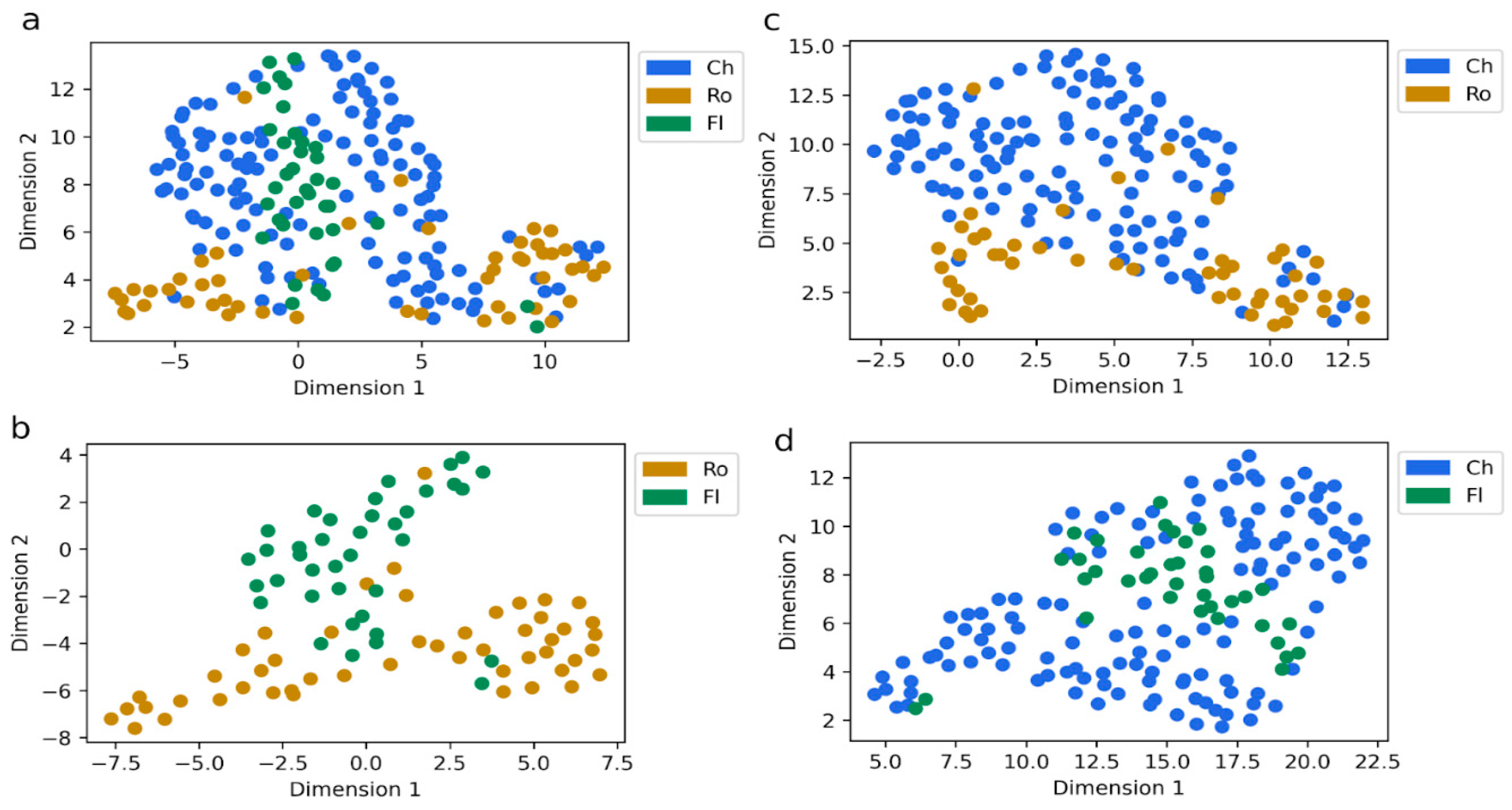
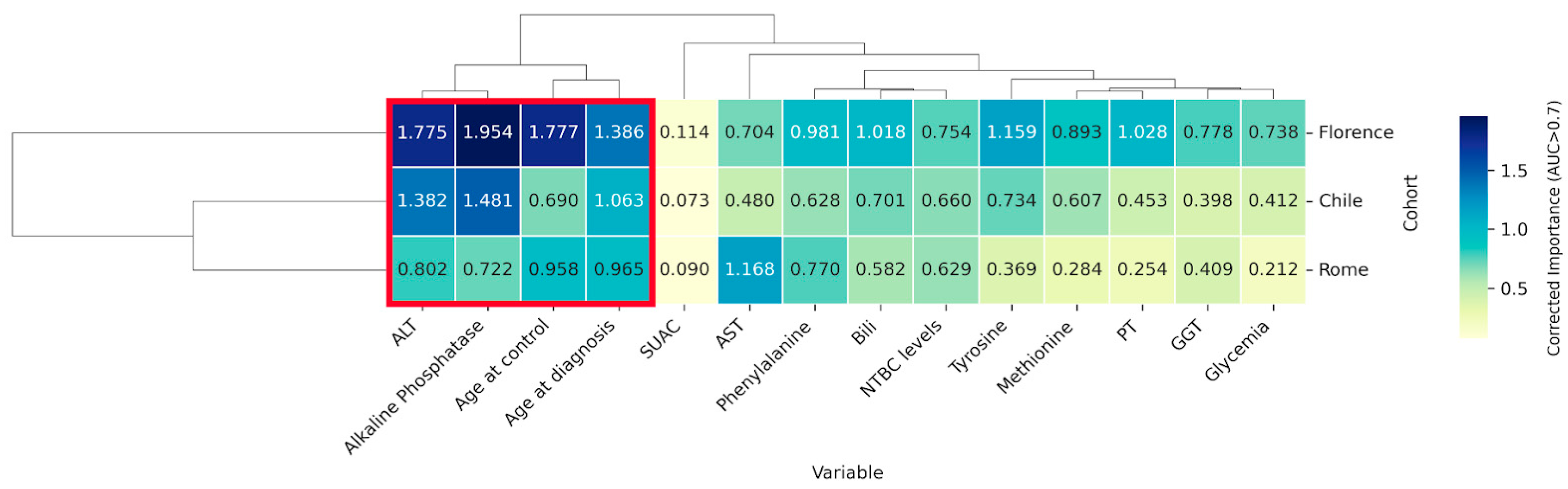
2.5. Identification of Key Variables Through Cross-Cohort Consensus Clustering
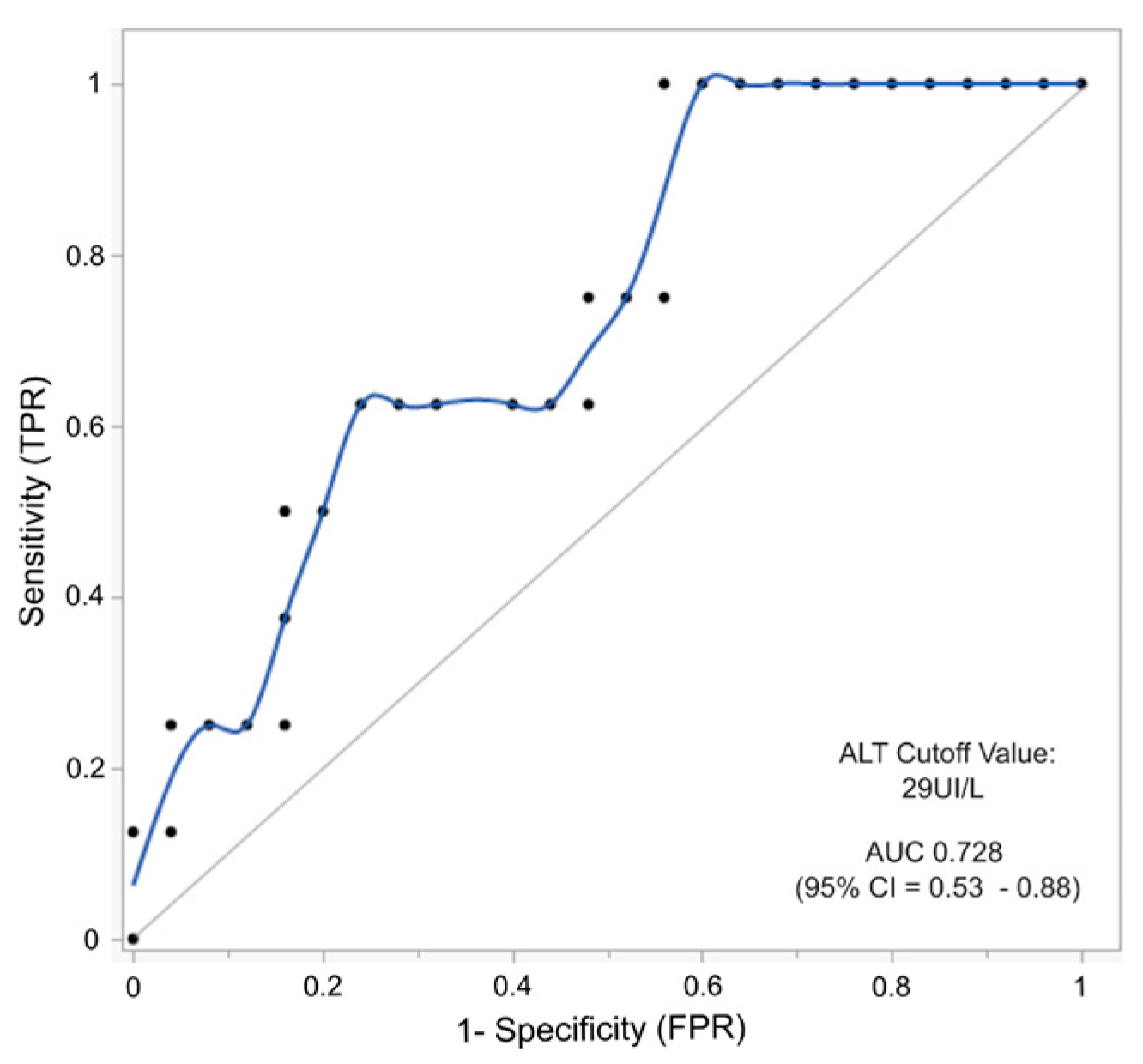
2.6. AFP and ALT as Combined Biomarkers to Predict the Risk of Hepatocellular Carcinoma in HT-1 Patients
3. Discussion
4. Materials and Methods
4.1. Patient Cohort and Eligibility Criteria
4.2. Dataset and Statistical Analysis
4.3. Missing Data Handling and Unsupervised Analysis
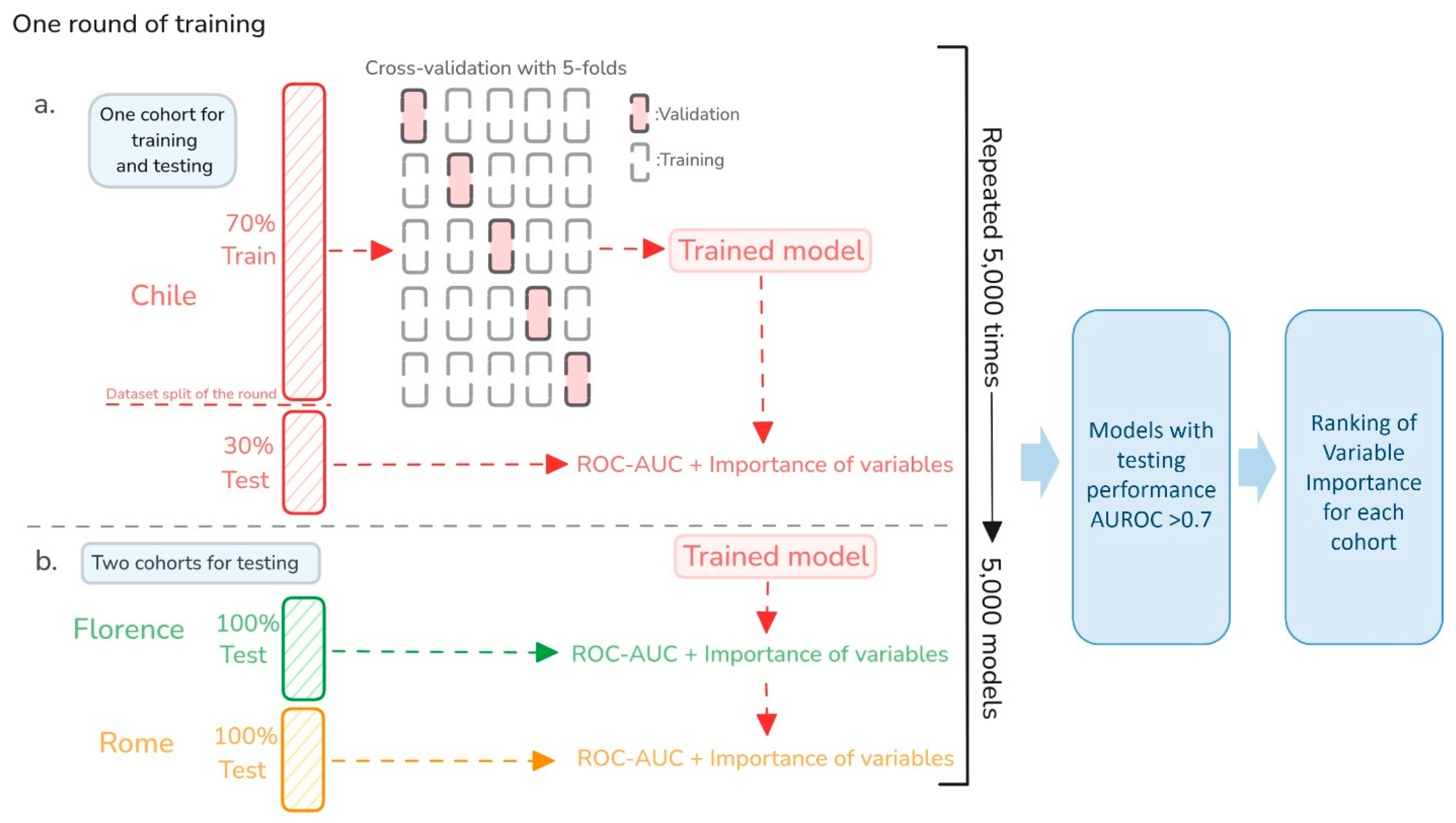
4.4. Predictive Model
4.5. Multi-Model Approach for Robust Generalization and Explainability
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFP | Alpha-fetoprotein |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| AUC | Area under the curve |
| Bili | Total bilirubin |
| DBS | Dried blood spot |
| GGT | Gamma-glutamyl transferase |
| HCC | Hepatocellular carcinoma |
| HT-1 | Hereditary tyrosinemia type 1 |
| Met | Methionine |
| ML | Machine learning |
| NBS | Newborn screening |
| NTBC | Nitisinone |
| Phe | Phenylalanine |
| PT | Prothrombin time |
| ROC | Receiver operating characteristic curve |
| Tyr | Tyrosine |
| SUAC | Succinylacetone |
Appendix A
References
- van Spronsen, F.J.; Thomasse, Y.; Smit, G.P.; Leonard, J.V.; Clayton, P.T.; Fidler, V.; Berger, R.; Heymans, H.S. Hereditary tyrosinemia type I: A new clinical classification with difference in prognosis on dietary treatment. Hepatol. Baltim. Md. 1994, 20, 1187–1191. [Google Scholar] [CrossRef]
- de Laet, C.; Dionisi-Vici, C.; Leonard, J.V.; McKiernan, P.; Mitchell, G.; Monti, L.; de Baulny, H.O.; Pintos-Morell, G.; Spiekerkötter, U. Recommendations for the management of tyrosinaemia type 1. Orphanet J. Rare Dis. 2013, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Schulz, A.; Ort, O.; Beyer, P.; Kleinig, H. SC-0051, a 2-benzoyl-cyclohexane-1,3-dione bleaching herbicide, is a potent inhibitor of the enzyme p-hydroxyphenylpyruvate dioxygenase. FEBS Lett. 1993, 318, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Chinsky, J.M.; Singh, R.; Ficicioglu, C.; van Karnebeek, C.D.M.; Grompe, M.; Mitchell, G.; Waisbren, S.E.; Gucsavas-Calikoglu, M.; Wasserstein, M.P.; Coakley, K.; et al. Diagnosis and treatment of tyrosinemia type I: A US and Canadian consensus group review and recommendations. Genet. Med. 2017, 19, 1380–1395. [Google Scholar] [CrossRef] [PubMed]
- Lindstedt, S.; Holme, E.; Lock, E.A.; Hjalmarson, O.; Strandvik, B. Treatment of hereditary tyrosinaemia type I by inhibition of 4-hydroxyphenylpyruvate dioxygenase. Lancet 1992, 340, 813–817. [Google Scholar] [CrossRef]
- Holme, E.; Lindstedt, S. Tyrosinaemia type I and NTBC (2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione). J. Inherit. Metab. Dis. 1998, 21, 507–517. [Google Scholar] [CrossRef]
- Hajji, H.; Imbard, A.; Spraul, A.; Taibi, L.; Barbier, V.; Habes, D.; Brassier, A.; Arnoux, J.B.; Bouchereau, J.; Pichard, S.; et al. Initial presentation, management and follow-up data of 33 treated patients with hereditary tyrosinemia type 1 in the absence of newborn screening. Mol. Genet. Metab. Rep. 2022, 33, 100933. [Google Scholar] [CrossRef] [PubMed]
- van Ginkel, W.G.; Rodenburg, I.L.; Harding, C.O.; Hollak, C.E.M.; Heiner-Fokkema, M.R.; van Spronsen, F.J. Long-Term Outcomes and Practical Considerations in the Pharmacological Management of Tyrosinemia Type 1. Pediatr. Drugs 2019, 21, 413–426. [Google Scholar] [CrossRef]
- Larochelle, J.; Alvarez, F.; Bussières, J.F.; Chevalier, I.; Dallaire, L.; Dubois, J.; Faucher, F.; Fenyves, D.; Goodyer, P.; Grenier, A.; et al. Effect of nitisinone (NTBC) treatment on the clinical course of hepatorenal tyrosinemia in Québec. Mol. Genet. Metab. 2012, 107, 49–54. [Google Scholar] [CrossRef]
- Zeybek, A.C.A.; Kiykim, E.; Soyucen, E.; Cansever, S.; Altay, S.; Zubarioglu, T.; Erkan, T.; Aydin, A. Hereditary tyrosinemia type 1 in Turkey: Twenty-year single-center experience. Pediatr. Int. Off. J. Jpn. Pediatr. Soc. 2015, 57, 281–289. [Google Scholar]
- Spiekerkoetter, U.; Couce, M.L.; Das, A.M.; de Laet, C.; Dionisi-Vici, C.; Lund, A.M.; Schiff, M.; Spada, M.; Sparve, E.; Szamosi, J.; et al. Long-term safety and outcomes in hereditary tyrosinaemia type 1 with nitisinone treatment: A 15-year non-interventional, multicentre study. Lancet Diabetes Endocrinol. 2021, 9, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Koelink, C.J.L.; van Hasselt, P.; van der Ploeg, A.; van den Heuvel-Eibrink, M.M.; Wijburg, F.A.; Bijleveld, C.M.; van Spronsen, F.J. Tyrosinemia type I treated by NTBC: How does AFP predict liver cancer? Mol. Genet. Metab. 2006, 89, 310–315. [Google Scholar] [CrossRef]
- van Ginkel, W.G.; Gouw, A.S.H.; van der Jagt, E.J.; de Jong, K.P.; Verkade, H.J.; van Spronsen, F.J. Hepatocellular carcinoma in tyrosinemia type 1 without clear increase of AFP. Pediatrics 2015, 135, e749–e752. [Google Scholar] [CrossRef]
- Choi, J.; Kim, G.A.; Han, S.; Lee, W.; Chun, S.; Lim, Y.S. Longitudinal Assessment of Three Serum Biomarkers to Detect Very Early-Stage Hepatocellular Carcinoma. Hepatology 2019, 69, 1983–1994. [Google Scholar] [CrossRef]
- Debes, J.D.; Romagnoli, P.A.; Prieto, J.; Arrese, M.; Mattos, A.Z.; Boonstra, A. Serum Biomarkers for the Prediction of Hepatocellular Carcinoma. Cancers 2021, 13, 1681. [Google Scholar] [CrossRef] [PubMed]
- Almuqbil, M.; Knoll, J.; Chinsky, J.M. Late Development of Hepatocellular Carcinoma in Tyrosinemia Type 1 Despite Nitisinone (NTBC) Treatment. J. Pediatr. Gastroenterol. Nutr. 2020, 71, e73–e75. [Google Scholar] [CrossRef]
- Bhushan, S.; Noble, C.; Balouch, F.; Lewindon, P.; Lampe, G.; Hodgkinson, P.; McGill, J.; Ee, L. Hepatocellular carcinoma requiring liver transplantation in hereditary tyrosinemia type 1 despite nitisinone therapy and α1-fetoprotein normalization. Pediatr. Transpl. 2022, 26, e14334. [Google Scholar] [CrossRef] [PubMed]
- Karaca, C.A.; Yilmaz, C.; Farajov, R.; Iakobadze, Z.; Aydogdu, S.; Kilic, M. Live donor liver transplantation for type 1 tyrosinemia: An analysis of 15 patients. Pediatr. Transpl. 2019, 23, e13498. [Google Scholar] [CrossRef]
- Van den Bulcke, T.; Vanden Broucke, P.; Van Hoof, V.; Wouters, K.; Vanden Broucke, S.; Smits, G.; Smits, E.; Proesmans, S.; Van Genechten, T.; Eyskens, F. Data mining methods for classification of Medium-Chain Acyl-CoA dehydrogenase deficiency (MCADD) using non-derivatized tandem MS neonatal screening data. J. Biomed. Inform. 2011, 44, 319–325. [Google Scholar] [CrossRef]
- Peng, G.; Tang, Y.; Cowan, T.M.; Enns, G.M.; Zhao, H.; Scharfe, C. Reducing False-Positive Results in Newborn Screening Using Machine Learning. Int. J. Neonatal Screen. 2020, 6, 16. [Google Scholar] [CrossRef]
- Zhu, Z.; Gu, J.; Genchev, G.Z.; Cai, X.; Wang, Y.; Guo, J.; Tian, G.; Lu, H. Improving the Diagnosis of Phenylketonuria by Using a Machine Learning-Based Screening Model of Neonatal MRM Data. Front. Mol. Biosci. 2020, 7, 115. [Google Scholar] [CrossRef] [PubMed]
- Subhashini, P.; Jaya Krishna, S.; Usha Rani, G.; Sushma Chander, N.; Maheshwar Reddy, G.; Naushad, S.M. Application of machine learning algorithms for the differential diagnosis of peroxisomal disorders. J. Biochem. 2019, 165, 67–73. [Google Scholar] [CrossRef]
- Baumgartner, C.; Böhm, C.; Baumgartner, D.; Marini, G.; Weinberger, K.; Olgemöller, B.; Liebl, B.; Roscher, A.A. Supervised machine learning techniques for the classification of metabolic disorders in newborns. Bioinforma 2004, 20, 2985–2996. [Google Scholar] [CrossRef]
- Shchelochkov, O.A.; Manoli, I.; Juneau, P.; Sloan, J.L.; Ferry, S.; Myles, J.; Schoenfeld, M.; Pass, A.; McCoy, S.; Van Ryzin, C.; et al. Severity modeling of propionic acidemia using clinical and laboratory biomarkers. Genet. Med. Off. J. Am. Coll. Med. Genet. 2021, 23, 1534–1542. [Google Scholar] [CrossRef] [PubMed]
- Leal-Witt, M.J.; Rojas-Agurto, E.; Muñoz-González, M.; Peñaloza, F.; Arias, C.; Fuenzalida, K.; Bunout, D.; Cornejo, V.; Acevedo, A. Risk of Developing Insulin Resistance in Adult Subjects with Phenylketonuria: Machine Learning Model Reveals an Association with Phenylalanine Concentrations in Dried Blood Spots. Metabolites 2023, 13, 677. [Google Scholar] [CrossRef] [PubMed]
- Friedemann, C.; Heneghan, C.; Mahtani, K.; Thompson, M.; Perera, R.; Ward, A.M. Cardiovascular disease risk in healthy children and its association with body mass index: Systematic review and meta-analysis. BMJ 2012, 345, e4759. [Google Scholar] [CrossRef]
- Chen, G.; Liu, C.; Yao, J.; Jiang, Q.; Chen, N.; Huang, H.; Liang, J.; Li, L.; Lin, L. Overweight, obesity, and their associations with insulin resistance and β-cell function among Chinese: A cross-sectional study in China. Metabolism 2010, 59, 1823–1832. [Google Scholar] [CrossRef]
- Giannini, E.G.; Sammito, G.; Farinati, F.; Ciccarese, F.; Pecorelli, A.; Rapaccini, G.L.; Di Marco, M.; Caturelli, E.; Zoli, M.; Borzio, F.; et al. Determinants of alpha-fetoprotein levels in patients with hepatocellular carcinoma: Implications for its clinical use. Cancer 2014, 120, 2150–2157. [Google Scholar] [CrossRef]
- Chan, S.L.; Mo, F.; Johnson, P.J.; Siu, D.Y.; Chan, M.H.; Lau, W.Y.; Lai, P.B.; Lam, C.W.; Yeo, W.; Yu, S.C. Performance of serum α-fetoprotein levels in the diagnosis of hepatocellular carcinoma in patients with a hepatic mass. HPB 2014, 16, 366–372. [Google Scholar] [CrossRef]
- Lok, A.S.; Lai, C.L. alpha-Fetoprotein monitoring in Chinese patients with chronic hepatitis B virus infection: Role in the early detection of hepatocellular carcinoma. Hepatology 1989, 9, 110–115. [Google Scholar] [CrossRef]
- Fuenzalida, K.; Leal-Witt, M.J.; Guerrero, P.; Hamilton, V.; Salazar, M.F.; Peñaloza, F.; Arias, C.; Cornejo, V. NTBC Treatment Monitoring in Chilean Patients with Tyrosinemia Type 1 and Its Association with Biochemical Parameters and Liver Biomarkers. J. Clin. Med. 2021, 10, 5832. [Google Scholar] [CrossRef] [PubMed]
- Couce, M.L.; Sánchez-Pintos, P.; Aldámiz-Echevarría, L.; Vitoria, I.; Navas, V.; Martín-Hernández, E.; García-Volpe, C.; Pintos, G.; Peña-Quintana, L.; Hernández, T.; et al. Evolution of tyrosinemia type 1 disease in patients treated with nitisinone in Spain. Medicine 2019, 98, e17303. [Google Scholar] [CrossRef] [PubMed]
- Mayorandan, S.; Meyer, U.; Gokcay, G.; Segarra, N.G.; de Baulny, H.O.; van Spronsen, F.; Zeman, J.; de Laet, C.; Spiekerkoetter, U.; Thimm, E.; et al. Cross-sectional study of 168 patients with hepatorenal tyrosinaemia and implications for clinical practice. Orphanet J. Rare Dis. 2014, 9, 107. [Google Scholar] [CrossRef]
- Bzdok, D.; Altman, N.; Krzywinski, M. Statistics versus machine learning. Nat. Methods. 2018, 15, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Liu, Y.; Jia, Y.; Zhao, T. Deep Learning and Machine Learning Applications in Biomedicine. Appl. Sci. 2024, 14, 307. [Google Scholar] [CrossRef]
- Stahlschmidt, S.R.; Ulfenborg, B.; Synnergren, J. Multimodal deep learning for biomedical data fusion: A review. Brief Bioinform. 2022, 23, bbab569. [Google Scholar] [CrossRef]
- Zierk, J.; Arzideh, F.; Haeckel, R.; Cario, H.; Frühwald, M.C.; Groß, H.J.; Gscheidmeier, T.; Hoffmann, R.; Krebs, A.; Lichtinghagen, R.; et al. Pediatric reference intervals for alkaline phosphatase. Clin. Chem. Lab. Med. CCLM 2017, 55, 102–110. [Google Scholar] [CrossRef]
- Tamber, S.S.; Bansal, P.; Sharma, S.; Singh, R.B.; Sharma, R. Biomarkers of liver diseases. Mol. Biol. Rep. 2023, 50, 7815–7823. [Google Scholar] [CrossRef]
- Kwo, P.Y.; Cohen, S.M.; Lim, J.K. ACG Clinical Guideline: Evaluation of Abnormal Liver Chemistries. Am. J. Gastroenterol. 2017, 112, 18–35. [Google Scholar] [CrossRef]
- Colantonio, D.A.; Kyriakopoulou, L.; Chan, M.K.; Daly, C.H.; Brinc, D.; Venner, A.A.; Pasic, M.D.; Armbruster, D.; Adeli, K. Closing the gaps in pediatric laboratory reference intervals: A CALIPER database of 40 biochemical markers in a healthy and multiethnic population of children. Clin. Chem. 2012, 58, 854–868. [Google Scholar] [CrossRef]
- Vos, M.B.; Abrams, S.H.; Barlow, S.E.; Caprio, S.; Daniels, S.R.; Kohli, R.; Mouzaki, M.; Sathya, P.; Schwimmer, J.B.; Sundaram, S.S.; et al. NASPGHAN Clinical Practice Guideline for the Diagnosis and Treatment of Nonalcoholic Fatty Liver Disease in Children. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. Mach Learn PYTHON. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. Available online: https://dl.acm.org/doi/10.1145/2939672.2939785 (accessed on 31 January 2024).
- Zhang, Y.; Feng, T.; Wang, S.; Dong, R.; Yang, J.; Su, J.; Wang, B. A Novel XGBoost Method to Identify Cancer Tissue-of-Origin Based on Copy Number Variations. Front. Genet. 2020, 11, 585029. [Google Scholar] [CrossRef]
- Panagiotopoulos, K.; Korfiati, A.; Theofilatos, K.; Hurwitz, P.; Deriu, M.A.; Mavroudi, S. MEvA-X: A hybrid multiobjective evolutionary tool using an XGBoost classifier for biomarkers discovery on biomedical datasets. Bioinformatics 2023, 39, btad384. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS’17), Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 4768–4777. [Google Scholar]


| Variable | Spearman’s ρ | p-Value | N° of Samples |
|---|---|---|---|
| ALT | 0.534 | <0.0001 | 228 |
| AST | 0.509 | <0.0001 | 223 |
| Age at Diagnosis | 0.4973 | <0.0001 | 231 |
| GGT | 0.449 | <0.0001 | 211 |
| Prothrombin Time | 0.299 | 0.0002 | 206 |
| Alkaline Phosphatase | 0.297 | <0.0001 | 200 |
| Total Billirrubin | 0.208 | 0.0034 | 196 |
| Methionine | 0.0853 | 0.216 | 212 |
| Glycemia | −0.0328 | 0.627 | 222 |
| Age at Control | −0.0598 | 0.3796 | 218 |
| NTBC Levels | −0.052 | 0.4456 | 217 |
| Phenylalanine | −0.1204 | 0.0818 | 210 |
| Tyrosine | −0.149 | 0.0298 | 212 |
| AUROC | |||
|---|---|---|---|
| Logistic Regression Models | Training | Validation | Testing |
| Model–ALT | 0.6954 | 0.5625 | 0.6745 |
| Model–ALKP | 0.7722 | 0.5463 | 0.7993 |
| Model–Age | 0.7909 | 0.5208 | 0.8246 |
| Model–Age at Diagnosis | 0.7892 | 0.5677 | 0.7833 |
| Complete Model | 0.8157 | 0.6563 | 0.8000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuenzalida, K.; Leal-Witt, M.J.; Acevedo, A.; Muñoz, M.; Gudenschwager, C.; Arias, C.; Cabello, J.F.; La Marca, G.; Rizzo, C.; Pietrobattista, A.; et al. Integrating Machine Learning and Follow-Up Variables to Improve Early Detection of Hepatocellular Carcinoma in Tyrosinemia Type 1: A Multicenter Study. Int. J. Mol. Sci. 2025, 26, 3839. https://doi.org/10.3390/ijms26083839
Fuenzalida K, Leal-Witt MJ, Acevedo A, Muñoz M, Gudenschwager C, Arias C, Cabello JF, La Marca G, Rizzo C, Pietrobattista A, et al. Integrating Machine Learning and Follow-Up Variables to Improve Early Detection of Hepatocellular Carcinoma in Tyrosinemia Type 1: A Multicenter Study. International Journal of Molecular Sciences. 2025; 26(8):3839. https://doi.org/10.3390/ijms26083839
Chicago/Turabian StyleFuenzalida, Karen, María Jesús Leal-Witt, Alejandro Acevedo, Manuel Muñoz, Camila Gudenschwager, Carolina Arias, Juan Francisco Cabello, Giancarlo La Marca, Cristiano Rizzo, Andrea Pietrobattista, and et al. 2025. "Integrating Machine Learning and Follow-Up Variables to Improve Early Detection of Hepatocellular Carcinoma in Tyrosinemia Type 1: A Multicenter Study" International Journal of Molecular Sciences 26, no. 8: 3839. https://doi.org/10.3390/ijms26083839
APA StyleFuenzalida, K., Leal-Witt, M. J., Acevedo, A., Muñoz, M., Gudenschwager, C., Arias, C., Cabello, J. F., La Marca, G., Rizzo, C., Pietrobattista, A., Spada, M., Dionisi-Vici, C., & Cornejo, V. (2025). Integrating Machine Learning and Follow-Up Variables to Improve Early Detection of Hepatocellular Carcinoma in Tyrosinemia Type 1: A Multicenter Study. International Journal of Molecular Sciences, 26(8), 3839. https://doi.org/10.3390/ijms26083839






